The packing of the title compound features C—H⋯O hydrogen bonds, C—F⋯π interactions, aromatic π–π stacking and short Br⋯O contacts.
Keywords: crystal structure, C—F⋯π interaction, π–π stacking interaction, Hirshfeld surface analysis
Abstract
In the title compound, C14H8Br2FN3O2, the nitro-substituted benzene ring and the 4-fluorophenyl ring form a dihedral angle of 65.73 (7)°. In the crystal, molecules are linked into chains by C—H⋯O hydrogen bonds running parallel to the c-axis direction. The crystal packing is consolidated by C—F⋯π interactions and π–π stacking interactions, and short Br⋯O [2.9828 (13) Å] contacts are observed. The Hirshfeld surface analysis of the crystal structure indicates that the most important contributions to the crystal packing are from H⋯H (17.4%), O⋯H/H⋯O (16.3%), Br⋯H/H⋯Br (15.5%), Br⋯C/C⋯Br (10.1%) and F⋯H/H⋯F (8.1%) contacts.
Chemical context
Azo dyes are chemical compounds with the general formula R—N=N—R′, where R and R′ can be either aryl, hetrocycle or alkyl functional groups. They find many applications such as molecular switches, optical data storage, antimicrobial agent, colour-changing materials, non-linear optics, molecular recognition, dye-sensitized solar cells, liquid crystals, catalysis, etc. (see, for example, Kopylovich et al., 2012 ▸; MacLeod et al., 2012 ▸; Viswanathan et al., 2019 ▸). Both E/Z isomerization and azo-to-hydrazo tautomerization of azo dyes is an important feature in the synthesis and design of new functional materials (Mahmudov et al., 2012 ▸, 2020 ▸; Mizar et al., 2012 ▸). On the other hand, the attachment of non-covalent bond-donor or acceptor centres to the azo dyes can be used as a synthetic strategy for the improvement of the functional properties of this class of organic compounds (Gurbanov et al., 2020a
▸,b
▸).
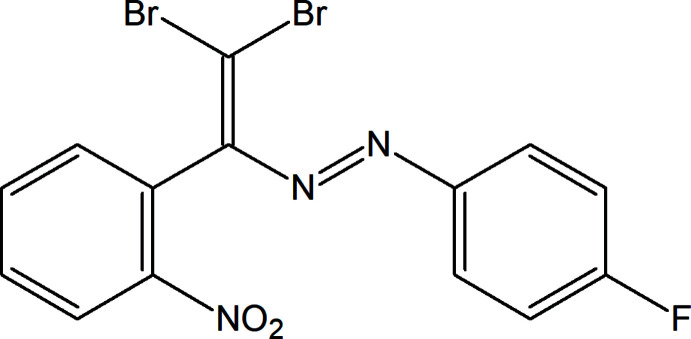
As part of our ongoing work in this area we have attached –F, –Br and –NO2 functional groups and aryl rings to the —N=N— moiety, leading to the title compound, C14H8Br2FN3O2, and determined its crystal structure.
Structural commentary
As shown in Fig. 1 ▸, the molecular conformation of the title compound is not planar, the nitro-substituted benzene ring and the 4-fluorophenyl ring forming a dihedral angle of 65.73 (7)°. There is a slight twist about the C1=C2 double bond with the dihedral angle between C1/Br1/Br2 and C2/C3/N2 being 3.35 (15)°, perhaps to minimize steric repulsion between Br2 and H8. Considered together, the N3/N2/C2/C1/Br1/Br2 moiety subtends dihedral angles of 70.40 (7) and 14.14 (7)° with the C3–C8 and C9–C14 rings, respectively. In the molecule, the aromatic ring and olefin synthon adopt a trans-configuration with respect to the N=N double bond and are almost coplanar with a C2—N2=N3—C9 torsion angle of −178.50 (11)°. All of the other bond lengths and angles in the title compound are similar to those in the related azo compounds reported in the Database survey.
Figure 1.
The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level.
