The molecule of the title compound contains an essentially planar indole ring system and a phenyl ring. In the crystal, the molecules are linked by a weak intermolecular C—H⋯O hydrogen bond and C—H⋯π interactions, forming a two-dimensional network structure.
Keywords: crystal structure, indole, Schiff base, bidentate ligand, C—H⋯π interactions
Abstract
The molecule of the title compound, C16H14N2O, contains an essentially planar indole ring system and a phenyl ring. In the crystal, the molecules are linked by a weak intermolecular C—H⋯O hydrogen bond and C—H⋯π interactions, forming a one-dimensional column structure along the b-axis direction. These columns are linked by other C—H⋯π interactions, forming a two-dimensional network structure.
Chemical context
Indole and its derivatives are useful starting compounds to derive pharmaceutical (Nalli et al., 2020 ▸) and biological materials (Arumugam et al., 2021 ▸). Indole can function as a hydrogen-bond donor because of the high acidity of the hydrogen atom at position 1. The introduction of a hydrogen-bond acceptor to position 2 of the indole ring forms a five-to-seven-membered intramolecular hydrogen-bonded ring (Nosenko, et al., 2008 ▸). In this work, a Schiff base including an indole ring, N-(indol-2-ylmethylidene)-4-methoxyaniline, was newly synthesized. Similar Schiff bases such as salicylideneamines often function as bidentate ligands (Wang et al., 2018 ▸). Whereas salicylideneamines form intramolecular hydrogen bonds between coordination site atoms, such intramolecular interactions are absent from the crystal structure of the title compound. We report herein on its molecular and crystal structure.
Structural commentary
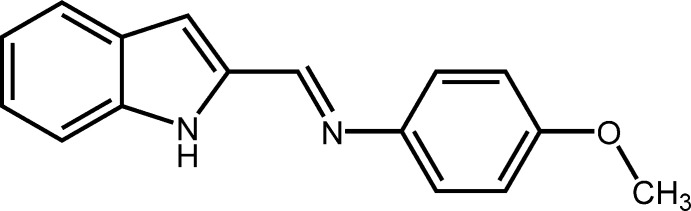
The molecular structure of the title compound is shown in Fig. 1 ▸. The C=N double bond adopts an E configuration. The indole moiety is almost planar with an r.m.s. deviation of 0.009 (1) Å. The bond lengths and angles in the title molecule are normal and agree with those in other indole imine compounds (IWIGUS; Suresh et al., 2016 ▸; KEVLON; Ho et al., 2006 ▸). The dihedral angle between the indole system and the benzene ring is 9.89 (5)°. In the related compound IWIGUS, the dihedral angles between the indole system and the benzene ring disordered over two sets of sites are widened to 81.8 (3) and 85.2 (3)° due to two isopropyl substituents in the benzene ring. There is no intramolecular hydrogen bond in the title compound, because the N2—H2⋯N3 angle is as small as 94.4 (10)°; however, the N2⋯N3 distance is 2.8633 (16) Å, and the N2—C4—C12—N3 torsion angle is 3.94 (19)°. Although no intramolecular hydrogen bond is observed, a broad peak assigned for the N—H proton is seen in the 1H NMR spectrum of the title compound in a CDCl3 medium and this suggests that the compound forms an intramolecular hydrogen bond in solution (see Synthesis and crystallization).
Figure 1.
The molecular structure of the title compound with atom labelling. Displacement ellipsoids are drawn at the 50% probability level. H atoms are represented by spheres of arbitrary radius.
