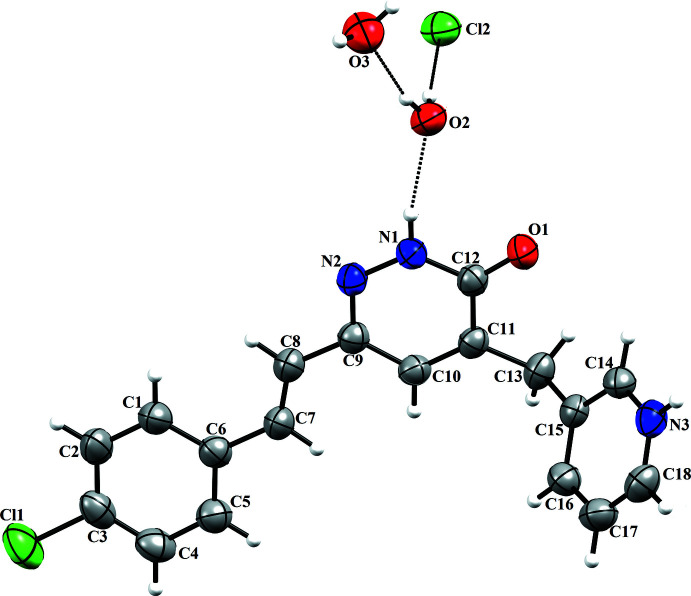
Three intramolecular hydrogen bonds are observed in the title compound. In the crystal, molecules are connected by C—H⋯Cl and N—H⋯O hydrogen bonds.
Keywords: crystal structure, pyridazine, pyridazinone derivative, hydrogen bonding, Hirshfeld surfaces
Abstract
In the title compound, C18H15ClN3O+·Cl−·2H2O, three intramolecular hydrogen bonds are observed, N—H⋯O, O—H⋯Cl and O—H⋯O. In the crystal, molecules are connected by C—H⋯Cl and N—H⋯O hydrogen bonds. Strong C—H⋯Cl, N—H⋯O, O—H⋯Cl and O—H⋯O hydrogen-bonding interactions are implied by the Hirshfeld surface analysis, which indicate that H⋯H contacts make the largest contribution to the overall crystal packing at 33.0%.
Chemical context
Pyridazine derivatives are an important class of heterocyclic chemicals that exhibit a wide range of biological actions. For example, their biological activity and antimicrobial properties have been researched extensively (Neumann et al., 2018 ▸). As a result, the pyridazine ring can be found in a range of commercial medicinal compounds, including Cadralazine and Hydralazine, Minaprine, Pipofezine and others (Abu-Hashem et al., 2020 ▸). Pyridazine derivatives can be found also in the backbones of several organic light-emitting diodes (OLEDs) (Liu et al., 2017 ▸), organic solar cells (OSCs) (Knall et al., 2021 ▸), chemosensors (Peng et al., 2020 ▸), trifluoroacetic acid (TFA) sensors (Li et al., 2018 ▸), bioconjugates (Bahou et al., 2021 ▸), low carbon steel corrosion inhibitors (Khadiria et al., 2016 ▸), and several other materials. They have also been used as starting materials in organic synthesis (Llona-Minguez et al., 2017 ▸), acylating agents (Kung et al., 2002 ▸), precursors for N-heterocyclic carbenes (NHCs) (Liu et al., 2012 ▸) and metallocarbene precursors. An overview of arylglyoxal monohydrates-based one-pot multi-component synthesis of potentially biologically active pyridazines is given by Mousavi (2022 ▸).

Structural commentary
A perspective view of the title molecule is shown in Fig. 1 ▸. The pyridazine and pyridine rings subtend a dihedral angle of 57.27 (5)°. The other two rings, pyridazine and chlorobenzene, are almost planar, making an angle of 8.54 (11)°. The lengths of the C=C [1.349 (3) Å], C=N [1.313 (2) Å], N—N [1.351 (2) Å] and C=O [1.237 (2) Å] bonds are comparable with values published for other pyridazinones (see the Database survey section). Three intramolecular hydrogen bonds are observed, N2—H2C⋯O2, O2—H2A⋯Cl2 and O2—H2B⋯O3 (Table 1 ▸).
Figure 1.
Perspective view and atom labelling of the molecule. Displacement ellipsoids are drawn at the 50% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C10—H10⋯Cl2i | 0.93 | 2.72 | 3.6387 (19) | 168 |
| C18—H18⋯Cl2ii | 0.93 | 2.94 | 3.622 (2) | 132 |
| N3—H3⋯O2iii | 0.80 (3) | 2.35 (3) | 2.965 (2) | 135 (2) |
| N3—H3⋯O1iii | 0.80 (3) | 2.25 (3) | 2.855 (2) | 133 (3) |
| N2—H2C⋯O2 | 0.86 (2) | 1.97 (2) | 2.801 (2) | 161 (2) |
| O2—H2A⋯Cl2 | 0.83 (2) | 2.35 (2) | 3.170 (2) | 175 (3) |
| O2—H2B⋯O3 | 0.84 (2) | 1.92 (2) | 2.739 (3) | 167 (3) |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 .