Supramolecular features
In the crystal, molecules are linked by C—H⋯O hydrogen bonds into chains propagating parallel to the c axis (Table 1 ▸; Fig. 2 ▸). The crystal packing is consolidated by weak C—F⋯π [F1⋯Cg1(x, 1 − y, −
 + z) = 3.4095 (12) Å; C—F⋯Cg1 = 136.95 (9)°] interactions and weak aromatic π–π stacking [Cg2⋯Cg2(1 − x, y,
+ z) = 3.4095 (12) Å; C—F⋯Cg1 = 136.95 (9)°] interactions and weak aromatic π–π stacking [Cg2⋯Cg2(1 − x, y,
 − z) = 3.9694 (9) Å], where Cg1 and Cg2 are the centroids of the C3–C8 and C9–C14 rings, respectively (Fig. 2 ▸). In addition, short bromine–oxygen contacts [Br2⋯O2(
− z) = 3.9694 (9) Å], where Cg1 and Cg2 are the centroids of the C3–C8 and C9–C14 rings, respectively (Fig. 2 ▸). In addition, short bromine–oxygen contacts [Br2⋯O2(
 − x,
− x,
 + y, z) = 2.9828 (13) Å; van der Waals contact distance = 3.37 Å] are observed.
+ y, z) = 2.9828 (13) Å; van der Waals contact distance = 3.37 Å] are observed.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C6—H6⋯O1i | 0.95 | 2.51 | 3.3244 (18) | 144 |
Symmetry code: (i)
 .
.
Figure 2.
View of the C—H⋯O, C—F⋯π and π–π stacking interactions in the title compound.
Hirshfeld surface analysis
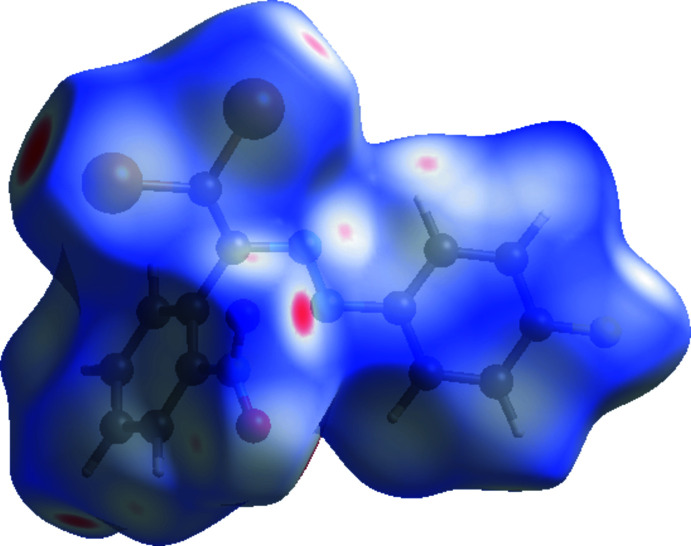
CrystalExplorer17 (Turner et al., 2017 ▸) was used to calculate the Hirshfeld surfaces for the title compound and generate the two-dimensional fingerprint plots. On the d norm surface, red, white, and blue regions indicate contacts with distances shorter, longer, and roughly equal to the van der Waals radii for the title compound (Fig. 3 ▸, Tables 1 ▸ and 2 ▸).
Figure 3.
The three-dimensional Hirshfeld surface of the title compound plotted over d norm in the range −0.24 to 1.44 a.u.
Table 2. Summary of short interatomic contacts (Å) in the title salt.
| Contact | Distance | Symmetry operation |
|---|---|---|
| H8⋯Br1 | 2.99 | 1 − x, y,
 − z
− z
|
| O1⋯H11 | 2.68 |
 − x,
− x,
 + y, z
+ y, z
|
| Br1⋯Br2 | 3.6164 |
x, 2 − y, −
 + z
+ z
|
| H7⋯Br2 | 3.19 | 1 − x, 2 − y, 1 − z |
| H13⋯F1 | 2.82 | 1 − x, 1 − y, −z |
| F1⋯H10 | 2.67 |
x, 1 − y, −
 + z
+ z
|
| O1⋯H6 | 2.51 |
 − x,
− x,
 − y, −
− y, −
 + z
+ z
|
| O2⋯H8 | 2.77 |
 + x,
+ x,
 − y, 1 − z
− y, 1 − z
|
| H7⋯H6 | 2.47 | 1 − x, y,
 − z
− z
|
The overall two-dimensional fingerprint plot (Fig. 4 ▸ a) and those delineated into H⋯H, O⋯H/H⋯O, Br⋯H/H⋯Br, Br⋯C/C⋯Br and F⋯H/H⋯F contacts (McKinnon et al., 2007 ▸) are illustrated in Fig. 4 ▸ b–f, respectively. The most important interaction is H⋯H, contributing 17.4% to the overall surface, which is reflected in Fig. 4 ▸ b as widely scattered points of high density due to the large hydrogen content of the molecule, with the tip at d e = d i = 1.15 Å. The reciprocal O⋯H/H⋯O interactions appear as two symmetrical broad wings with d e + d i ≃ 2.40 Å and contribute 16.3% to the Hirshfeld surface (Fig. 4 ▸ c). In the Br⋯H/H⋯Br fingerprint plot, there are two symmetrical wings with d e + d i ≃ 2.85 Å and they contribute 15.5% to the Hirshfeld surface (Fig. 4 ▸ d). The pair of characteristic wings in the fingerprint plot delineated into Br⋯C/C⋯Br contacts (Fig. 8e; 10.1% contribution to the Hirshfeld surface), have the tips at d e + d i ≃ 3.80 Å, while for F⋯H/H⋯F contacts (Fig. 4 ▸ f; 8.1% contribution to the Hirshfeld surface), they have the tips at d e + d i ≃ 2.60 Å. The remaining contributions from the other different interatomic contacts to the Hirshfeld surfaces are listed in Table 3 ▸. The dominance of H-atom contacts suggest that van der Waals interactions play the major role in establishing the crystal packing for the title compound (Hathwar et al., 2015 ▸).
