Abstract
Background
Increasing evidence has suggested an association of adiponectin gene polymorphisms rs1501299, rs2241766, rs266729 and rs3774261 with risk of nonalcoholic fatty liver disease (NAFLD). This correlation has been extensively meta-analyzed for the first two polymorphisms, but not the second two.
Methods
The PubMed, EMBASE, Google Scholar, and China National Knowledge Infrastructure databases were searched for relevant literature. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated.
Results
A total of 10 case-control studies on rs266729 (2,619 cases and 1,962 controls) and 3 case-control studies on rs3774261 (562 cases and 793 controls) were included. Meta-analysis showed that rs266729 was associated with significantly higher NAFLD risk based on the following five models: allelic, OR 1.72, 95% CI 1.34-2.21, P < 0.001; recessive, OR 2.35, 95% CI 1.86-2.95, P < 0.001; dominant, OR 1.84, 95% CI 1.34-2.53, P < 0.001; homozygous, OR 2.69, 95% CI 1.84-3.92, P < 0.001; and heterozygous, OR 1.72, 95% CI 1.28-2.32, P < 0.001. This association between rs266729 and NAFLD risk remained significant for all five models among studies with Asian, Chinese and Caucasian samples. The rs2241766 polymorphism was associated with significantly higher NAFLD risk according to the recessive model (OR 1.87, 95% CI 1.15-3.04, P = 0.01).
Conclusion
Polymorphisms rs266729 and rs3774261 in the adiponectin gene may be risk factors for NAFLD. These findings may pave the way for novel therapeutic strategies, but they should be verified in large, well-designed studies.
Keywords: adiponectin, polymorphism, nonalcoholic fatty liver disease, system review, meta-analysis
Introduction
Nonalcoholic fatty liver disease (NAFLD), also known as metabolism-associated fatty liver disease (1), is rapidly becoming the most common liver disease worldwide. The primary characteristic of NAFLD is hepatocellular macrovesicular steatosis. NAFLD can progress to hepatic injury, which can range from simple steatosis or nonalcoholic steatohepatitis (NASH), to fibrosis, cirrhosis, and even hepatocellular carcinoma or end-stage liver disease (2–6). NAFLD and its progression have been linked to diet (7), insulin resistance (8, 9), lipotoxicity (10), inflammation (11, 12), genetic predisposition and increases in compounds produced by gut microbes (13, 14). Genetic factors, for example, can alter hepatic lipid metabolism. In this way, NAFLD is a complex metabolic state to which lifestyle and genetic factors contribute (15, 16).
Adiponectin is a protein specific to adipose tissue that regulates insulin sensitivity, glucose homeostasis, and lipid metabolism (17). Decreased levels of adiponectin in plasma are associated with NAFLD as well as obesity, type 2 diabetes, and coronary artery disease (18, 19). Adiponectin is encoded by the 16-kb AMP1 gene on human chromosome 3q27, and it consists of three exons and two introns. Genetic and epigenetic changes in the adiponectin gene may reduce adiponectin levels in plasma and dysregulate hepatic lipid metabolism, which may help explain differences in NAFLD risk among individuals (20, 21). Thus, single-nucleotide polymorphisms (SNPs) in the adiponectin gene may alter levels of the protein in circulation, in turn affecting lipid metabolism and NAFLD risk.
The two adiponectin SNPs most thoroughly investigated for their association with NAFLD risk are rs2241766, which leads to genomic mutation T45G, and rs1501299, which leads to mutation G276T (22–31). Indeed, these two associations have been extensively reviewed and meta-analyzed (32–35). In contrast, much less is known about potential associations of the polymorphisms rs266729 (-11377C>G) and rs3774261 with NAFLD risk (36–47).
Thus, we meta-analyzed here the relevant literature on potential associations of rs266729 and rs3774261 with NAFLD risk.
Material and Methods
Search Strategy
The PubMed, EMBASE, Google Scholar, Web of Science and China National Knowledge Infrastructure (CNKI) databases were searched up to October 20, 2021 without language restrictions using the following search terms: (a) adiponectin, ADIPOQ, APMI, -11377, -11377C>G, rs266729 and rs3774261; (b) those seven terms in combination with polymorphisms, SNP, variant, variants, variation, genotype, genetic or mutation; and (c) all of the above terms in combination with nonalcoholic fatty liver disease or NAFLD. Only studies involving humans were considered. Reference lists in original and review articles were searched manually to identify additional studies. In the case of multiple studies involving overlapping samples, only the largest study was retained.
Inclusion and Exclusion Criteria of the Studies
Studies were included if they met the following criteria: (a) studies had a case-control design to assess the association of adiponectin rs266729 or rs3774261 with NAFLD risk; (b) all patients were diagnosed with NAFLD based on the following diagnostic criteria: abnormal levels of aspartate aminotransferase and alanine aminotransferase persisting for at least 6 months, or evidence of fatty liver based on ultrasonography and/or evidence of diffuse fatty liver based on other imaging examinations, or liver histology; (c) the full text was available and it reported genotype frequencies in cases and controls, or sufficient data to estimate odds ratios (ORs) and 95% confidence intervals (CIs).
Studies were excluded if they: (a) were not a case-control study; (b) did not report precise genotypes; (c) were duplicate publications of data from the same study; (d) were meta-analyses, letters, reviews, or editorial articles; (e) investigated other polymorphisms of adiponectin.
Data Extraction
Two authors (YTZ and LYL) independently selected eligible studies and extracted the following data: first author’s name, year of publication, ethnicity, country, sample size, type of controls, genotyping method, genotype distribution, P value for Hardy-Weinberg equilibrium among controls, and matched parameters.
Assessment of Methodological Quality
The quality of included studies was assessed independently by two investigators (YTZ and LYL) using the Newcastle–Ottawa Scale (48). Scores of 0-4 were considered to indicate poor methodological quality; scores of 5-9, high quality (49). Any disagreements about scoring were resolved through comprehensive reassessment by the other authors. Only high-quality studies were included in the meta-analysis.
Statistical Analysis
The strength of association of rs266729 and rs3774261 with NAFLD risk was calculated in terms of unadjusted ORs with 95% CIs based on genotype frequencies in cases and controls. The significance of pooled ORs was determined using the Z test, with P < 0.05 defined as significant. Meta-analysis was conducted using a fixed-effect model when P > 0.10 for the Q test, indicating lack of heterogeneity among studies; otherwise, meta-analysis was conducted using a random-effect model. All statistical tests for meta-analyses were performed using Review Manager 5.3 (Cochrane Collaboration). Publication bias was assessed using Begg’s funnel plot and Egger’s weighted regression in Stata 12.0 (Stata Corp, College Station, TX, USA), with P < 0.05 considered statistically significant.