.
Supramolecular features
The water molecules and chloride anions are located in channels between the organic cations and are connected by O—H⋯O and O—H⋯Cl hydrogen bonds (Table 1 ▸) into chains, which are further connected via N—H⋯O and C—H⋯Cl hydrogen bonds into a three-dimensional supramolecular architecture. Fig. 2 ▸ a shows a view of the hydrogen bonds along the b-axis direction. π–π interactions are present (Fig. 2 ▸ b) between the pyridazine rings [centroid–centroid distance = 3.4902 (12) Å], and also between the pyridine and benzene rings [3.7293 (13) and 3.8488 (13) Å], forming sheets.
Figure 2.
(a)View along the b axis of the unit cell showing the molecular sheets. (b) π–π interactions.
Database survey
There are no direct precedents for the structure of the title compound in the crystallographic literature. A search of the Cambridge Structural Database (ConQuest version 2021.3.0; Groom et al., 2016 ▸) for the 2,3-dihydropyridazin-4-yl moiety gave various hits, four of them for similar pyridazine compounds but with different substituents on the pyridazine ring: 5-(2-chlorobenzyl)-6-methyl-3(2H)pyridazinone (ZAYJIS; Moreau et al., 1995 ▸), 2-{4-[(5-chloro- 1-benzofuran-2-yl)methyl]-3-methyl-6- oxo-1,6-dihydropyridazin-1-yl}acetate (XULSEE; Boukharsa et al., 2015 ▸) , 4-[3-(trifluoromethyl)phenyl]-5,6,7,8-tetrahydrocinnolin-3(2H)-one (GISZAK; Wang et al., 2008 ▸) and 5-(2-Chlorobenzyl)-2-(2-hydroxyethyl)-6-methylpyridazin-3(2H)-one (IJEMOZ; Abourichaa et al., 2003 ▸). In ZAYJIS, the lengths of the C=C [1.343 (3) Å], C=N [1.301 (4) Å], N—N [1.357 (3) Å] and C=O [1.255 (3) Å] bonds in the pyridazinone ring are very similar to those in the title compound. In XULSEE, te Cl—C1 bond length is 1.742 (2) Å while in the pyridazine ring, the N1—N2 bond length is 1.365 (2) Å and O2=C2 is 1.228 (2) Å. In GISZAK, the N1—N2 bond is 1.343 (5) Å whereas the C8=O1 bond is 1.246 (5) Å. In IJEMOZ, the pyridazinone ring has a similar value for the N4—N5 bond of 1.367 (2) Å.
Hirshfeld surface analysis
To investigate the effect of the molecular interactions on the crystal packing, the Hirshfeld surface (Fig. 3 ▸) and fingerprint plots of the organic cation were analysed (Turner et al., 2017 ▸). In Fig. 4 ▸ a, the circular depressions (deep red) on the Hirshfeld surface imply strong hydrogen-bonding interactions of types C—H⋯Cl, N—H⋯O, O—H⋯Cl and O—H⋯O. In the shape-index map (Fig. 4 ▸ b), the π–π interactions are indicated by the red and blue triangles. Fig. 4 ▸ c and Fig. 4 ▸ d show d i and d e surfaces and Fig. 4 ▸ e and 4f the curvedness and fragment path surfaces. Fig. 5 ▸ a shows the overall two-dimensional fingerprint plot. The fingerprint plot delineated into H⋯H contacts (33.0% contribution, Fig. 5 ▸ b) has a point with the tip at d e + d i = 2.05 Å. The pair of wings in the fingerprint plot defined into H⋯C/C⋯H contacts (19.3 percent contribution to the HS), Fig.5c, has a pair of thin edges at d e + d i ∼2.99 Å while the pair of wings for the H⋯Cl/Cl⋯H contacts (15.9% contribution, Fig. 5 ▸ d) are seen as two spikes with the points at d e + d i = 2.97 Å and d e + d i = 2.41 Å. The fingerprint plot for H⋯O/O⋯H contacts (11.5% contribution, Fig. 5 ▸ e) has two spikes with the tips at d e + d i = 2.11 Å and d e + d i = 1.83 Å. As seen in Fig. 5 ▸ f the C⋯C contacts (7.4%) have an arrow-shaped distribution of points with tips at d e + d i = 3.37 Å. The contributions of the N⋯H/H⋯N contacts to the Hirshfeld surface (5.8%) are less important (Fig. 5 ▸ g). Fig. 6 ▸ shows a pie chart of all interactions with their percentage contributions.