Supramolecular features
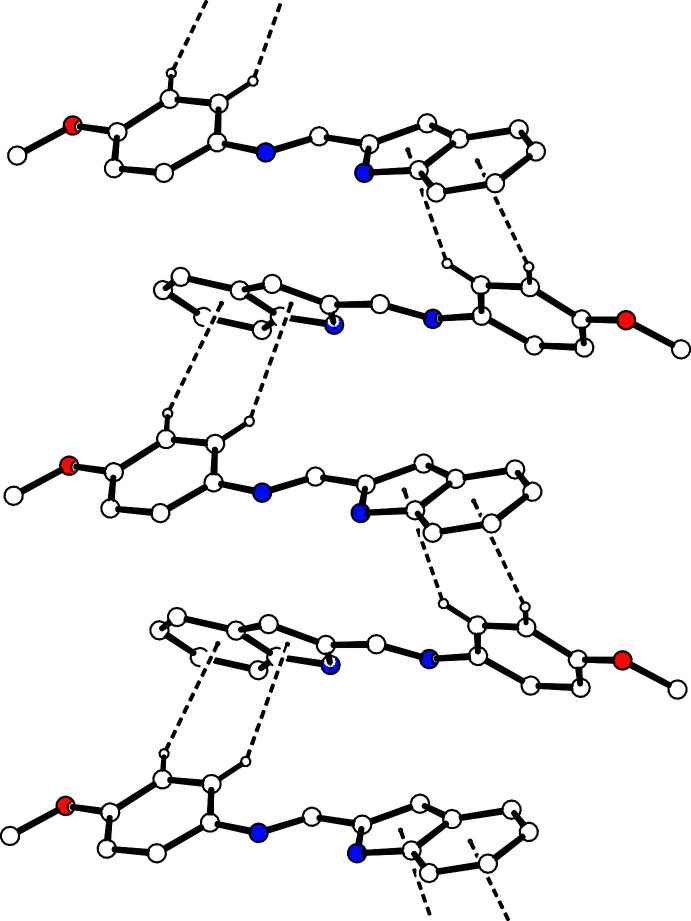
The title compound contains an N—H group, which is a hydrogen-bond donor, and an imino group, which is a hydrogen-bond acceptor, but neither of them forms an intermolecular hydrogen bond in the crystal. Compounds containing a similar indol-2-ylmethylidene-aniline fragment with a cis-conformation of the C—C single bond between the N atoms often form dimers by intermolecular N—H⋯N hydrogen bonds (see Database survey). However, in the crystal the molecules of the title compound are linked by a weak intermolecular C10—H10⋯O1i hydrogen bond and C—H⋯π interactions [C17—H17⋯Cg1i and C19—H19C⋯Cg2i; Cg1 is the centroid of the N2/C4–C6/C11 ring and Cg2 is the centroid of the C6–C11 ring; symmetry code: (i)
 − x,
− x,
 + y,
+ y,
 − z], forming columns along the b-axis direction (Fig. 2 ▸, Table 1 ▸). Besides this, the molecules belonging to different columns are joined by other C—H⋯π interactions [C14—H14⋯Cg1ii and C15—H15⋯Cg2ii; symmetry code: (ii)
− z], forming columns along the b-axis direction (Fig. 2 ▸, Table 1 ▸). Besides this, the molecules belonging to different columns are joined by other C—H⋯π interactions [C14—H14⋯Cg1ii and C15—H15⋯Cg2ii; symmetry code: (ii)
 − x, −
− x, −
 + y,
+ y,
 − z] (Fig. 3 ▸, Table 1 ▸). As a result, the intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions form a two-dimensional network structure (Fig. 4 ▸).
− z] (Fig. 3 ▸, Table 1 ▸). As a result, the intermolecular C—H⋯O hydrogen bonds and C—H⋯π interactions form a two-dimensional network structure (Fig. 4 ▸).
Figure 2.
One-dimensional column structure in the crystal of the title compound viewed along the a axis. The C—H⋯O hydrogen bonds and the C—H⋯π interactions are shown as dashed lines. H atoms not involved in these interactions are omitted for clarity.
Table 1. Hydrogen-bond geometry (Å, °).
Cg1 and Cg2 are the centroids of the N2/C4–C6/C11 and C6–C11 rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C10—H10⋯O1i | 0.93 | 2.58 | 3.2479 (16) | 129 |
| C14—H14⋯Cg1ii | 0.93 | 2.81 | 3.6006 (14) | 143 |
| C15—H15⋯Cg2ii | 0.93 | 2.79 | 3.5153 (14) | 136 |
| C17—H17⋯Cg1i | 0.93 | 2.89 | 3.5718 (14) | 131 |
| C19—H19C⋯Cg2i | 0.96 | 2.97 | 3.7716 (16) | 142 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 .
.
Figure 3.
Part of the crystal structure of the title compound showing the formation of ribbons along the b-axis direction. The C—H⋯π interactions are shown as dashed lines. H atoms not involved in these interactions are omitted for clarity.
Figure 4.