Results
Characteristics of Primary Studies
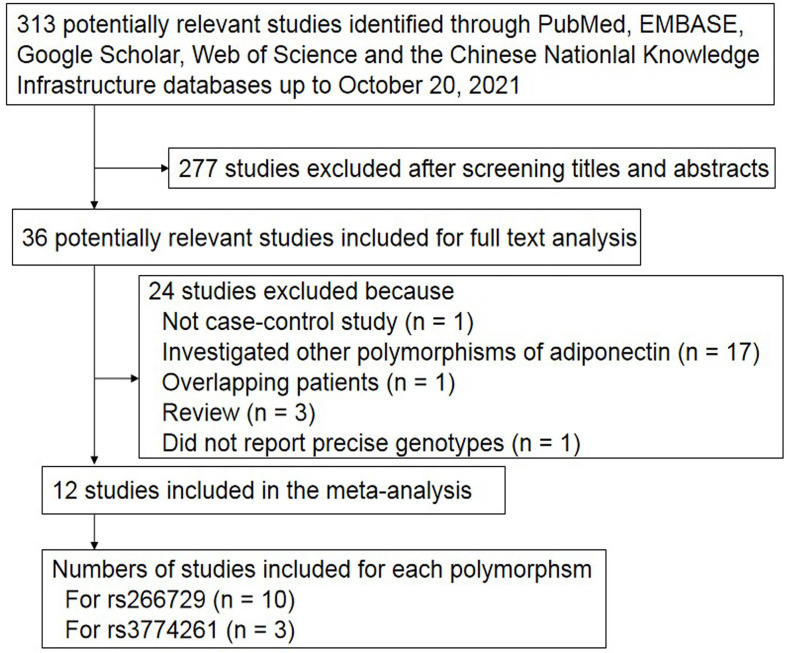
The search strategy retrieved 313 potentially relevant studies, 277 of which were excluded on the basis of titles and abstracts ( Figure 1 ). Another 17 studies were excluded because they investigated other polymorphisms of the adiponectin gene, one study was excluded because it enrolled only cases (50), three studies were excluded because they were review articles (51–53), and one study was excluded because it did not report precise genotypes (30). Two publications were based on the same participants, so they were considered as one study (38, 54). Ultimately, 12 case-control studies (36–47) were included in the meta-analysis ( Table 1 ).
Figure 1.
Flowchart of study selection.
Table 1.
Characteristics of the included studies and genotype distributions.
| First author | Year | Ethnicity | Country | Sample size | Genotyping method | P for HWE | Source of controls | No. of cases | No. of controls | NOS score | Matched parameters | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | |||||||||||||||
| rs266729 | CC | CG | GG | CC | CG | GG | ||||||||||
| Gupta (36) | 2012 | Asian | India | 137 | 250 | PCR | 0.003 | HB | 77 | 53 | 7 | 156 | 92 | 2 | 7 | Age, Sex, BMI |
| Hashemi (37) | 2013 | Asian | India | 83 | 93 | Tetra ARMS-PCR | 0.107 | PB | 27 | 53 | 3 | 49 | 41 | 3 | 6 | Undetermined |
| Ye (38) | 2014 | Asian | China | 130 | 130 | PCR-RFLP | 0.458 | HB | 77 | 47 | 6 | 83 | 40 | 7 | 7 | Age, Sex |
| Hsieh (39) | 2015 | Asian | Taiwan, China | 350 | 209 | TaqMan | 0.545 | T2DM without NAFLD | 175 | 126 | 49 | 113 | 79 | 17 | 7 | Age, Sex |
| Cheng (40) | 2015 | Asian | China | 338 | 280 | PCR | 0.715 | HB | 158 | 149 | 31 | 164 | 102 | 14 | 7 | Age, Sex, Height |
| Zhang (41) | 2016 | Asian | China | 302 | 310 | PCR-RFLP | 0.619 | HB | 152 | 126 | 24 | 197 | 102 | 11 | 7 | Age, Sex, Drinking, Smoking |
| Zhang (42) | 2016 | Asian | China | 600 | 200 | PCR-RFLP | <0.001 | HB | 180 | 202 | 218 | 143 | 28 | 29 | 7 | Age, Sex, Ethnicity, Birthplace |
| Du (43) | 2016 | Asian | China | 493 | 304 | PCR | 0.068 | HB | 278 | 175 | 40 | 219 | 73 | 12 | 7 | Age, Sex |
| Mahmoud (44) | 2019 | Caucasian | Egypt | 100 | 100 | PCR | 0.539 | HB | 38 | 44 | 18 | 47 | 45 | 8 | 7 | Age, Sex |
| Hasan (45) | 2021 | Caucasian | Egypt | 86 | 86 | PCR-RFLP | 0.236 | HB | 28 | 53 | 5 | 44 | 38 | 4 | 6 | Age |
| rs3774261 | AA | AG | GG | AA | AG | GG | ||||||||||
| Zhang (46) | 2012 | Asian | China | 119 | 350 | PCR-RFLP | 0.808 | PB | 41 | 50 | 28 | 107 | 171 | 72 | 7 | Sex |
| Li (47) | 2015 | Asian | China | 357 | 357 | PCR-RFLP | 0.044 | HB | 48 | 179 | 130 | 131 | 155 | 71 | 7 | Age, Sex |
| Hasan (45) | 2021 | Caucasian | Egypt | 86 | 86 | PCR-RFLP | 0.954 | HB | 11 | 55 | 20 | 39 | 38 | 9 | 6 | Age |
T2DM, diabetes mellitus type 2; NAFLD, nonalcoholic fatty liver disease risk; ARMS, amplification refractory mutation system; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; HB, hospital-based; PB, population-based; BMI, body mass index; HWE, Hardy-Weinberg equilibrium; NOS, Newcastle–Ottawa Scale.
Ten studies (36–45) focused on rs266729 and three (45–47) on rs3774261. The distribution of genotypes in controls was consistent with Hardy-Weinberg equilibrium in all but three studies (36, 42, 47). The mean Newcastle-Ottawa score for the 12 studies was 6.83 (range, 6-7). Thus the overall quality of the included studies was adequate.