Figure 4.
The full two-dimensional fingerprint plot (a) for the title compound and those delineated into (b) H⋯H (17.4%), (c) O⋯H/H⋯O (16.3%), (d) Br⋯H/H⋯Br (15.5%), (e) Br⋯C/C⋯Br (10.1%) and (f) F⋯H/H⋯F (8.1%) interactions.
Table 3. Percentage contributions of interatomic contacts to the Hirshfeld surface for the title salt.
| Contact | Percentage contribution |
|---|---|
| H⋯H | 17.4 |
| O⋯H/H⋯O | 16.3 |
| Br⋯H/H⋯Br | 15.5 |
| Br.·C/C⋯Br | 10.1 |
| F⋯H/H⋯F | 8.1 |
| C⋯H/H⋯C | 7.0 |
| N⋯H/H⋯N | 5.5 |
| C⋯C | 4.7 |
| Br.·O/O⋯Br | 4.2 |
| F⋯C/C⋯F | 3.5 |
| Br⋯Br | 3.1 |
| N⋯C/C⋯N | 1.4 |
| Br⋯F/F⋯Br | 1.1 |
| N⋯N | 0.9 |
| O⋯C/C⋯O | 0.1 |
| F⋯O/O⋯F | 0.6 |
| F⋯N/N⋯F | 0.5 |
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.42, update of September 2021; Groom et al., 2016 ▸) for the (E)-1-(2,2-dichloro-1-phenylethenyl)-2-phenyldiazene unit gave 26 hits. Seven compounds are closely related to the title compound, viz. CSD refcode GUPHIL (I) (Özkaraca et al., 2020 ▸), HONBUK (II) (Akkurt et al., 2019 ▸), HONBOE (III) (Akkurt et al., 2019 ▸), HODQAV (IV) (Shikhaliyev et al., 2019 ▸), XIZREG (V) (Atioğlu et al., 2019 ▸), LEQXOX (VI) (Shikhaliyev et al., 2018 ▸) and LEQXIR (VII) (Shikhaliyev et al., 2018 ▸).
In the crystal of (I), molecules are linked into inversion dimers via short halogen–halogen contacts [Cl1⋯Cl1 = 3.3763 (9) Å C16—Cl1⋯Cl1 = 141.47 (7)°] compared to the van der Waals radius sum of 3.50 Å for two chlorine atoms. No other directional contacts could be identified and the shortest aromatic-ring-centroid separation is greater than 5.25 Å. In the crystals of (II) and (III), the aromatic rings form dihedral angles of 64.1 (2) and 60.9 (2)°, respectively. Molecules are linked through weak X⋯Cl contacts [X = Cl for (II) and Br for (III)], C—H⋯Cl and C—Cl⋯π interactions into sheets lying parallel to the ab plane. In the crystal of (IV), the planes of the benzene rings make a dihedral angle of 56.13 (13)°. Molecules are stacked in columns along the a-axis direction via weak C—H⋯Cl hydrogen bonds and face-to-face π–π stacking interactions. The crystal packing is further consolidated by short Cl⋯Cl contacts. In (V), the benzene rings form a dihedral angle of 63.29 (8)°. Molecules are linked by C—H⋯O hydrogen bonds into zigzag chains running along the c-axis direction. The crystal packing also features C—Cl⋯π, C—F⋯π and N—O⋯π interactions. In the crystals of (VI) and (VII), the dihedral angles between the aromatic rings are 60.31 (14) and 56.18 (12) °, respectively. In (VI) C—H⋯N and short Cl⋯Cl contacts are observed and in (VII), C—H⋯N and C—H⋯O hydrogen bonds and short Cl⋯O contacts occur.