A packing diagram of the title compound viewed along the c axis, showing the two-dimensional network. The C—H⋯O hydrogen bonds and C—H⋯π interactions are shown as dashed lines. H atoms not involved in these interactions are omitted for clarity.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.42, update of May 2021; Groom et al., 2016 ▸) using ConQuest (Bruno et al., 2002 ▸) for indole derivatives gave 5272 hits, and for the (1H-indol-2-yl)methanimine skeleton gave 86 hits. Among these, the imino N atom bonded to an H atom gave one hit, to an N atom gave 24 hits, and to a C atom gave 61 hits. A search for the indol-2-ylmethylidene-aniline fragment gave 30 hits, and those containing a (1H-indol-2-yl)methylidene-aniline fragment with a cis-conformation of the C—C single bond gave seven hits. These seven compounds include five examples of dimers linked by complementary N—H⋯N hydrogen bonds (FORJAA; Li et al., 2019 ▸; IWIGUS; Suresh et al., 2016 ▸; KEZCUQ; Ariyasu et al., 2016 ▸; VACKES; Gadekar et al., 2016 ▸; WAGCEP; Tian et al., 2016 ▸), one example of a one-dimensional-chain structure (UWUSAI; Kalalbandi & Seetharamappa, 2016 ▸), and one example of a monomer protected from hydrogen bonding by steric hindrance (KEVLON; Ho et al., 2006 ▸). These structures contain intermolecular or intramolecular hydrogen bonds involving the N—H or the imino groups. Of these structures, the compounds most closely related to the title compound are N-(2,6-diisopropylphenyl)-1-(1H-indol-2-yl)methanimine (IWIGUS; Suresh et al., 2016 ▸), 4,6-dimethoxy-3-methyl-2,7-bis[(phenylimino)methyl]indole (KEVLON; Ho et al., 2006 ▸) and 2-(phenyl-N-oxidoiminomethyl)-3-phenylaminoindole (CIPWED; Greci & Sgarabotto, 1984 ▸). In the crystal of IWIGUS, which features a large dihedral angle between the indole and benzene rings, two neighbouring molecules are associated through pairs of N—H⋯N intermolecular hydrogen bonds, forming a centrosymmetric dimer. The crystal structure of an indol-2-ylmethylidene-aniline compound without a hydrogen bond between the N—H and imino groups has not yet been reported. In an almost planar molecule without a bulky substituent such as the tile compound, the formation of a dimer by intermolecular N—H⋯N hydrogen bonding is probably not appropriate for the crystal packing.
Synthesis and crystallization
Indole-2-carbaldehyde (145 mg, 1.00 mmol) and p-anisidine (148 mg, 1.20 mmol) were dissolved in toluene (20 mL), and the solution was refluxed under inert gas for 6 h, followed by evaporation. The residue was purified by recrystallization from a solvent mixture of acetone and n-hexane (1:1), and the title compound was then obtained (212 mg, 0.848 mmol, 84.8%) as a pale-red powder. The recrystallization of the title compound from a mixture of acetone and methanol afforded single crystals suitable for X-ray structure analysis. 