Quantitative Data Synthesis
rs266729 and NAFLD Risk
Meta-analysis of data from 2,619 cases and 1,962 controls indicated that rs266729 was associated with significantly increased NAFLD risk according to the following five models: allelic, OR 1.72, 95% CI 1.34-2.21, P < 0.001; recessive, OR 2.35, 95% CI 1.86-2.95, P < 0.001; dominant, OR 1.84, 95% CI 1.34-2.53, P < 0.001; homozygous, OR 2.69, 95% CI 1.84-3.92, P < 0.001; and heterozygous, OR 1.72, 95% CI 1.28-2.32, P < 0.001 ( Table 2 and Figures 2A–E ).
Table 2.
Meta-analysis of associations of rs266729 or rs3774261 with risk of nonalcoholic fatty liver disease.
| Genetic model | OR [95% CI] | Z (P value) | Heterogeneity of study design | Meta-analysis model | ||
|---|---|---|---|---|---|---|
| χ2 | df (P value) | I2 (%) | ||||
| Adiponectin rs266729 polymorphism | ||||||
| Adiponectin rs266729 polymorphism in total population from 10 case control studies (36–45) (2,619 cases and 1,962 controls) | ||||||
| Allelic model (G-allele vs. C-allele) | 1.72 [1.34, 2.21] | 4.26 (<0.001) | 53.95 | 9 (<0.001) | 83 | Random |
| Recessive model (GG vs. CG + CC) | 2.35 [1.86, 2.95] | 7.25 (<0.001) | 10.29 | 9 (0.33) | 13 | Fixed |
| Dominant model (CG + GG vs. CC) | 1.84 [1.34, 2.53] | 3.74 (<0.001) | 55.59 | 9 (<0.001) | 84 | Random |
| Homozygous model (GG vs. CC) | 2.69 [1.84, 3.92] | 5.11 (<0.001) | 18.42 | 9 (0.03) | 51 | Random |
| Heterozygous model (CG vs. CC) | 1.72 [1.28, 2.32] | 3.55 (<0.001) | 42.95 | 9 (<0.001) | 79 | Random |
| Adiponectin rs266729 polymorphism in Asian population from 8 case-control studies (36–43) (2,433 cases and 1,776 controls) | ||||||
| Allelic model (G-allele vs. C-allele) | 1.76 [1.31, 2.37] | 3.75 (<0.001) | 53.01 | 7 (<0.001) | 87 | Random |
| Recessive model (GG vs. CG + CC) | 2.38 [1.86, 3.03] | 6.98 (<0.001) | 9.48 | 7 (0.22) | 26 | Fixed |
| Dominant model (CG + GG vs. CC) | 1.85 [1.28, 2.69] | 3.26 (0.001) | 54.56 | 7 (<0.001) | 87 | Random |
| Homozygous model (GG vs. CC) | 2.70 [1.73, 4.23] | 4.35 (0.001) | 18.00 | 7 (0.01) | 61 | Random |
| Heterozygous model (CG vs. CC) | 1.75 [1.23, 2.47] | 3.15 (0.002) | 41.11 | 7 (<0.001) | 83 | Random |
| Adiponectin rs266729 polymorphism in Chinese population from 6 case-control studies (38–43) (2,213 cases and 1,433 controls) | ||||||
| Allelic model (G-allele vs. C-allele) | 1.74 [1.20, 2.52] | 2.94 (0.003) | 52.49 | 5 (<0.001) | 90 | Random |
| Recessive model (GG vs. CG + CC) | 2.35 [1.83, 3.01] | 6.72 (<0.001) | 7.01 | 5 (0.22) | 29 | Fixed |
| Dominant model (CG + GG vs. CC) | 1.91 [1.21, 3.00] | 2.78 (0.005) | 50.92 | 5 (<0.001) | 90 | Random |
| Homozygous model (GG vs. CC) | 2.58 [1.57, 4.24] | 3.75 (<0.001) | 16.55 | 5 (0.005) | 70 | Random |
| Heterozygous model (CG vs. CC) | 1.79 [1.18, 2.73] | 2.71 (0.007) | 37.20 | 5 (<0.001) | 87 | Random |
| Adiponectin rs266729 polymorphism in Caucasian population from 2 case-control studies (44, 45) (186 cases and 186 controls) | ||||||
| Allelic model (G-allele vs. C-allele) | 1.55 [1.14, 2.10] | 2.79 (0.005) | 0.02 | 1 (0.90) | 0 | Fixed |
| Recessive model (GG vs. CG + CC) | 2.07 [0.99, 4.30] | 1.94 (0.05) | 0.70 | 1 (0.40) | 0 | Fixed |
| Dominant model (CG + GG vs. CC) | 1.74 [1.15, 2.63] | 2.61 (0.009) | 0.90 | 1 (0.34) | 0 | Fixed |
| Homozygous model (GG vs. CC) | 2.51 [1.16, 5.44] | 2.33 (0.02) | 0.16 | 1 (0.68) | 0 | Fixed |
| Heterozygous model (CG vs. CC) | 1.60 [1.04, 2.46] | 2.14 (0.03) | 1.80 | 1 (0.18) | 45 | Fixed |
| Adiponectin rs3774261 polymorphism | ||||||
| Adiponectin rs3774261 polymorphism in total population from 3 case-control studies (45–47) (562 cases and 793 controls) | ||||||
| Allelic model (G-allele vs. A-allele) | 1.76 [0.98, 3.18] | 1.86 (0.06) | 22.72 | 2 (<0.001) | 91 | Random |
| Recessive model (GG vs. AG + AA) | 1.87 [1.15, 3.04] | 2.51 (0.01) | 5.20 | 2 (0.07) | 62 | Random |
| Dominant model (AG + GG vs. AA) | 2.55 [0.81, 7.98] | 1.60 (0.11) | 31.99 | 2 (<0.001) | 94 | Random |
| Homozygous model (GG vs. AA) | 3.29 [0.97, 11.15] | 1.91 (0.06) | 22.63 | 2 (<0.001) | 91 | Random |
| Heterozygous model (AG vs. AA) | 2.25 [0.75, 6.76] | 1.45 (0.15) | 26.19 | 2 (<0.001) | 92 | Random |
| Adiponectin rs3774261 polymorphism in Chinese population from 2 case-control studies (46, 47) (476 cases and 707 controls) | ||||||
| Allelic model (G-allele vs. A-allele) | 1.49 [0.66, 3.35] | 0.97 (0.33) | 19.79 | 1 (<0.001) | 95 | Random |
| Recessive model (GG vs. AG + AA) | 1.70 [0.89, 3.25] | 1.60 (0.11) | 4.69 | 1 (0.03) | 79 | Random |
| Dominant model (AG + GG vs. AA) | 1.78 [0.41, 7.68] | 0.77 (0.44) | 25.73 | 1 (<0.001) | 96 | Random |
| Homozygous model (GG vs. AA) | 2.28 [0.48, 10.85] | 1.03 (0.30) | 19.02 | 1 (<0.001) | 95 | Random |
| Heterozygous model (AG vs. AA) | 1.56 [0.39, 6.27] | 0.63 (0.53) | 20.11 | 1 (<0.001) | 95 | Random |
OR, odds ratio; 95% CI, 95% confidence interval.