Figure 3.
Intermolecular interactions with d norm surface.
Figure 4.
Graphical depictions of the molecular Hirshfeld surfaces; (a) d norm, (b)shape-index, (c) d i, (d) d e,(e) curvedness and (f) fragment-path.
Figure 5.

Fingerprint plots of the interactions involving the organic cation. (a) All contributions and decomposed into the main contributions: (b) H⋯H, (c) H⋯C/C⋯H, (d) H⋯Cl/Cl⋯H, (e) H⋯O/O⋯H, (f) C⋯C and (g) N⋯H/H⋯N interactions
Figure 6.
All interactions with percentage contributions.
Synthesis and crystallization
The title compound was synthesized according to a previously published procedure (Daoui et al., 2019 ▸, 2021 ▸). To a solution of (E)-6-(4-chlorostyryl)-4,5-dihydropyridazin-3(2H)-one (0.23 g, 1 mmol) and nicotinaldehyde (0.107 g, 1 mmol) in 30 ml of ethanol, sodium ethanoate (0.23 g, 2.8 mmol) was added. The mixture was refluxed for 3 h. The reaction mixture was cooled, diluted with cold water and acidified with concentrated hydrochloric acid. The precipitate was filtered, washed with water, dried and recrystallized from ethanol. White single crystals were obtained by slow evaporation at room temperature, yield 86%; m.p. 453 K; FT–IR (KBr): ν 3322 (NH), 1651 (C=O), 1584 cm−1 (C=N); 1H NMR (300 MHz, DMSO-d 6) δ 13.20 (s, 1H, H-pyridyl) , 8.98 (d, J = 1.8 Hz, 1H, H-pyridyl), 8.83 (d, J = 5.6 Hz, 1H, H-pyridyl), 8.57 (dt, J = 8.1, 1.8 Hz, 1H, H-pyridyl), 8.05 (s, 1H, H-pyridazinone) 8.02 (dd, J = 8.1, 5.6 Hz, 1H, H-pyridyl), 7.65 (d, J = 8.4 Hz, 2H, H1, H-Ar), 7.45 (d, J = 8.4 Hz, 2H, H 4, H-Ar), 7.36 (d, J = 16.7 Hz, 1H, CH=CH), 7.08 (,d J = 16.7 Hz, 1H, CH=CH), 4.09 ppm (s, 2H, CH2); 13C NMR (75 MHz, DMSO-d 6) δ 160.43, 145.98, 143.89, 141.87, 140.05, 139.25, 137.97, 134.90, 132.84,130.85, 128.82, 128.62, 128.54, 126.80, 125.08, 32.33 ppm. ESI-MS: m/z = 324.08 [M+H]+.
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All C-bound H atoms were placed in calculated positions (C—H = 0.93–0.98 Å) and thereafter treated as riding. A torsional parameter was refined for the methyl group. The positions of N- and O-bound H atoms were refined freely (distances are in Table 1 ▸). For all H atoms, U iso(H) = 1.2 U eq(C,N,O).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C18H15ClN3O+·Cl−·2H2O |
| M r | 396.26 |
| Crystal system, space group | Monoclinic, I2/a |
| Temperature (K) | 296 |
| a, b, c (Å) | 19.6562 (14), 7.5587 (3), 26.4903 (16) |
| β (°) | 109.762 (5) |
| V (Å3) | 3704.0 (4) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.37 |
| Crystal size (mm) | 0.68 × 0.41 × 0.16 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Numerical (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.818, 0.961 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 13762, 5273, 3083 |
| R int | 0.064 |
| (sin θ/λ)max (Å−1) | 0.702 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.050, 0.142, 0.98 |
| No. of reflections | 5273 |
| No. of parameters | 265 |
| No. of restraints | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.26, −0.43 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022003346/jq2014sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022003346/jq2014Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989022003346/jq2014Isup3.cml
CCDC reference: 2161716
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge the Faculty of Arts and Sciences, Ondokuz Mayıs University, Turkey, for the use of the Stoe IPDS 2 diffractometer. The authors’ contributions are as follows. Conceptualization, SD, EBÇ, ND, and ES; methodology, KK, EBÇ, and ND; investigation, NB and ND; writing (original draft), EBÇ and SD; writing (review and editing of the manuscript), SD, NB, ES, KK and EBÇ; visualization, EBÇ, and KK; funding acquisition, ND; resources, ND and KK; supervision, SD and NB.