1H NMR (CDCl3, 400 MHz) δ = 3.84 (s, 3H, OCH3), 6.93–6.97 (m, 3H, ArH), 7.13 (td, 1H, Jortho = 7.5 Hz, Jmeta = 1.0 Hz, ArH), 7.25–7.31 (m, 3H, ArH), 7.40 (dd, 1H, Jortho = 8.3 Hz, Jmeta = 0.9 Hz, ArH), 7.66 (d, 1H, Jortho = 8.0 Hz, ArH), 8.48 (s, 1H, N=CH), 9.25 (br, 1H, NH). HR–MS (m/z): calculated for [C16H15N2O]+, m/z = 251.1179; found, 251.1192.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The H atom attached to N2 was located in a difference-Fourier map and freely refined. The C-bound H atoms were positioned geometrically and refined using a riding model: C—H = 0.93–0.96 Å with U iso(H) = 1.2U eq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C16H14N2O |
| M r | 250.29 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 123 |
| a, b, c (Å) | 5.87685 (19), 7.5999 (3), 28.4578 (11) |
| β (°) | 90.604 (3) |
| V (Å3) | 1270.95 (8) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.08 |
| Crystal size (mm) | 0.30 × 0.20 × 0.10 |
| Data collection | |
| Diffractometer | Rigaku AFC10 Saturn70 area detector |
| Absorption correction | Multi-scan CrysAlis PRO; Rigaku OD, 2018 ▸) |
| T min, T max | 0.608, 0.992 |
| No. of measured, independent and observed [F 2 > 2.0σ(F 2)] reflections | 11128, 2907, 2525 |
| R int | 0.056 |
| (sin θ/λ)max (Å−1) | 0.649 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.049, 0.127, 1.05 |
| No. of reflections | 2907 |
| No. of parameters | 178 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.39, −0.29 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989022002973/yk2166sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022002973/yk2166Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989022002973/yk2166Isup3.cml
CCDC reference: 2159620
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C16H14N2O | F(000) = 528.00 |
| Mr = 250.29 | Dx = 1.308 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 5.87685 (19) Å | Cell parameters from 6329 reflections |
| b = 7.5999 (3) Å | θ = 2.8–30.8° |
| c = 28.4578 (11) Å | µ = 0.08 mm−1 |
| β = 90.604 (3)° | T = 123 K |
| V = 1270.95 (8) Å3 | Prism, colourless |
| Z = 4 | 0.30 × 0.20 × 0.10 mm |
Data collection
| Rigaku AFC10 Saturn70 area detector diffractometer | 2907 independent reflections |
| Radiation source: rotating anode X-ray generator, micromax007 | 2525 reflections with F2 > 2.0σ(F2) |
| Multi-layer mirror optics monochromator | Rint = 0.056 |
| Detector resolution: 28.5714 pixels mm-1 | θmax = 27.5°, θmin = 2.8° |
| ω scans | h = −7→7 |
| Absorption correction: multi-scan CrysAlisPro; Rigaku OD, 2018) | k = −8→9 |
| Tmin = 0.608, Tmax = 0.992 | l = −36→35 |
| 11128 measured reflections |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.049 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.127 | w = 1/[σ2(Fo2) + (0.0686P)2 + 0.3253P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 2907 reflections | Δρmax = 0.39 e Å−3 |
| 178 parameters | Δρmin = −0.29 e Å−3 |
| 0 restraints | Extinction correction: SHELXL |