Figure 2.
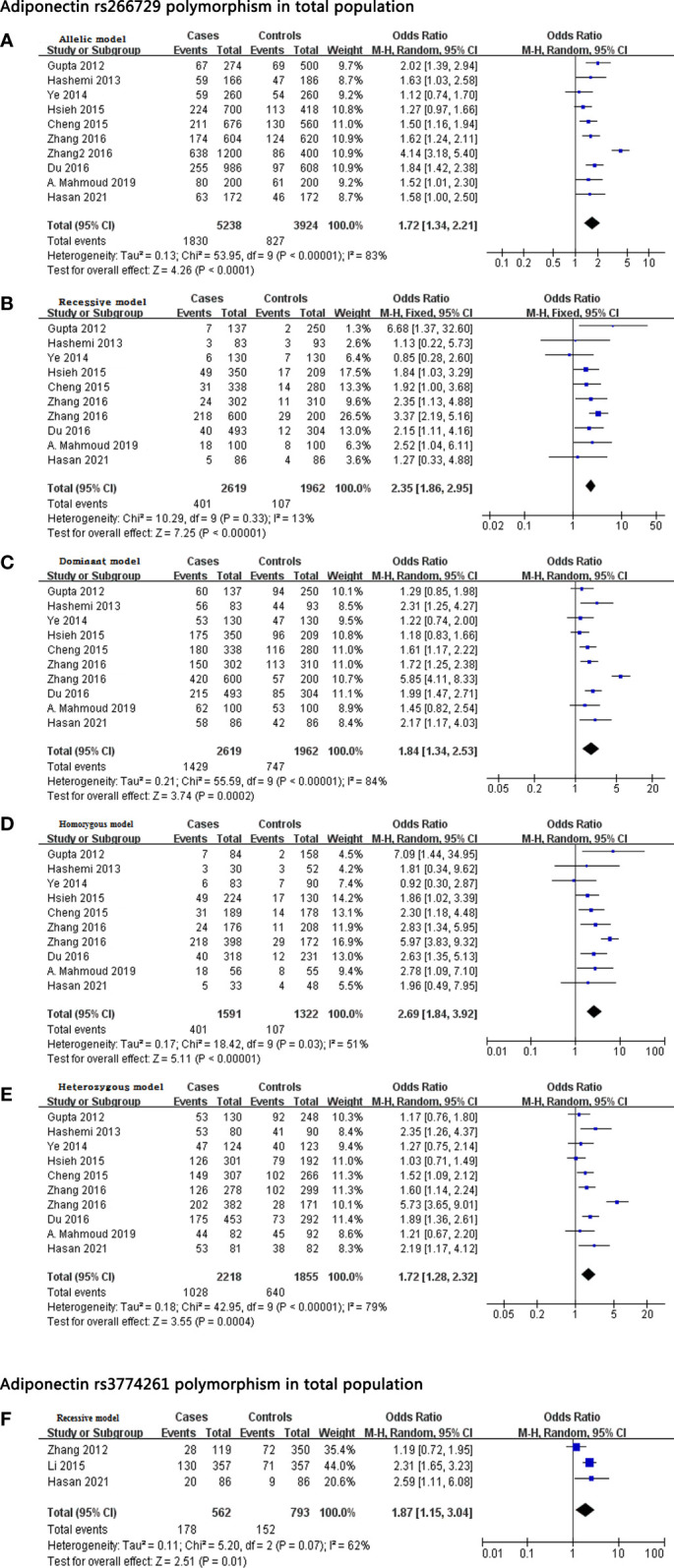
(A–E) Forest plot showing the relationship between rs266729 polymorphism and NAFLD risk in the total population according to different genetic models: (A) allelic (G-allele vs. C-allele), (B) recessive (GG vs. CG + CC), (C) dominant (CG + GG vs. CC), (D) homozygous (GG vs. CC), or (E) heterozygous (CG vs. CC). (F) Forest plot showing the relationship between rs3774261 polymorphism and NAFLD risk in the total population according to the recessive model (GG vs. AG + AA). CI, confidence interval; df, degree of freedom; M-H, Mantel-Haenszel; NAFLD, nonalcoholic fatty liver disease.
This association remained significant when we meta-analyzed only the eight studies involving 2,433 Asian cases and 1,776 Asian controls (36–43). Again, significance was obtained with all five models: allelic, OR 1.76, 95% CI 1.31-2.37, P < 0.001; recessive, OR 2.38, 95% CI 1.86-3.03, P < 0.001; dominant, OR 1.85, 95% CI 1.28-2.69, P = 0.001; homozygous, OR 2.70, 95% CI 1.73-4.23, P = 0.001; and heterozygous, OR 1.75, 95% CI 1.23-2.47, P = 0.002 ( Table 2 ).
Next, this association remained significant when we meta-analyzed only the eight studies involving 2,213 Chinese cases and 1,433 Chinese controls (38–43). Again, significance was obtained with all five models: allelic, OR 1.74, 95% CI 1.20-2.52, P = 0.003; recessive, OR 2.35, 95% CI 1.83-3.01, P < 0.001; dominant, OR 1.91, 95% CI 1.21-3.00, P = 0.005; homozygous, OR 2.58, 95% CI 1.57-4.24, P < 0.001; and heterozygous, OR 1.79, 95% CI 1.18-2.73, P = 0.007 ( Table 2 ).