Synthesis and crystallization
A 20 ml screw-neck vial was charged with DMSO (10 ml), (E)-1-(4-fluorophenyl)-2-(2-nitrobenzylidene)hydrazine (1 mmol), tetramethylethylenediamine (TMEDA) (295 mg, 2.5 mmol), CuCl (2 mg, 0.02 mmol) and CBr4 (4.5 mmol). After 1–3h (until TLC analysis showed complete consumption of corresponding Schiff base) the reaction mixture was poured into a ∼0.01 M solution of HCl (100 ml, pH = 2–3), and extracted with dichloromethane (3 × 20 ml). The combined organic phase was washed with water (3 × 50 ml), brine (30 ml), dried over anhydrous Na2SO4 and concentrated in vacuo using a rotary evaporator. The residue was purified by column chromatography on silica gel using appropriate mixtures of hexane and dichloromethane (3/1–1/1). Crystals suitable for X-ray analysis were obtained by slow evaporation of an ethanol solution. Light-orange solid (52%); m.p. 377 K. Analysis calculated for C14H8Br2FN3O2 (M = 429.04): C 39.19, H 1.88, N 9.79; found: C 39.14, H 1.87, N 9.73%. 1H NMR (300MHz, CDCl3) δ 7.86–7.14 (8H, Ar–H). 13C NMR (75MHz, CDCl3) δ 165.02, 163.23, 163.01, 149.72, 133.01, 132.10, 129.70, 124.98, 124.87, 124.80, 124.29, 116.07, 115.91, 86.88. ESI–MS: m/z: 430.02 [M + H]+.
Refinement details
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. All H atoms were positioned geometrically [C—H = 0.95 Å] and refined using a riding model with U iso(H) = 1.2U eq(C).
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C14H8Br2FN3O2 |
| M r | 429.05 |
| Crystal system, space group | Orthorhombic, P b c n |
| Temperature (K) | 100 |
| a, b, c (Å) | 14.8700 (4), 15.2915 (4), 13.1030 (4) |
| V (Å3) | 2979.42 (14) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 5.46 |
| Crystal size (mm) | 0.59 × 0.26 × 0.20 |
| Data collection | |
| Diffractometer | Bruker AXS D8 QUEST Photon III detector |
| Absorption correction | Multi-scan (SADABS; Krause et al., 2015 ▸) |
| T min, T max | 0.047, 0.115 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 87568, 5429, 4773 |
| R int | 0.041 |
| (sin θ/λ)max (Å−1) | 0.758 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.023, 0.057, 1.06 |
| No. of reflections | 5429 |
| No. of parameters | 199 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.83, −0.46 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698902200278X/hb8012sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902200278X/hb8012Isup2.hkl
Supporting information file. DOI: 10.1107/S205698902200278X/hb8012Isup3.cml
CCDC reference: 2158375
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors’ contributions are as follows. Conceptualization, NQS, MA and AB; synthesis, NAM and GTS; X-ray analysis, STÇ, VNK and MA; writing (review and editing of the manuscript) STÇ, MA and AB; funding acquisition, NQS, NAM and GTS; supervision, NQS, MA and AB.