supplementary crystallographic information
Crystal data
| C18H15ClN3O+·Cl−·2H2O | F(000) = 1648 |
| Mr = 396.26 | Dx = 1.421 Mg m−3 |
| Monoclinic, I2/a | Mo Kα radiation, λ = 0.71073 Å |
| a = 19.6562 (14) Å | Cell parameters from 18653 reflections |
| b = 7.5587 (3) Å | θ = 1.6–30.3° |
| c = 26.4903 (16) Å | µ = 0.37 mm−1 |
| β = 109.762 (5)° | T = 296 K |
| V = 3704.0 (4) Å3 | Prism, colorless |
| Z = 8 | 0.68 × 0.41 × 0.16 mm |
Data collection
| Stoe IPDS 2 diffractometer | 5273 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 3083 reflections with I > 2σ(I) |
| Plane graphite monochromator | Rint = 0.064 |
| Detector resolution: 6.67 pixels mm-1 | θmax = 29.9°, θmin = 1.6° |
| rotation method scans | h = −21→27 |
| Absorption correction: numerical (X-RED32; Stoe & Cie, 2002) | k = −8→10 |
| Tmin = 0.818, Tmax = 0.961 | l = −36→36 |
| 13762 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: mixed |
| wR(F2) = 0.142 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.98 | w = 1/[σ2(Fo2) + (0.0709P)2] where P = (Fo2 + 2Fc2)/3 |
| 5273 reflections | (Δ/σ)max < 0.001 |
| 265 parameters | Δρmax = 0.26 e Å−3 |
| 2 restraints | Δρmin = −0.43 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl2 | 0.43892 (4) | 0.44826 (8) | 0.29544 (2) | 0.06204 (18) | |
| Cl1 | 0.16095 (4) | 0.93975 (11) | 0.67565 (3) | 0.0831 (2) | |
| O2 | 0.51631 (9) | 0.7860 (3) | 0.36086 (6) | 0.0569 (4) | |
| O1 | 0.63332 (8) | 0.6580 (2) | 0.47656 (6) | 0.0603 (4) | |
| N2 | 0.52423 (9) | 0.7727 (2) | 0.46837 (7) | 0.0440 (4) | |
| N1 | 0.46811 (9) | 0.8166 (2) | 0.48443 (6) | 0.0437 (4) | |
| O3 | 0.47043 (12) | 1.0366 (3) | 0.28189 (9) | 0.0724 (5) | |
| N3 | 0.83161 (10) | 0.6802 (3) | 0.61940 (8) | 0.0521 (4) | |
| C11 | 0.58620 (10) | 0.6148 (3) | 0.54755 (7) | 0.0414 (4) | |
| C9 | 0.47235 (10) | 0.7645 (3) | 0.53269 (7) | 0.0427 (4) | |
| C12 | 0.58492 (10) | 0.6822 (3) | 0.49587 (7) | 0.0434 (4) | |
| C15 | 0.71539 (10) | 0.5767 (3) | 0.61025 (7) | 0.0420 (4) | |
| C6 | 0.34431 (11) | 0.8182 (3) | 0.61458 (8) | 0.0470 (5) | |
| C10 | 0.53148 (11) | 0.6600 (3) | 0.56490 (7) | 0.0441 (4) | |
| H10 | 0.5323 | 0.6223 | 0.5985 | 0.053* | |
| C8 | 0.41189 (11) | 0.8140 (3) | 0.54971 (8) | 0.0477 (5) | |
| H8 | 0.3747 | 0.8785 | 0.5256 | 0.057* | |
| C7 | 0.40518 (11) | 0.7752 (3) | 0.59642 (8) | 0.0481 (5) | |
| H7 | 0.4434 | 0.7136 | 0.6206 | 0.058* | |
| C14 | 0.76951 (11) | 0.6075 (3) | 0.58944 (8) | 0.0479 (5) | |
| H14 | 0.7626 | 0.5772 | 0.5540 | 0.057* | |
| C13 | 0.64570 (11) | 0.4898 (3) | 0.57732 (8) | 0.0496 (5) | |
| H13A | 0.6554 | 0.4116 | 0.5515 | 0.060* | |
| H13B | 0.6288 | 0.4173 | 0.6009 | 0.060* | |