Lastly, this association remained significant when we meta-analyzed only the eight studies involving 186 Caucasian cases and 186 Caucasian controls (44, 45). Again, significance was obtained with all five models: allelic, OR 1.55, 95% CI 1.14-2.10, P = 0.005; recessive, OR 2.07, 95% CI 0.99-4.30, P = 0.05; dominant, OR 1.74, 95% CI 1.15-2.63, P = 0.009; homozygous, OR 2.51, 95% CI 1.16-5.44, P = 0.02; and heterozygous, OR 1.60, 95% CI 1.04-2.46, P = 0.03 ( Table 2 ).
rs3774261 and NAFLD risk
Meta-analysis of three studies (45–47) involving 562 cases and 793 controls showed that rs3774261 was associated with significantly increased NAFLD risk according to the recessive model (OR 1.87, 95% CI 1.15-3.04, P = 0.01; Table 2 and Figure 2F ). But this association could not be found in the Chinese population ( Table 2 ).
Sensitivity Analysis
To assess the reliability of the outcomes in the meta-analysis, we repeated the meta-analysis after excluding, one by one, three studies in which the P value associated with Hardy-Weinberg equilibrium was less than 0.05 (36, 42, 47).
After excluding the study by Gupta et al. (36), the results did not differ substantially either in total or in Asian population for rs266729 polymorphism ( Supplementary Table S1 ).
After excluding the study by Zhang et al. (42), the results did not differ substantially in total, Asian or Chinese population for rs266729 polymorphism ( Supplementary Table S2 ).
After excluding the study by Li et al. (47), the results were altered in recessive model in total population for rs3774261 polymorphism ( Supplementary Table S3 ). Therefore, the results for rs3774261 polymorphism should be interpreted with caution.
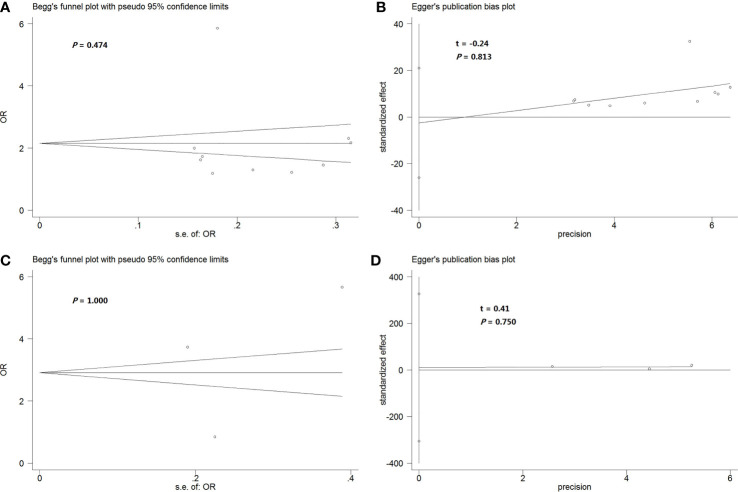
Publication Bias
Begg’s funnel plot and Egger’s test were performed to detect potential publication bias in our meta-analysis. Funnel plots showed no obvious asymmetry in the dominant model of the rs266729 polymorphism ( Figure 3A ), and the result for Egger’s test was not significant ( Figure 3B ). Similar results were obtained with the dominant model of the rs3774261 polymorphism ( Figures 3C, D ).
Figure 3.
(A, B) Begg’s funnel plot (A) and Egger’s test (B) to assess publication bias in the meta-analysis of the association between rs266729 polymorphism and NAFLD risk in the total population according to the dominant genetic model. (C, D) Begg’s funnel plot (C) and Egger’s test (D) to assess publication bias in the meta-analysis of the association between rs3774261polymorphism and NAFLD risk in the total population according to the dominant genetic model. NAFLD, nonalcoholic fatty liver disease; OR, odds ratio.
Discussion
The physiological roles of adiponectin remain unclear, but it has been associated with obesity, insulin resistance, type 2 diabetes, atherosclerosis, hypertension, coronary artery disease, various inflammatory diseases, metabolic syndrome and NAFLD (18, 19, 55, 56). In fact, high levels of adiponectin may protect against NAFLD (56), perhaps by activating AMPK and peroxisome proliferator-activated receptor γ to improve insulin sensitivity, reduce fatty acid synthesis and enhance fatty acid oxidation (57). Here we provide additional evidence that adiponectin levels may influence onset of NAFLD by demonstrating associations between two SNPs in the adiponectin gene and risk of the disorder.
We found that rs266729 was significantly related to elevated NAFLD risk across all ethnic groups examined, as well as specifically in Asian, Chinese and Caucasian populations. Consistent with our findings, a previous meta-analysis (35) of three case-control studies (30, 36, 37) suggested a similar association among Asians. We included two of those case-control studies in the present meta-analysis but not one (30) because it did not report precise genotypes. Another Chinese study (43) reported an association between rs266729 and elevated NAFLD risk, as well as elevated risk of coronary artery disease among NAFLD patients. Our results extend the findings of a previous study linking rs266729 to elevated NAFLD risk in a southeastern Iranian population (37). However, our results contrast with a study (38) that failed to link rs266729 to NAFLD risk among Han Chinese. The relatively large sample in our meta-analysis may make our findings more reliable.
We also found that rs3774261 was significantly related to elevated NAFLD risk across all ethnic groups examined. The fact that our meta-analysis contained only three case-control studies involving 562 cases and 793 controls emphasizes the need for further research. Indeed, further research is needed into the potential association of the adiponectin SNPs rs17300539 (G–11391A) (24, 58) and rs822393 (42) and risk of NAFLD. We were unable to include those SNPs in our meta-analysis because of the limited data available.
Our results should be interpreted with caution in light of several limitations. First, the controls in one study (39) had diabetes mellitus type 2, so they may not be comparable to healthy controls in other studies. Second, the P value associated with Hardy-Weinberg equilibrium was < 0.001 in three studies (36, 42, 47), suggesting a lack of generalizability to the broader population. Nevertheless, excluding each of those studies one at a time did not substantially alter the meta-analysis. Third, the robustness of our meta-analysis may be reduced by the fact that studies used genotyping methods differing in sensitivity and specificity, and by confounding due to sex, age, insulin resistance, family history of type 2 diabetes, obesity, coronary artery disease, hypertension and metabolic syndrome. We were unable to account for those factors in our meta-analysis because the original studies either did not report their frequencies or they aggregated the factors in different ways.