| Primary atom site location: structure-invariant direct methods | Extinction coefficient: 0.259 (14) |
| Secondary atom site location: difference Fourier map |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.06455 (16) | 0.60985 (13) | 0.93970 (3) | 0.0292 (3) | |
| N2 | 0.57779 (18) | 0.68608 (14) | 0.66418 (4) | 0.0237 (3) | |
| N3 | 0.40410 (18) | 0.64748 (14) | 0.75724 (4) | 0.0248 (3) | |
| C4 | 0.7005 (2) | 0.61479 (16) | 0.70151 (4) | 0.0240 (3) | |
| C5 | 0.9058 (2) | 0.55340 (17) | 0.68542 (5) | 0.0249 (3) | |
| H5 | 1.0199 | 0.5007 | 0.7034 | 0.030* | |
| C6 | 0.9115 (2) | 0.58536 (16) | 0.63609 (4) | 0.0221 (3) | |
| C7 | 1.0717 (2) | 0.55168 (17) | 0.60070 (5) | 0.0253 (3) | |
| H7 | 1.2079 | 0.4950 | 0.6079 | 0.030* | |
| C8 | 1.0237 (2) | 0.60388 (17) | 0.55530 (5) | 0.0272 (3) | |
| H8 | 1.1287 | 0.5822 | 0.5318 | 0.033* | |
| C9 | 0.8174 (2) | 0.68975 (17) | 0.54398 (5) | 0.0266 (3) | |
| H9 | 0.7899 | 0.7250 | 0.5131 | 0.032* | |
| C10 | 0.6548 (2) | 0.72278 (17) | 0.57776 (4) | 0.0253 (3) | |
| H10 | 0.5182 | 0.7780 | 0.5701 | 0.030* | |
| C11 | 0.7037 (2) | 0.67011 (16) | 0.62387 (4) | 0.0219 (3) | |
| C12 | 0.6095 (2) | 0.60207 (17) | 0.74822 (4) | 0.0245 (3) | |
| H12 | 0.7019 | 0.5599 | 0.7724 | 0.029* | |
| C13 | 0.3193 (2) | 0.62794 (16) | 0.80344 (4) | 0.0231 (3) | |
| C14 | 0.4210 (2) | 0.52631 (17) | 0.83908 (5) | 0.0264 (3) | |
| H14 | 0.5511 | 0.4612 | 0.8327 | 0.032* | |
| C15 | 0.3296 (2) | 0.52215 (17) | 0.88355 (5) | 0.0266 (3) | |
| H15 | 0.3993 | 0.4548 | 0.9069 | 0.032* | |
| C16 | 0.1332 (2) | 0.61828 (16) | 0.89388 (4) | 0.0233 (3) | |
| C17 | 0.0233 (2) | 0.71292 (17) | 0.85845 (5) | 0.0253 (3) | |
| H17 | −0.1111 | 0.7729 | 0.8645 | 0.030* | |
| C18 | 0.1179 (2) | 0.71653 (17) | 0.81370 (5) | 0.0247 (3) | |
| H18 | 0.0446 | 0.7798 | 0.7900 | 0.030* | |
| C19 | −0.1427 (3) | 0.6969 (2) | 0.95149 (5) | 0.0367 (4) | |
| H19A | −0.1793 | 0.6722 | 0.9836 | 0.044* | |
| H19B | −0.2632 | 0.6554 | 0.9313 | 0.044* | |
| H19C | −0.1247 | 0.8215 | 0.9474 | 0.044* | |
| H2 | 0.438 (3) | 0.737 (2) | 0.6678 (5) | 0.032 (4)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0294 (5) | 0.0304 (5) | 0.0279 (5) | 0.0010 (4) | 0.0024 (4) | −0.0028 (4) |
| N2 | 0.0196 (5) | 0.0258 (6) | 0.0258 (5) | 0.0019 (4) | −0.0007 (4) | −0.0004 (4) |
| N3 | 0.0257 (5) | 0.0218 (6) | 0.0270 (6) | −0.0006 (4) | −0.0012 (4) | 0.0001 (4) |
| C4 | 0.0223 (6) | 0.0226 (6) | 0.0270 (7) | −0.0032 (5) | −0.0028 (5) | −0.0007 (5) |
| C5 | 0.0216 (6) | 0.0242 (6) | 0.0288 (7) | −0.0008 (5) | −0.0046 (5) | 0.0014 (5) |
| C6 | 0.0194 (6) | 0.0180 (6) | 0.0289 (7) | −0.0027 (4) | −0.0030 (5) | −0.0015 (5) |
| C7 | 0.0192 (6) | 0.0228 (6) | 0.0339 (7) | 0.0008 (5) | −0.0006 (5) | −0.0029 (5) |
| C8 | 0.0247 (6) | 0.0259 (7) | 0.0310 (7) | −0.0034 (5) | 0.0029 (5) | −0.0056 (5) |
| C9 | 0.0289 (6) | 0.0249 (7) | 0.0258 (6) | −0.0022 (5) | −0.0032 (5) | −0.0005 (5) |