supplementary crystallographic information
Crystal data
| C14H8Br2FN3O2 | Dx = 1.913 Mg m−3 |
| Mr = 429.05 | Mo Kα radiation, λ = 0.71073 Å |
| Orthorhombic, Pbcn | Cell parameters from 9970 reflections |
| a = 14.8700 (4) Å | θ = 2.5–34.3° |
| b = 15.2915 (4) Å | µ = 5.46 mm−1 |
| c = 13.1030 (4) Å | T = 100 K |
| V = 2979.42 (14) Å3 | Block, light orange |
| Z = 8 | 0.59 × 0.26 × 0.20 mm |
| F(000) = 1664 |
Data collection
| Bruker AXS D8 QUEST Photon III detector diffractometer | 5429 independent reflections |
| Radiation source: fine-focus sealed X-Ray tube | 4773 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.041 |
| Detector resolution: 7.31 pixels mm-1 | θmax = 32.6°, θmin = 2.5° |
| φ and ω shutterless scans | h = −22→22 |
| Absorption correction: multi-scan (SADABS; Krause et al., 2015) | k = −23→23 |
| Tmin = 0.047, Tmax = 0.115 | l = −19→19 |
| 87568 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.023 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.057 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.0274P)2 + 1.6564P] where P = (Fo2 + 2Fc2)/3 |
| 5429 reflections | (Δ/σ)max = 0.005 |
| 199 parameters | Δρmax = 0.83 e Å−3 |
| 0 restraints | Δρmin = −0.46 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > 2sigma(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | 0.62265 (2) | 0.95963 (2) | 0.16792 (2) | 0.02193 (4) | |
| Br2 | 0.64744 (2) | 1.02154 (2) | 0.39424 (2) | 0.02154 (4) | |
| F1 | 0.61074 (8) | 0.42468 (7) | 0.03550 (8) | 0.0350 (2) | |
| O1 | 0.79994 (8) | 0.81157 (8) | 0.37024 (8) | 0.0279 (2) | |
| O2 | 0.82768 (8) | 0.70323 (8) | 0.47212 (9) | 0.0279 (2) | |
| N1 | 0.78022 (8) | 0.76388 (8) | 0.44249 (9) | 0.0204 (2) | |
| N2 | 0.61421 (8) | 0.77830 (8) | 0.26409 (9) | 0.0173 (2) | |
| N3 | 0.61871 (8) | 0.70235 (8) | 0.30197 (9) | 0.0180 (2) | |
| C1 | 0.62788 (9) | 0.92776 (9) | 0.30578 (10) | 0.0172 (2) | |
| C2 | 0.62325 (9) | 0.84437 (9) | 0.33887 (9) | 0.0163 (2) | |
| C3 | 0.62473 (9) | 0.82188 (9) | 0.44946 (10) | 0.0160 (2) | |
| C4 | 0.69718 (9) | 0.78201 (9) | 0.49868 (10) | 0.0167 (2) | |
| C5 | 0.69526 (10) | 0.75800 (9) | 0.60064 (10) | 0.0198 (2) | |
| H5 | 0.745761 | 0.730610 | 0.631466 | 0.024* | |
| C6 | 0.61793 (10) | 0.77479 (10) | 0.65687 (10) | 0.0216 (3) | |
| H6 | 0.615605 | 0.760257 | 0.727309 | 0.026* | |
| C7 | 0.54428 (10) | 0.81279 (10) | 0.60985 (10) | 0.0221 (3) | |
| H7 | 0.491119 | 0.823434 | 0.648092 | 0.026* | |
| C8 | 0.54741 (10) | 0.83559 (9) | 0.50689 (10) | 0.0195 (2) | |
| H8 | 0.495992 | 0.860845 | 0.475544 | 0.023* | |
| C9 | 0.61222 (9) | 0.63404 (9) | 0.22921 (10) | 0.0173 (2) | |
| C10 | 0.62860 (10) | 0.55039 (9) | 0.26748 (11) | 0.0214 (3) | |
| H10 | 0.640277 | 0.542361 | 0.338142 | 0.026* | |
| C11 | 0.62783 (11) | 0.47892 (10) | 0.20219 (13) | 0.0253 (3) | |