Conclusion
The available evidence suggests that SNPs rs266729 and rs3774261 in the adiponectin gene are risk factors for NAFLD. If our results can be verified in large, well-designed studies, they may help pave the way for novel therapeutic strategies.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Author Contributions
Designed the study: L-YL and Y-TZ. Searched databases and collected full-text papers: T-MX and C-XW. Extracted and analyzed the data: L-YL and Y-TZ. Statistical analyses: J-YC. Wrote the manuscript: Y-TZ. All authors reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2022.798417/full#supplementary-material
References
- 1. Zheng KI, Fan JG, Shi JP, Wong VW, Eslam M, George J, et al. From NAFLD to MAFLD: A “Redefining” Moment for Fatty Liver Disease. Chin Med J-Peking (2020) 133(19):2271. doi: 10.1097/CM9.0000000000000981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bellentani S. The Epidemiology of Non-Alcoholic Fatty Liver Disease. Liver Int (2017) 37:81–4. doi: 10.1111/liv.13299 [DOI] [PubMed] [Google Scholar]
- 3. Fazel Y, Koenig AB, Sayiner M, Goodman ZD, Younossi ZM. Epidemiology and Natural History of Non-Alcoholic Fatty Liver Disease. Metabolism (2016) 65(8):1017–25. doi: 10.1016/j.metabol.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 4. Reccia I, Kumar J, Akladios C, Virdis F, Pai M, Habib N, et al. Non-Alcoholic Fatty Liver Disease: A Sign of Systemic Disease. Metabolism (2017) 72:94–108. doi: 10.1016/j.metabol.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 5. Lazarus JV, Mark HE, Anstee QM, Arab JP, Batterham RL, Castera L, et al. Advancing the Global Public Health Agenda for NAFLD: A Consensus Statement. Nat Rev Gastro Hepat (2021) 19(1):60–78. doi: 10.1038/s41575-021-00523-4 [DOI] [PubMed] [Google Scholar]
- 6. Morandi A, Di Sessa A, Zusi C, Umano GR, El Mazloum D, Fornari E, et al. Nonalcoholic Fatty Liver Disease and Estimated Insulin Resistance in Obese Youth: A Mendelian Randomization Analysis. J Clin Endocrinol Metab (2020) 105(11):e4046–54. doi: 10.1210/clinem/dgaa583 [DOI] [PubMed] [Google Scholar]
- 7. Chung M, Ma J, Patel K, Berger S, Lau J, Lichtenstein AH. Fructose, High-Fructose Corn Syrup, Sucrose, and Nonalcoholic Fatty Liver Disease or Indexes of Liver Health: A Systematic Review and Meta-Analysis. Am J Clin Nutr (2014) 100(3):833– 49. doi: 10.3945/ajcn.114.086314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Utzschneider KM, Kahn SE. The Role of Insulin Resistance in Nonalcoholic Fatty Liver Disease. J Clin Endocr Metab (2006) 91(12):4753–61. doi: 10.1210/jc.2006-0587 [DOI] [PubMed] [Google Scholar]
- 9. Wong VW, Hui AY, Tsang SW, Chan JL, Wong GL, Chan AW, et al. Prevalence of Undiagnosed Diabetes and Postchallenge Hyperglycaemia in Chinese Patients With Non-Alcoholic Fatty Liver Disease. Aliment Pharmacol Ther (2006) 24(8):1215–22. doi: 10.1111/j.1365-2036.2006.03112.x [DOI] [PubMed] [Google Scholar]
- 10. Lake AD, Novak P, Hardwick RN, Flores-Keown B, Zhao F, Klimecki WT, et al. The Adaptive Endoplasmic Reticulum Stress Response to Lipotoxicity in Progressive Human Nonalcoholic Fatty Liver Disease. Toxicol Sci (2014) 137(1):26–35. doi: 10.1093/toxsci/kft230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Obika M, Noguchi H. Diagnosis and Evaluation of Nonalcoholic Fatty Liver Disease. Exp Diabetes Res (2011) 2012:12. doi: 10.1155/2012/145754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tarantino G, Colao A, Capone D, Tarantino M, Grimaldi E, Savastano S. Circulating Levels of Cytochrome C, Gamma-Glutamyl Transferase, Triglycerides and Unconjugated Bilirubin in Overweight/Obese Patients With Non-Alcoholic Fatty Liver Disease. J Biol Reg Homeos Ag (2011) 25(1):47–56. [PubMed] [Google Scholar]
- 13. Buzzetti E, Pinzani M, Tsochatzis EA. The Multiple-Hit Pathogenesis of non-Alcoholic Fatty Liver Disease (NAFLD). Metabolism (2016) 65(8):1038–48. doi: 10.1016/j.metabol.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 14. Polyzos SA, Kountouras J, Zavos C. The Multi-Hit Process and the Antagonistic Roles of Tumor Necrosis Factor-Alpha and Adiponectin in Non Alcoholic Fatty Liver Disease. Hippokratia (2009) 13(2):127. [PMC free article] [PubMed] [Google Scholar]
- 15. Day CP. Genetic and Environmental Susceptibility to Non-Alcoholic Fatty Liver Disease. Digest Dis (2010) 28(1):255–60. doi: 10.1159/000282098 [DOI] [PubMed] [Google Scholar]
- 16. Licastro F, Chiappelli M, Porcellini E, Campo G, Buscema M, Grossi E, et al. Gene-Gene and Gene-Clinical Factors Interaction in Acute Myocardial Infarction: A New Detailed Risk Chart. Curr Pharm Design (2010) 16(7):783–8. doi: 10.2174/138161210790883543 [DOI] [PubMed] [Google Scholar]
- 17. Cnop M, Havel PJ, Utzschneider KM, Carr DB, Sinha MK, Boyko EJ, et al. Relationship of Adiponectin to Body Fat Distribution, Insulin Sensitivity and Plasma Lipoproteins: Evidence for Independent Roles of Age and Sex. Diabetologia (2003) 46(4):459–69. doi: 10.1007/s00125-003-1074-z [DOI] [PubMed] [Google Scholar]
- 18. Yang WS, Chuang LM. Human Genetics of Adiponectin in the Metabolic Syndrome. J Mol Med (Berl) (2006) 84(2):112–21. doi: 10.1007/s00109-005-0011-7 [DOI] [PubMed] [Google Scholar]