| C5 | 0.34973 (12) | 0.7804 (3) | 0.66698 (9) | 0.0540 (5) | |
| H5 | 0.3919 | 0.7288 | 0.6898 | 0.065* | |
| C16 | 0.72876 (12) | 0.6223 (3) | 0.66349 (8) | 0.0514 (5) | |
| H16 | 0.6936 | 0.6025 | 0.6792 | 0.062* | |
| C18 | 0.84516 (12) | 0.7257 (3) | 0.67006 (9) | 0.0577 (5) | |
| H18 | 0.8892 | 0.7768 | 0.6897 | 0.069* | |
| C3 | 0.23208 (13) | 0.8927 (3) | 0.65221 (9) | 0.0566 (6) | |
| C2 | 0.22442 (12) | 0.9330 (3) | 0.60014 (9) | 0.0583 (6) | |
| H2 | 0.1820 | 0.9840 | 0.5776 | 0.070* | |
| C1 | 0.28082 (12) | 0.8966 (3) | 0.58179 (9) | 0.0561 (5) | |
| H1 | 0.2762 | 0.9252 | 0.5466 | 0.067* | |
| C17 | 0.79392 (13) | 0.6969 (3) | 0.69313 (9) | 0.0593 (6) | |
| H17 | 0.8029 | 0.7274 | 0.7288 | 0.071* | |
| C4 | 0.29405 (13) | 0.8174 (3) | 0.68616 (9) | 0.0600 (6) | |
| H4 | 0.2986 | 0.7917 | 0.7215 | 0.072* | |
| H3 | 0.8616 (16) | 0.701 (4) | 0.6061 (11) | 0.070 (8)* | |
| H2C | 0.5201 (13) | 0.802 (3) | 0.4362 (10) | 0.053 (6)* | |
| H2A | 0.4937 (17) | 0.700 (3) | 0.3444 (12) | 0.094 (11)* | |
| H2B | 0.5030 (16) | 0.874 (3) | 0.3409 (10) | 0.079 (9)* | |
| H3A | 0.495 (3) | 1.018 (6) | 0.2630 (17) | 0.127 (16)* | |
| H3B | 0.466 (2) | 1.141 (6) | 0.2847 (14) | 0.095 (13)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl2 | 0.0694 (4) | 0.0648 (4) | 0.0496 (3) | 0.0006 (3) | 0.0170 (2) | 0.0021 (2) |
| Cl1 | 0.0642 (4) | 0.1042 (6) | 0.0982 (5) | −0.0103 (4) | 0.0502 (4) | −0.0206 (4) |
| O2 | 0.0539 (9) | 0.0660 (12) | 0.0463 (8) | 0.0028 (9) | 0.0111 (7) | 0.0035 (8) |
| O1 | 0.0471 (8) | 0.0848 (12) | 0.0534 (8) | 0.0146 (8) | 0.0229 (7) | 0.0071 (8) |
| N2 | 0.0415 (8) | 0.0494 (10) | 0.0429 (8) | 0.0012 (8) | 0.0168 (7) | 0.0023 (7) |
| N1 | 0.0375 (8) | 0.0469 (10) | 0.0463 (8) | 0.0001 (7) | 0.0138 (7) | 0.0001 (7) |
| O3 | 0.0801 (14) | 0.0676 (14) | 0.0748 (12) | 0.0046 (11) | 0.0331 (10) | 0.0102 (10) |
| N3 | 0.0397 (9) | 0.0596 (12) | 0.0591 (10) | 0.0003 (9) | 0.0195 (8) | 0.0078 (8) |
| C11 | 0.0363 (9) | 0.0416 (10) | 0.0427 (9) | −0.0032 (8) | 0.0089 (7) | −0.0017 (7) |
| C9 | 0.0394 (9) | 0.0448 (11) | 0.0431 (9) | −0.0026 (9) | 0.0128 (7) | −0.0010 (8) |
| C12 | 0.0385 (9) | 0.0455 (11) | 0.0454 (9) | −0.0018 (8) | 0.0130 (8) | −0.0034 (8) |
| C15 | 0.0373 (9) | 0.0417 (11) | 0.0445 (9) | 0.0049 (8) | 0.0107 (7) | 0.0040 (7) |
| C6 | 0.0431 (10) | 0.0513 (12) | 0.0468 (10) | −0.0051 (9) | 0.0153 (8) | −0.0065 (8) |
| C10 | 0.0424 (10) | 0.0486 (12) | 0.0396 (9) | −0.0033 (9) | 0.0116 (8) | 0.0009 (8) |
| C8 | 0.0402 (10) | 0.0529 (12) | 0.0479 (10) | 0.0024 (9) | 0.0123 (8) | −0.0003 (8) |
| C7 | 0.0390 (10) | 0.0570 (13) | 0.0463 (10) | 0.0018 (9) | 0.0119 (8) | −0.0015 (8) |