| H11 | 0.639299 | 0.421601 | 0.226931 | 0.030* | |
| C12 | 0.60996 (11) | 0.49353 (11) | 0.10055 (12) | 0.0245 (3) | |
| C13 | 0.59060 (10) | 0.57525 (10) | 0.06081 (11) | 0.0224 (3) | |
| H13 | 0.577105 | 0.582364 | −0.009565 | 0.027* | |
| C14 | 0.59139 (10) | 0.64622 (9) | 0.12606 (10) | 0.0194 (2) | |
| H14 | 0.577851 | 0.703011 | 0.101002 | 0.023* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.02940 (8) | 0.02164 (7) | 0.01475 (6) | −0.00253 (5) | −0.00260 (5) | 0.00271 (5) |
| Br2 | 0.02785 (8) | 0.01832 (7) | 0.01843 (6) | −0.00056 (5) | 0.00111 (5) | −0.00383 (5) |
| F1 | 0.0542 (7) | 0.0220 (5) | 0.0287 (5) | 0.0035 (5) | −0.0047 (4) | −0.0101 (4) |
| O1 | 0.0232 (5) | 0.0403 (6) | 0.0201 (5) | 0.0021 (5) | 0.0046 (4) | 0.0009 (4) |
| O2 | 0.0267 (5) | 0.0279 (6) | 0.0292 (6) | 0.0120 (5) | −0.0039 (4) | −0.0073 (4) |
| N1 | 0.0184 (5) | 0.0245 (6) | 0.0183 (5) | 0.0033 (5) | −0.0014 (4) | −0.0066 (4) |
| N2 | 0.0197 (5) | 0.0170 (5) | 0.0152 (5) | 0.0005 (4) | 0.0008 (4) | −0.0012 (4) |
| N3 | 0.0200 (5) | 0.0178 (5) | 0.0161 (5) | 0.0002 (4) | 0.0002 (4) | −0.0008 (4) |
| C1 | 0.0205 (6) | 0.0179 (6) | 0.0134 (5) | 0.0009 (5) | 0.0000 (4) | −0.0017 (4) |
| C2 | 0.0170 (5) | 0.0182 (6) | 0.0136 (5) | 0.0015 (5) | 0.0003 (4) | −0.0012 (4) |
| C3 | 0.0185 (6) | 0.0154 (5) | 0.0140 (5) | 0.0012 (5) | −0.0003 (4) | −0.0006 (4) |
| C4 | 0.0181 (6) | 0.0160 (5) | 0.0161 (5) | 0.0014 (5) | −0.0004 (4) | −0.0026 (4) |
| C5 | 0.0236 (6) | 0.0192 (6) | 0.0166 (5) | 0.0031 (5) | −0.0031 (5) | −0.0001 (4) |
| C6 | 0.0278 (7) | 0.0218 (6) | 0.0151 (5) | 0.0021 (5) | 0.0006 (5) | 0.0022 (5) |
| C7 | 0.0235 (7) | 0.0262 (7) | 0.0165 (6) | 0.0037 (5) | 0.0046 (5) | 0.0027 (5) |
| C8 | 0.0192 (6) | 0.0229 (6) | 0.0162 (5) | 0.0036 (5) | 0.0011 (4) | 0.0022 (5) |
| C9 | 0.0183 (6) | 0.0173 (6) | 0.0162 (5) | 0.0002 (5) | 0.0001 (4) | −0.0007 (4) |
| C10 | 0.0258 (7) | 0.0192 (6) | 0.0191 (6) | 0.0013 (5) | −0.0022 (5) | 0.0009 (5) |
| C11 | 0.0321 (8) | 0.0180 (6) | 0.0259 (7) | 0.0029 (6) | −0.0028 (6) | −0.0009 (5) |
| C12 | 0.0289 (7) | 0.0213 (6) | 0.0232 (7) | 0.0004 (6) | −0.0013 (5) | −0.0063 (5) |
| C13 | 0.0273 (7) | 0.0225 (7) | 0.0175 (6) | −0.0003 (6) | −0.0012 (5) | −0.0027 (5) |
| C14 | 0.0224 (6) | 0.0189 (6) | 0.0168 (5) | −0.0006 (5) | 0.0001 (5) | 0.0001 (4) |
Geometric parameters (Å, º)
| Br1—C1 | 1.8725 (13) | C6—C7 | 1.384 (2) |
| Br2—C1 | 1.8667 (13) | C6—H6 | 0.9500 |
| F1—C12 | 1.3546 (17) | C7—C8 | 1.3942 (18) |
| O1—N1 | 1.2304 (17) | C7—H7 | 0.9500 |
| O2—N1 | 1.2284 (16) | C8—H8 | 0.9500 |
| N1—C4 | 1.4641 (18) | C9—C10 | 1.3953 (19) |
| N2—N3 | 1.2647 (16) | C9—C14 | 1.3992 (19) |
| N2—C2 | 1.4138 (17) | C10—C11 | 1.388 (2) |
| N3—C9 | 1.4175 (17) | C10—H10 | 0.9500 |
| C1—C2 | 1.3486 (19) | C11—C12 | 1.376 (2) |