- 19. Cooney MT, Dudina A, de Bacquer D, Wilhelmsen L, Sans S, Menotti A. HDL Cholesterol Protects Against Cardiovascular Disease in Both Genders, at All Ages and at All Levels of Risk. Atherosclerosis (2009) 206(2):611–6. doi: 10.1016/j.atherosclerosis.2009.02.041 [DOI] [PubMed] [Google Scholar]
- 20. Thompson JF, Durham LK, Lira ME, Shear C, Milos PM. CETP Polymorphisms Associated With HDL Cholesterol May Differ From Those Associated With Cardiovascular Disease. Atherosclerosis (2005) 181(1):45–53. doi: 10.1016/j.atherosclerosis.2005.01.015 [DOI] [PubMed] [Google Scholar]
- 21. Ridker PM, Pare G, Parker AN, Zee RY, Miletich JP, Chasman DI. Polymorphism in the CETP Gene Region, HDL Cholesterol, and Risk of Future Myocardial Infarction: Genomewide Analysis Among 18 245 Initially Healthy Women From the Women’s Genome Health Study. Circ Cardiovasc Gene (2009) 2(1):26–33. doi: 10.1161/CIRCGENETICS.108.817304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gong LQ, Li L, Zhao XG, Zhang YH, Jiang Y, Gu YJ, et al. Association of Single Nucleotide Polymorphisms of the Adiponectin Gene and Plasma Levels of Adiponectin With Risk of Nonalcoholic Fatty Liver Disease. J Xinjiang Med Univ (2013) 36:860–4. [Google Scholar]
- 23. Mohseni F, Moghbelinejad S, Najafipour R. Major Components of Metabolic Parameters and Nutritional Intakes in Different Genotypes of Adiponectin+ 276 G> T Gene Polymorphism in Non-Diabetes and Non-Alcoholic Iranian Fatty Liver Patients. Avicenna J Med Biotechnol (2017) 9(3):155. [PMC free article] [PubMed] [Google Scholar]
- 24. Yu K, Gorshunska M, Tyzhnenko T, Krasova N, Dunaeva I, Gladkih A, et al. Adiponectin Gene Single-Nucleotide Polymorphisms in Patients With Type 2 Diabetes Mellitus and Nonalcoholic Fatty Liver Disease. Int J Endocrinol (2017) 13(4):229–37. [Google Scholar]
- 25. Huang CM, Li YY, Zhou YJ, Nie YQ, Du YL. Correlation Between Adiponectin Gene Polymorphisms and Susceptibility of Non-Alcoholic Fatty Liver Disease. Acad J Guangzhou Med Col (2010) 38:1–5. [Google Scholar]
- 26. Musso G, Gambino R, De Michieli F, Durazzo M, Pagano G, Cassader M. Adiponectin Gene Polymorphisms Modulate Acute Adiponectin Response to Dietary Fat: Possible Pathogenetic Role in NASH. Hepatology (2008) 47(4):1167–77. doi: 10.1002/hep.22142 [DOI] [PubMed] [Google Scholar]
- 27. Shi SL, Nie YQ, Li YY, Zhou YJ, Du YL. Association of T45G Polymorphism in Exon 2 of Adiponectin Gene With Nonalcoholic Fatty Liver Disease. Acad J Guangzhou Med Col (2005) 33(6):1–3. [Google Scholar]
- 28. Tokushige K, Hashimoto E, Noto H, Yatsuji S, Taniai M, Torii N, et al. Influence of Adiponectin Gene Polymorphisms in Japanese Patients With Non-Alcoholic Fatty Liver Disease. J Gastroenterol (2009) 44(9):976–82. doi: 10.1007/s00535-009-0085-z [DOI] [PubMed] [Google Scholar]
- 29. Wang ZL, Xia B, Shrestha U, Jiang L, Ma CW, Chen Q, et al. Correlation Between Adiponectin Polymorphisms and Non-Alcoholic Fatty Liver Disease With or Without Metabolic Syndrome in Chinese Population. J Endocrinol Invest (2008) 31(12):1086–91. doi: 10.1007/BF03345657 [DOI] [PubMed] [Google Scholar]
- 30. Wong VW, Wong GL, Tsang SW, Hui AY, Chan AW, Choi PC, et al. Genetic Polymorphisms of Adiponectin and Tumor Necrosis Factor-Alpha and Nonalcoholic Fatty Liver Disease in Chinese People. J Gastroen Hepatol (2008) 23(6):914–21. doi: 10.1111/j.1440-1746.2008.05344.x [DOI] [PubMed] [Google Scholar]
- 31. Zhou YJ, Li YY, Nie YQ, Yang H, Zhan Q, Huang J, et al. Influence of Polygenetic Polymorphisms on the Susceptibility to Non-Alcoholic Fatty Liver Disease of Chinese People. J Gastroen Hepatol (2010) 25(4):772–7. doi: 10.1111/j.1440-1746.2009.06144.x [DOI] [PubMed] [Google Scholar]
- 32. Wang BF, Wang Y, Ao R, Tong J, Wang BY. Adipoq T45 G and G276 T Polymorphisms and Susceptibility to Nonalcoholic Fatty Liver Disease Among Asian Populations: A Meta-Analysis and Meta-Regression. J Clin Lab Anal (2016) 30(1):47–57. doi: 10.1002/jcla.21814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J, Xing J, Wang B, Wei C, Yang R, Zhu Y, et al. Correlation Between Adiponectin Gene Rs1501299 Polymorphism and Nonalcoholic Fatty Liver Disease Susceptibility: A Systematic Review and Meta-Analysis. Med Sci Monitor (2019) 25:1078. doi: 10.12659/MSM.912737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu M, Liu S, Shang M, Liu X, Wang Y, Li Q, et al. Association Between ADIPOQ G276T and C11377G Polymorphisms and the Risk of Non-Alcoholic Fatty Liver Disease: An Updated Meta-Analysis. Mol Genet Genom Med (2019) 7(5):e624. doi: 10.1002/mgg3.624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J, Guo XF, Yu SJ, Song J, Zhang JX, Cao Z, et al. Adiponectin Polymorphisms and non-Alcoholic Fatty Liver Disease Risk: A Meta-Analysis. J Gastroen Hepatol (2014) 29(7):1396–405. doi: 10.1111/jgh.12562 [DOI] [PubMed] [Google Scholar]
- 36. Gupta AC, Misra R, Sakhuja P, Singh Y, Basir SF, Sarin SK, et al. Association of Adiponectin Gene Functional Polymorphisms (-11377C/G and +45T/G) With Nonalcoholic Fatty Liver Disease. Gene (2012) 496(1):63–7. doi: 10.1016/j.gene.2011.12.023 [DOI] [PubMed] [Google Scholar]