| C14 | 0.0458 (11) | 0.0560 (12) | 0.0423 (9) | 0.0039 (10) | 0.0154 (8) | 0.0037 (8) |
| C13 | 0.0397 (10) | 0.0481 (12) | 0.0552 (10) | 0.0008 (9) | 0.0085 (9) | 0.0019 (9) |
| C5 | 0.0495 (11) | 0.0632 (14) | 0.0496 (11) | −0.0041 (11) | 0.0171 (9) | −0.0003 (9) |
| C16 | 0.0483 (11) | 0.0615 (13) | 0.0473 (10) | 0.0002 (10) | 0.0200 (9) | 0.0003 (9) |
| C18 | 0.0437 (11) | 0.0615 (14) | 0.0594 (12) | −0.0045 (10) | 0.0062 (9) | 0.0012 (10) |
| C3 | 0.0494 (11) | 0.0620 (14) | 0.0662 (13) | −0.0148 (11) | 0.0297 (10) | −0.0187 (10) |
| C2 | 0.0414 (11) | 0.0720 (16) | 0.0589 (12) | −0.0006 (11) | 0.0133 (9) | −0.0128 (11) |
| C1 | 0.0500 (11) | 0.0731 (15) | 0.0453 (10) | 0.0025 (11) | 0.0163 (9) | −0.0048 (10) |
| C17 | 0.0588 (13) | 0.0703 (16) | 0.0449 (10) | −0.0028 (12) | 0.0125 (10) | −0.0049 (10) |
| C4 | 0.0611 (14) | 0.0736 (16) | 0.0527 (12) | −0.0123 (12) | 0.0290 (11) | −0.0046 (10) |
Geometric parameters (Å, º)
| Cl1—C3 | 1.748 (2) | C6—C1 | 1.389 (3) |
| O2—H2A | 0.825 (18) | C6—C7 | 1.469 (3) |
| O2—H2B | 0.837 (18) | C10—H10 | 0.9300 |
| O1—C12 | 1.237 (2) | C8—C7 | 1.321 (3) |
| N2—N1 | 1.351 (2) | C8—H8 | 0.9300 |
| N2—C12 | 1.354 (3) | C7—H7 | 0.9300 |
| N2—H2C | 0.86 (2) | C14—H14 | 0.9300 |
| N1—C9 | 1.313 (2) | C13—H13A | 0.9700 |
| O3—H3A | 0.81 (5) | C13—H13B | 0.9700 |
| O3—H3B | 0.80 (4) | C5—C4 | 1.382 (3) |
| N3—C18 | 1.322 (3) | C5—H5 | 0.9300 |
| N3—C14 | 1.329 (3) | C16—C17 | 1.376 (3) |
| N3—H3 | 0.80 (3) | C16—H16 | 0.9300 |
| C11—C10 | 1.349 (3) | C18—C17 | 1.361 (3) |
| C11—C12 | 1.453 (3) | C18—H18 | 0.9300 |
| C11—C13 | 1.503 (3) | C3—C4 | 1.369 (4) |
| C9—C10 | 1.426 (3) | C3—C2 | 1.370 (3) |
| C9—C8 | 1.455 (3) | C2—C1 | 1.380 (3) |
| C15—C14 | 1.373 (3) | C2—H2 | 0.9300 |
| C15—C16 | 1.388 (3) | C1—H1 | 0.9300 |
| C15—C13 | 1.504 (3) | C17—H17 | 0.9300 |
| C6—C5 | 1.386 (3) | C4—H4 | 0.9300 |
| H2A—O2—H2B | 107 (3) | N3—C14—C15 | 120.65 (18) |
| N1—N2—C12 | 128.25 (16) | N3—C14—H14 | 119.7 |
| N1—N2—H2C | 116.0 (16) | C15—C14—H14 | 119.7 |
| C12—N2—H2C | 115.7 (16) | C11—C13—C15 | 115.12 (17) |
| C9—N1—N2 | 116.31 (16) | C11—C13—H13A | 108.5 |
| H3A—O3—H3B | 109 (4) | C15—C13—H13A | 108.5 |
| C18—N3—C14 | 122.87 (19) | C11—C13—H13B | 108.5 |
| C18—N3—H3 | 118 (2) | C15—C13—H13B | 108.5 |
| C14—N3—H3 | 119 (2) | H13A—C13—H13B | 107.5 |
| C10—C11—C12 | 118.06 (18) | C4—C5—C6 | 121.6 (2) |
| C10—C11—C13 | 123.32 (18) | C4—C5—H5 | 119.2 |
| C12—C11—C13 | 118.51 (17) | C6—C5—H5 | 119.2 |
| N1—C9—C10 | 121.28 (17) | C17—C16—C15 | 120.08 (19) |
| N1—C9—C8 | 115.79 (17) | C17—C16—H16 | 120.0 |
| C10—C9—C8 | 122.88 (17) | C15—C16—H16 | 120.0 |
| O1—C12—N2 | 120.86 (17) | N3—C18—C17 | 119.2 (2) |