| C2—C3 | 1.4895 (18) | C11—H11 | 0.9500 |
| C3—C8 | 1.3900 (19) | C12—C13 | 1.384 (2) |
| C3—C4 | 1.3958 (18) | C13—C14 | 1.382 (2) |
| C4—C5 | 1.3859 (18) | C13—H13 | 0.9500 |
| C5—C6 | 1.390 (2) | C14—H14 | 0.9500 |
| C5—H5 | 0.9500 | ||
| O2—N1—O1 | 123.62 (13) | C6—C7—H7 | 119.7 |
| O2—N1—C4 | 117.94 (13) | C8—C7—H7 | 119.7 |
| O1—N1—C4 | 118.41 (12) | C3—C8—C7 | 120.92 (13) |
| N3—N2—C2 | 112.28 (11) | C3—C8—H8 | 119.5 |
| N2—N3—C9 | 114.15 (11) | C7—C8—H8 | 119.5 |
| C2—C1—Br2 | 122.32 (10) | C10—C9—C14 | 120.52 (13) |
| C2—C1—Br1 | 123.65 (10) | C10—C9—N3 | 114.96 (12) |
| Br2—C1—Br1 | 113.92 (7) | C14—C9—N3 | 124.52 (12) |
| C1—C2—N2 | 117.24 (12) | C11—C10—C9 | 119.93 (14) |
| C1—C2—C3 | 122.03 (12) | C11—C10—H10 | 120.0 |
| N2—C2—C3 | 120.71 (12) | C9—C10—H10 | 120.0 |
| C8—C3—C4 | 117.01 (12) | C12—C11—C10 | 118.05 (14) |
| C8—C3—C2 | 118.66 (12) | C12—C11—H11 | 121.0 |
| C4—C3—C2 | 124.19 (12) | C10—C11—H11 | 121.0 |
| C5—C4—C3 | 123.04 (13) | F1—C12—C11 | 118.75 (14) |
| C5—C4—N1 | 116.88 (12) | F1—C12—C13 | 117.83 (13) |
| C3—C4—N1 | 120.08 (12) | C11—C12—C13 | 123.42 (14) |
| C4—C5—C6 | 118.63 (13) | C14—C13—C12 | 118.34 (13) |
| C4—C5—H5 | 120.7 | C14—C13—H13 | 120.8 |
| C6—C5—H5 | 120.7 | C12—C13—H13 | 120.8 |
| C7—C6—C5 | 119.75 (12) | C13—C14—C9 | 119.68 (13) |
| C7—C6—H6 | 120.1 | C13—C14—H14 | 120.2 |
| C5—C6—H6 | 120.1 | C9—C14—H14 | 120.2 |
| C6—C7—C8 | 120.61 (13) | ||
| C2—N2—N3—C9 | −178.50 (11) | C3—C4—C5—C6 | −0.4 (2) |
| Br2—C1—C2—N2 | −175.87 (9) | N1—C4—C5—C6 | 179.45 (13) |
| Br1—C1—C2—N2 | 0.19 (18) | C4—C5—C6—C7 | 1.6 (2) |
| Br2—C1—C2—C3 | 5.92 (19) | C5—C6—C7—C8 | −1.0 (2) |
| Br1—C1—C2—C3 | −178.02 (10) | C4—C3—C8—C7 | 2.0 (2) |
| N3—N2—C2—C1 | 173.96 (13) | C2—C3—C8—C7 | 177.74 (13) |
| N3—N2—C2—C3 | −7.80 (18) | C6—C7—C8—C3 | −0.9 (2) |
| C1—C2—C3—C8 | 75.95 (18) | N2—N3—C9—C10 | 172.10 (13) |
| N2—C2—C3—C8 | −102.20 (16) | N2—N3—C9—C14 | −7.8 (2) |
| C1—C2—C3—C4 | −108.63 (17) | C14—C9—C10—C11 | 2.6 (2) |
| N2—C2—C3—C4 | 73.22 (18) | N3—C9—C10—C11 | −177.28 (14) |
| C8—C3—C4—C5 | −1.4 (2) | C9—C10—C11—C12 | −0.5 (2) |
| C2—C3—C4—C5 | −176.84 (13) | C10—C11—C12—F1 | 178.69 (15) |
| C8—C3—C4—N1 | 178.78 (12) | C10—C11—C12—C13 | −1.7 (3) |
| C2—C3—C4—N1 | 3.3 (2) | F1—C12—C13—C14 | −178.71 (14) |
| O2—N1—C4—C5 | 26.58 (18) | C11—C12—C13—C14 | 1.6 (2) |
| O1—N1—C4—C5 | −151.37 (13) | C12—C13—C14—C9 | 0.5 (2) |
| O2—N1—C4—C3 | −153.54 (13) | C10—C9—C14—C13 | −2.6 (2) |
| O1—N1—C4—C3 | 28.51 (19) | N3—C9—C14—C13 | 177.24 (13) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6···O1i | 0.95 | 2.51 | 3.3244 (18) | 144 |
Symmetry code: (i) −x+3/2, −y+3/2, z+1/2.
Funding Statement
This work was funded by Science Development Foundation of the President of Azerbaijan grant EIF-BGM-4-RFTF-1/2017-21/13/4.