- 37. Hashemi M, Hanafifi BH, Eskandari NE, Hashemzehi NA, Shafieipour S, Ghavami S, et al. Association of Adiponectin Rs1501299 and Rs266729 Gene Polymorphisms With Nonalcoholic Fatty Liver Disease. Hepat Mon (2013) 13(5):e9527. doi: 10.5812/hepatmon.9527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ye Y, Yu J, Wang J, Lu G. Risk Factors of Non-Alcoholic Fatty Liver and Their Relationship With Polymorphisms of Adiponectin Gene Promoter Region in the Han Population in Guilin City. Guangdong Med J (2014) 35:2115–9. doi: 10.13820/j.cnki.gdyx.2014.13.054 [DOI] [Google Scholar]
- 39. Hsieh CJ, Wang PW, Hu TH. Association of Adiponectin Gene Polymorphism With Nonalcoholic Fatty Liver Disease in Taiwanese Patients With Type 2 Diabetes. PloS One (2015) 10(6):e0127521. doi: 10.1371/journal.pone.0127521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng Y, Jiang M, Xin Y, Baiquan AN, Xuan S. Association Between Adiponectin Polymorphisms and Nonalcoholic Fatty Liver Disease in Han Chinese Popu-Lation in Qingdao. J Pract Med (2015) 9):1421–4. [Google Scholar]
- 41. Zhang CX, Guo LK, Qin YM, Li GY. Association of Polymorphisms of Adiponectin Gene Promoter-11377C/G, Glutathione Peroxidase-1 Gene C594T, and Cigarette Smoking in Nonalcoholic Fatty Liver Disease. J Chin Med Assoc (2016) 79(4):195–204. doi: 10.1016/j.jcma.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 42. Zhang W, Zhu LQ, Huo XL, Qin J, Yuan GY. Association of Adiponectin Gene Polymorphisms and Additional Gene-Gene Interaction With Nonalcoholic Fatty Liver Disease in the Chinese Han Population. Hepatol Int (2016) 10(3):511–7. doi: 10.1007/s12072-015-9687-0 [DOI] [PubMed] [Google Scholar]
- 43. Du SX, Lu LL, Liu Y, Dong QJ, Xuan SY, Xin YN. Association of Adiponectin Gene Polymorphisms With the Risk of Coronary Artery Disease in Patients With Nonalcoholic Fatty Liver Disease in a Chinese Han Population. Hepat Mon (2016) 16(7):e37388. doi: 10.5812/hepatmon.37388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mahmoud AA, Moghazy HM, Yousef LM, Mohammad AN. Adiponectin Rs2241766 and Rs266729 Gene Polymorphisms in Non-Alcoholic Fatty Liver Disease. Gene Rep (2019) 15:100381. doi: 10.1016/j.genrep.2019.100381 [DOI] [Google Scholar]
- 45. Hasan EM, Abd AA, Sabry D, Badary HA, Gaber Y, Yosry A, et al. The Association of Adiponectin Gene Polymorphisms With Susceptibility and Progression of NAFLD in a Cohort of Egyptian Patients. Egyptian Liver J (2021) 11(1):1–11. doi: 10.1186/s43066-021-00103-w [DOI] [Google Scholar]
- 46. Zhang Z. The Link of Adiponectin Gene Polymorphisms to NAFLD Occurrence and Progression in a General Population of Guangdong Province. [Master’s Thesis]. Guangdong: Guangzhou Medical College; (2012). [Google Scholar]
- 47. Li HJ, Li CP, Zhang C, Zhong XL, Shi L. Association of Adiponectin Gene Polymorphisms and Nonalcoholic Fatty Liver Disease. Int J Clin Exp Med (2015) 8(9):16676. [PMC free article] [PubMed] [Google Scholar]
- 48. Wells GA, Shea B, O’Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-Analyses. Canada: University of Ottawa; (2018). Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. [Google Scholar]
- 49. Ownby RL, Crocco E, Acevedo A, John V, Loewenstein D. Depression and Risk for Alzheimer Disease: Systematic Review, Meta-Analysis. Arch Gen Psychiatry (2006) 63(5):530–8. doi: 10.1001/archpsyc.63.5.530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang Q, Wang R, Li D, Wang JY, Zhang L, He G, et al. Association of Phlegm-Dampness Constitution With Polymorphism of LEPR and ADIPQQ With Elderly Population of NAFLD. Chin J Integrated Traditional Western Med Liver Dis (2019) 29(5):390–4. doi: 10.3969/j.issn.1005-0264.2019.05.003 [DOI] [Google Scholar]
- 51. Boutari C, Perakakis N, Mantzoros CS. Association of Adipokines With Development and Progression of Nonalcoholic Fatty Liver Disease. Endocrinol Metab (2018) 33(1):33–43. doi: 10.3803/EnM.2018.33.1.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang C, Gong J, Wu H. Development of Gene Polymorphisms in Meditators of Nonalcoholic Fatty Liver Disease. Biomed Rep (2017) 7(2):95–104. doi: 10.3892/br.2017.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chi Z. Pathogenesis of Non-Alcoholic Fatty Liver Disease. World Chin J Digestology (2017) 25(8):670–83. doi: 10.11569/wcjd.v25.i8.670 [DOI] [Google Scholar]
- 54. Ye Y, Yu J, Wang J, Lu G, Hu Y. Nonalcoholic Fatty Liver Disease and Snps -11377C/G in Promoter Region of Adiponectin Gene: A Correlation Study in Han Elderly Population in Guilin. Chin Gen Prac (2013) 41:4135–9. [Google Scholar]
- 55. Robinson K, Prins J, Venkatesh B. Clinical Review: Adiponectin Biology and Its Role in Inflammation and Critical Illness. Crit Care (2011) 15(2):1–9. doi: 10.1186/cc10021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dıez JJ, Iglesias P. The Role of the Novel Adipocyte-Derived Hormone Adiponectin in Human Disease. Eur J Endocrinol (2003) 148(3):293–300. doi: 10.1530/eje.0.1480293 [DOI] [PubMed] [Google Scholar]
- 57. Chen H, Zhang L, Li X, Li X, Sun G, Yuan X, et al. Adiponectin Activates the AMPK Signaling Pathway to Regulate Lipid Metabolism in Bovine Hepatocytes. J Steroid Biochem (2013) 138:445–54. doi: 10.1016/j.jsbmb.2013.08.013 [DOI] [PubMed] [Google Scholar]
- 58. Rezaei F, Nezhadali M, Hedayati M. Association of Adiponectin Rs17300539 Gene Polymorphism With a Non-Alcoholic Fatty Liver Disease in an Iranian Population. Feyz J Kashan Univ Med Sci (2018) 22(4):379–386.59. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.