| C10 | 0.0235 (6) | 0.0238 (6) | 0.0283 (7) | 0.0021 (5) | −0.0040 (5) | 0.0000 (5) |
| C11 | 0.0194 (6) | 0.0189 (6) | 0.0273 (6) | −0.0015 (5) | −0.0009 (4) | −0.0023 (5) |
| C12 | 0.0237 (6) | 0.0239 (7) | 0.0259 (6) | −0.0032 (5) | −0.0042 (5) | −0.0007 (5) |
| C13 | 0.0221 (6) | 0.0200 (6) | 0.0270 (6) | −0.0023 (5) | −0.0017 (5) | −0.0005 (5) |
| C14 | 0.0216 (6) | 0.0242 (7) | 0.0335 (7) | 0.0047 (5) | 0.0029 (5) | 0.0042 (5) |
| C15 | 0.0250 (6) | 0.0243 (7) | 0.0305 (7) | 0.0019 (5) | −0.0022 (5) | 0.0053 (5) |
| C16 | 0.0239 (6) | 0.0192 (6) | 0.0266 (6) | −0.0043 (5) | −0.0009 (5) | −0.0025 (5) |
| C17 | 0.0206 (6) | 0.0212 (6) | 0.0340 (7) | 0.0012 (5) | −0.0017 (5) | −0.0023 (5) |
| C18 | 0.0227 (6) | 0.0222 (6) | 0.0289 (7) | 0.0009 (5) | −0.0054 (5) | 0.0016 (5) |
| C19 | 0.0341 (8) | 0.0398 (9) | 0.0364 (8) | 0.0040 (6) | 0.0069 (6) | −0.0064 (7) |
Geometric parameters (Å, º)
| O1—C16 | 1.3705 (15) | C9—H9 | 0.9300 |
| O1—C19 | 1.4287 (17) | C10—C11 | 1.3989 (17) |
| N2—C11 | 1.3769 (16) | C10—H10 | 0.9300 |
| N2—C4 | 1.3876 (16) | C12—H12 | 0.9300 |
| N2—H2 | 0.912 (17) | C13—C18 | 1.3950 (17) |
| N3—C12 | 1.2840 (17) | C13—C14 | 1.4035 (18) |
| N3—C13 | 1.4189 (17) | C14—C15 | 1.3803 (18) |
| C4—C5 | 1.3768 (18) | C14—H14 | 0.9300 |
| C4—C12 | 1.4413 (18) | C15—C16 | 1.4000 (18) |
| C5—C6 | 1.4252 (18) | C15—H15 | 0.9300 |
| C5—H5 | 0.9300 | C16—C17 | 1.3918 (18) |
| C6—C7 | 1.4094 (18) | C17—C18 | 1.3950 (18) |
| C6—C11 | 1.4205 (17) | C17—H17 | 0.9300 |
| C7—C8 | 1.3781 (19) | C18—H18 | 0.9300 |
| C7—H7 | 0.9300 | C19—H19A | 0.9600 |
| C8—C9 | 1.4111 (18) | C19—H19B | 0.9600 |
| C8—H8 | 0.9300 | C19—H19C | 0.9600 |
| C9—C10 | 1.3852 (19) | ||
| C16—O1—C19 | 117.50 (11) | C10—C11—C6 | 121.79 (12) |
| C11—N2—C4 | 108.89 (10) | N3—C12—C4 | 121.67 (12) |
| C11—N2—H2 | 128.4 (10) | N3—C12—H12 | 119.2 |
| C4—N2—H2 | 122.7 (10) | C4—C12—H12 | 119.2 |
| C12—N3—C13 | 119.83 (11) | C18—C13—C14 | 118.05 (12) |
| C5—C4—N2 | 109.15 (11) | C18—C13—N3 | 116.78 (11) |
| C5—C4—C12 | 128.24 (12) | C14—C13—N3 | 125.17 (12) |
| N2—C4—C12 | 122.51 (11) | C15—C14—C13 | 120.54 (12) |
| C4—C5—C6 | 107.42 (11) | C15—C14—H14 | 119.7 |
| C4—C5—H5 | 126.3 | C13—C14—H14 | 119.7 |
| C6—C5—H5 | 126.3 | C14—C15—C16 | 120.68 (12) |
| C7—C6—C11 | 119.10 (12) | C14—C15—H15 | 119.7 |
| C7—C6—C5 | 134.07 (12) | C16—C15—H15 | 119.7 |
| C11—C6—C5 | 106.83 (11) | O1—C16—C17 | 125.06 (12) |
| C8—C7—C6 | 119.08 (12) | O1—C16—C15 | 115.28 (11) |
| C8—C7—H7 | 120.5 | C17—C16—C15 | 119.65 (12) |
| C6—C7—H7 | 120.5 | C16—C17—C18 | 119.03 (12) |
| C7—C8—C9 | 120.94 (12) | C16—C17—H17 | 120.5 |
| C7—C8—H8 | 119.5 | C18—C17—H17 | 120.5 |
| C9—C8—H8 | 119.5 | C13—C18—C17 | 121.91 (11) |
| C10—C9—C8 | 121.49 (12) | C13—C18—H18 | 119.0 |
| C10—C9—H9 | 119.3 | C17—C18—H18 | 119.0 |
| C8—C9—H9 | 119.3 | O1—C19—H19A | 109.5 |
| C9—C10—C11 | 117.58 (12) | O1—C19—H19B | 109.5 |
| C9—C10—H10 | 121.2 | H19A—C19—H19B | 109.5 |
| C11—C10—H10 | 121.2 | O1—C19—H19C | 109.5 |