References
- Akkurt, M., Shikhaliyev, N. Q., Suleymanova, G. T., Babayeva, G. V., Mammadova, G. Z., Niyazova, A. A., Shikhaliyeva, I. M. & Toze, F. A. A. (2019). Acta Cryst. E75, 1199–1204. [DOI] [PMC free article] [PubMed]
- Atioğlu, Z., Akkurt, M., Shikhaliyev, N. Q., Suleymanova, G. T., Bagirova, K. N. & Toze, F. A. A. (2019). Acta Cryst. E75, 237–241. [DOI] [PMC free article] [PubMed]
- Bruker (2018). APEX3 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gurbanov, A. V., Kuznetsov, M. L., Demukhamedova, S. D., Alieva, I. N., Godjaev, N. M., Zubkov, F. I., Mahmudov, K. T. & Pombeiro, A. J. L. (2020a). CrystEngComm, 22, 628–633.
- Gurbanov, A. V., Kuznetsov, M. L., Mahmudov, K. T., Pombeiro, A. J. L. & Resnati, G. (2020b). Chem. Eur. J. 26, 14833–14837. [DOI] [PubMed]
- Hathwar, V. R., Sist, M., Jørgensen, M. R. V., Mamakhel, A. H., Wang, X., Hoffmann, C. M., Sugimoto, K., Overgaard, J. & Iversen, B. B. (2015). IUCrJ, 2, 563–574. [DOI] [PMC free article] [PubMed]
- Kopylovich, M. N., Mac Leod, T. C. O., Haukka, M., Amanullayeva, G. I., Mahmudov, K. T. & Pombeiro, A. J. L. (2012). J. Inorg. Biochem. 115, 72–77. [DOI] [PubMed]
- Krause, L., Herbst-Irmer, R., Sheldrick, G. M. & Stalke, D. (2015). J. Appl. Cryst. 48, 3–10. [DOI] [PMC free article] [PubMed]
- MacLeod, T. C. O., Kopylovich, M. N., Guedes da Silva, M. F. C., Mahmudov, K. T. & Pombeiro, A. J. L. (2012). Appl. Catal. Gen. 439–440, 15–23.
- Mahmudov, K. T., Guedes da Silva, M. F. C., Glucini, M., Renzi, M., Gabriel, K. C. P., Kopylovich, M. N., Sutradhar, M., Marchetti, F., Pettinari, C., Zamponi, S. & Pombeiro, A. J. L. (2012). Inorg. Chem. Commun. 22, 187–189.
- Mahmudov, K. T., Gurbanov, A. V., Aliyeva, V. A., Resnati, G. & Pombeiro, A. J. L. (2020). Coord. Chem. Rev. 418, 213381.
- McKinnon, J. J., Jayatilaka, D. & Spackman, M. A. (2007). Chem. Commun. pp. 3814–3816. [DOI] [PubMed]
- Mizar, A., Guedes da Silva, M. F. C., Kopylovich, M. N., Mukherjee, S., Mahmudov, K. T. & Pombeiro, A. J. L. (2012). Eur. J. Inorg. Chem. pp. 2305–2313.
- Özkaraca, K., Akkurt, M., Shikhaliyev, N. Q., Askerova, U. F., Suleymanova, G. T., Mammadova, G. Z. & Shadrack, D. M. (2020). Acta Cryst. E76, 1251–1254. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shikhaliyev, N. Q., Ahmadova, N. E., Gurbanov, A. V., Maharramov, A. M., Mammadova, G. Z., Nenajdenko, V. G., Zubkov, F. I., Mahmudov, K. T. & Pombeiro, A. J. L. (2018). Dyes Pigments, 150, 377–381.
- Shikhaliyev, N. Q., Kuznetsov, M. L., Maharramov, A. M., Gurbanov, A. V., Ahmadova, N. E., Nenajdenko, V. G., Mahmudov, K. T. & Pombeiro, A. J. L. (2019). CrystEngComm, 21, 5032–5038.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. The University of Western Australia.
- Viswanathan, A., Kute, D., Musa, A., Mani, S. K., Sipilä, V., Emmert-Streib, F., Zubkov, F. I., Gurbanov, A. V., Yli-Harja, O. & Kandhavelu, M. (2019). Eur. J. Med. Chem. 166, 291–303. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698902200278X/hb8012sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902200278X/hb8012Isup2.hkl
Supporting information file. DOI: 10.1107/S205698902200278X/hb8012Isup3.cml
CCDC reference: 2158375
Additional supporting information: crystallographic information; 3D view; checkCIF report