| O1—C12—C11 | 124.57 (18) | N3—C18—H18 | 120.4 |
| N2—C12—C11 | 114.55 (16) | C17—C18—H18 | 120.4 |
| C14—C15—C16 | 117.37 (19) | C4—C3—C2 | 121.6 (2) |
| C14—C15—C13 | 121.23 (17) | C4—C3—Cl1 | 119.49 (17) |
| C16—C15—C13 | 121.36 (18) | C2—C3—Cl1 | 118.91 (19) |
| C5—C6—C1 | 117.58 (18) | C3—C2—C1 | 118.8 (2) |
| C5—C6—C7 | 119.16 (19) | C3—C2—H2 | 120.6 |
| C1—C6—C7 | 123.26 (18) | C1—C2—H2 | 120.6 |
| C11—C10—C9 | 121.28 (17) | C2—C1—C6 | 121.6 (2) |
| C11—C10—H10 | 119.4 | C2—C1—H1 | 119.2 |
| C9—C10—H10 | 119.4 | C6—C1—H1 | 119.2 |
| C7—C8—C9 | 125.74 (19) | C18—C17—C16 | 119.8 (2) |
| C7—C8—H8 | 117.1 | C18—C17—H17 | 120.1 |
| C9—C8—H8 | 117.1 | C16—C17—H17 | 120.1 |
| C8—C7—C6 | 127.5 (2) | C3—C4—C5 | 118.8 (2) |
| C8—C7—H7 | 116.3 | C3—C4—H4 | 120.6 |
| C6—C7—H7 | 116.3 | C5—C4—H4 | 120.6 |
| C12—N2—N1—C9 | −0.4 (3) | C13—C15—C14—N3 | −178.22 (19) |
| N2—N1—C9—C10 | −3.0 (3) | C10—C11—C13—C15 | −100.2 (2) |
| N2—N1—C9—C8 | 179.47 (17) | C12—C11—C13—C15 | 83.7 (2) |
| N1—N2—C12—O1 | −177.03 (19) | C14—C15—C13—C11 | −92.5 (2) |
| N1—N2—C12—C11 | 4.6 (3) | C16—C15—C13—C11 | 90.2 (2) |
| C10—C11—C12—O1 | 176.3 (2) | C1—C6—C5—C4 | 0.5 (3) |
| C13—C11—C12—O1 | −7.4 (3) | C7—C6—C5—C4 | −179.8 (2) |
| C10—C11—C12—N2 | −5.4 (3) | C14—C15—C16—C17 | 0.6 (3) |
| C13—C11—C12—N2 | 170.88 (18) | C13—C15—C16—C17 | 178.0 (2) |
| C12—C11—C10—C9 | 2.6 (3) | C14—N3—C18—C17 | 0.3 (4) |
| C13—C11—C10—C9 | −173.49 (19) | C4—C3—C2—C1 | 0.0 (4) |
| N1—C9—C10—C11 | 1.8 (3) | Cl1—C3—C2—C1 | 179.66 (18) |
| C8—C9—C10—C11 | 179.15 (19) | C3—C2—C1—C6 | 0.9 (4) |
| N1—C9—C8—C7 | 179.9 (2) | C5—C6—C1—C2 | −1.1 (3) |
| C10—C9—C8—C7 | 2.5 (3) | C7—C6—C1—C2 | 179.2 (2) |
| C9—C8—C7—C6 | −178.2 (2) | N3—C18—C17—C16 | −0.5 (4) |
| C5—C6—C7—C8 | −174.2 (2) | C15—C16—C17—C18 | 0.0 (4) |
| C1—C6—C7—C8 | 5.4 (4) | C2—C3—C4—C5 | −0.5 (4) |
| C18—N3—C14—C15 | 0.3 (3) | Cl1—C3—C4—C5 | 179.77 (19) |
| C16—C15—C14—N3 | −0.8 (3) | C6—C5—C4—C3 | 0.3 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C10—H10···Cl2i | 0.93 | 2.72 | 3.6387 (19) | 168 |
| C18—H18···Cl2ii | 0.93 | 2.94 | 3.622 (2) | 132 |
| N3—H3···O2iii | 0.80 (3) | 2.35 (3) | 2.965 (2) | 135 (2) |
| N3—H3···O1iii | 0.80 (3) | 2.25 (3) | 2.855 (2) | 133 (3) |
| N2—H2C···O2 | 0.86 (2) | 1.97 (2) | 2.801 (2) | 161 (2) |
| O2—H2A···Cl2 | 0.83 (2) | 2.35 (2) | 3.170 (2) | 175 (3) |
| O2—H2B···O3 | 0.84 (2) | 1.92 (2) | 2.739 (3) | 167 (3) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x+1/2, y+1/2, z+1/2; (iii) −x+3/2, y, −z+1.
Funding Statement
This work was funded by Funding for this research was provided by: Ondokuz May?is University under Project No. PYO.FEN.1906.19.001.
References
- Abourichaa, S., Benchat, N., Anaflous, A., Melhaoui, A., Ben-Hadda, T., Oussaid, B., Mimouni, M., El Bali, B. & Bolte, M. (2003). Acta Cryst. E59, o1041–o1042.
- Abu-Hashem, A. A., Fathy, U. & Gouda, M. A. (2020). J. Heterocycl. Chem. 57, 3461–3474.
- Bahou, C., Szijj, P. A., Spears, R. J., Wall, A., Javid, F., Sattikar, A., Love, E. A., Baker, J. R. & Chudasama, V. (2021). Bioconjugate Chem. 32, 672–679. [DOI] [PMC free article] [PubMed]
- Boukharsa, Y., El Ammari, L., Taoufik, J., Saadi, M. & Ansar, M. (2015). Acta Cryst. E71, o291–o292. [DOI] [PMC free article] [PubMed]
- Daoui, S., Baydere, C., El Kalai, F., Saddik, R., Dege, N., Karrouchi, K. & Benchat, N. (2019). Acta Cryst. E75, 1734–1737. [DOI] [PMC free article] [PubMed]
- Daoui, S., Cinar, E. B., Dege, N., Chelfi, T., El Kalai, F., Abudunia, A., Karrouchi, K. & Benchat, N. (2021). Acta Cryst. E77, 23–27. [DOI] [PMC free article] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Khadiria, A., Saddik, R., Bekkouchea, K., Aouniti, A., Hammouti, B., Benchat, N., Bouachrine, M. & Solmaz, R. (2016). J. Taiwan Inst. Chem. Eng. 58, 552–564.
- Knall, A. C., Rabensteiner, S., Hoefler, S. F., Reinfelds, M., Hobisch, M., Ehmann, H. M. A., Pastukhova, N., Pavlica, E., Bratina, G., Honzu, I., Wen, S., Yang, R., Trimmel, G. & Rath, T. (2021). New J. Chem. 45, 1001–1009.
- Kung, Y. J., Chung, H. A., Kim, J. J. & Yoon, Y. J. (2002). Synthesis, 6, 733–738.
- Li, M., Yuan, Y. & Chen, Y. (2018). ACS Appl. Mater. Interfaces. 10, 1237–1243. [DOI] [PubMed]
- Liu, S., Zhang, X., Ou, C., Wang, S., Yang, X., Zhou, X., Mi, B., Cao, D. & Gao, Z. (2017). ACS Appl. Mater. Interfaces. 9, 26242–26251. [DOI] [PubMed]
- Liu, X. & Chen, W. (2012). Organometallics, 31, 6614–6622.
- Llona-Minguez, S., Höglund, A., Ghassemian, A., Desroses, M., Calderón, J. M., Valerie, N. C. K., Witta, E., Almlöf, I., Koolmeister, T., Mateus, A., Cazares-Körner, C., Sanjiv, K., Homan, E., Loseva, O., Baranczewski, P., Darabi, M., Mehdizadeh, A., Fayezi, S., Jemth, A. S., Berglund, U. W., Sigmundsson, K., Lundbäck, T., Jensen, A. J., Artursson, P., Scobie, M. & Helleday, T. J. (2017). Med. Chem. 60, 4279–4292.
- Macrae, C. F., Sovago, I., Cottrell, S. J., Galek, P. T. A., McCabe, P., Pidcock, E., Platings, M., Shields, G. P., Stevens, J. S., Towler, M. & Wood, P. A. (2020). J. Appl. Cryst. 53, 226–235. [DOI] [PMC free article] [PubMed]
- Moreau, S., Metin, J., Coudert, P. & Couquelet, J. (1995). Acta Cryst. C51, 1834–1836.
- Mousavi, H. (2022). J. Mol. Struct. 1251, 131742–131771.
- Neumann, K., Gambardella, A., Lilienkampf, A. & Bradley, M. (2018). Chem. Sci. 9, 7198–7203. [DOI] [PMC free article] [PubMed]
- Peng, S., Lv, J., Liu, G., Fan, C. & Pu, S. (2020). Tetrahedron, 76, 131618–131627.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Stoe & Cie. (2002). X-AREA and X-RED32. Stoe & Cie GmbH, Darmstadt, Germany.
- Turner, M. J., MacKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer 17.5. University of Western Australia. http://hirshfeldsurface.net.
- Wang, X., Zou, X.-M., Zhu, Y.-Q., Hu, X.-H. & Yang, H.-Z. (2008). Acta Cryst. E64, o464. [DOI] [PMC free article] [PubMed]
- Westrip, S. P. (2010). J. Appl. Cryst. 43, 920–925.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022003346/jq2014sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022003346/jq2014Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989022003346/jq2014Isup3.cml
CCDC reference: 2161716
Additional supporting information: crystallographic information; 3D view; checkCIF report