| N2—C11—C10 | 130.50 (11) | H19A—C19—H19C | 109.5 |
| N2—C11—C6 | 107.71 (11) | H19B—C19—H19C | 109.5 |
| C11—N2—C4—C5 | 0.24 (14) | C5—C6—C11—C10 | 179.05 (11) |
| C11—N2—C4—C12 | −176.34 (11) | C13—N3—C12—C4 | 178.13 (11) |
| N2—C4—C5—C6 | −0.69 (14) | C5—C4—C12—N3 | −171.94 (13) |
| C12—C4—C5—C6 | 175.63 (12) | N2—C4—C12—N3 | 3.94 (19) |
| C4—C5—C6—C7 | −179.42 (14) | C12—N3—C13—C18 | 165.19 (12) |
| C4—C5—C6—C11 | 0.87 (14) | C12—N3—C13—C14 | −15.23 (19) |
| C11—C6—C7—C8 | 0.81 (18) | C18—C13—C14—C15 | −3.23 (19) |
| C5—C6—C7—C8 | −178.88 (13) | N3—C13—C14—C15 | 177.21 (12) |
| C6—C7—C8—C9 | −0.08 (19) | C13—C14—C15—C16 | 0.4 (2) |
| C7—C8—C9—C10 | −0.8 (2) | C19—O1—C16—C17 | 3.66 (18) |
| C8—C9—C10—C11 | 0.89 (19) | C19—O1—C16—C15 | −176.29 (12) |
| C4—N2—C11—C10 | −179.44 (13) | C14—C15—C16—O1 | −177.25 (11) |
| C4—N2—C11—C6 | 0.32 (14) | C14—C15—C16—C17 | 2.80 (19) |
| C9—C10—C11—N2 | 179.59 (12) | O1—C16—C17—C18 | 177.07 (11) |
| C9—C10—C11—C6 | −0.14 (19) | C15—C16—C17—C18 | −2.98 (18) |
| C7—C6—C11—N2 | 179.51 (11) | C14—C13—C18—C17 | 3.04 (19) |
| C5—C6—C11—N2 | −0.73 (13) | N3—C13—C18—C17 | −177.36 (11) |
| C7—C6—C11—C10 | −0.71 (19) | C16—C17—C18—C13 | 0.05 (19) |
Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of the N2/C4–C6/C11 and C6–C11 rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| C10—H10···O1i | 0.93 | 2.58 | 3.2479 (16) | 129 |
| C14—H14···Cg1ii | 0.93 | 2.81 | 3.6006 (14) | 143 |
| C15—H15···Cg2ii | 0.93 | 2.79 | 3.5153 (14) | 136 |
| C17—H17···Cg1i | 0.93 | 2.89 | 3.5718 (14) | 131 |
| C19—H19C···Cg2i | 0.96 | 2.97 | 3.7716 (16) | 142 |
Symmetry codes: (i) −x+1/2, y+1/2, −z+3/2; (ii) −x+3/2, y−1/2, −z+3/2.
Funding Statement
This work was funded by Cooperative Research Program of Network Joint Research Center for Materials and Devices grant 20212018; Japan Society for the Promotion of Science grant JP21K02520.
References
- Altomare, A., Cascarano, G., Giacovazzo, C. & Guagliardi, A. (1993). J. Appl. Cryst. 26, 343–350.
- Ariyasu, K., Miyatake, R., Kumai, Y., Ohta, A. & Oda, M. (2016). Am. J. Org. Chem. 6, 93–101.
- Arumugam, N., Almansour, A. I., Kumar, R. S., Yeswanthkumar, S., Padmanaban, R., Arun, Y., Kansız, S., Dege, N., Manohar, T. S. & Venketesh, S. (2021). J. Mol. Struct. 1225, 129165–129166.
- Bruno, I. J., Cole, J. C., Edgington, P. R., Kessler, M., Macrae, C. F., McCabe, P., Pearson, J. & Taylor, R. (2002). Acta Cryst. B58, 389–397. [DOI] [PubMed]
- Gadekar, S. C., Reddy, B. K., Panchal, S. P. & Anand, V. G. (2016). Chem. Commun. 52, 4565–4568. [DOI] [PubMed]
- Greci, L. & Sgarabotto, P. (1984). J. Chem. Soc. Perkin Trans. 2, pp. 1281–1284.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Ho, J. H. H., Black, D. St C., Messerle, B. A., Clegg, J. K. & Turner, P. T. (2006). Organometallics, 25, 5800–5810.
- Kalalbandi, V. K. A. & Seetharamappa, J. (2016). Synth. Commun. 46, 626–635.
- Li, J., Wang, Z.-B., Xu, Y., Lu, X.-C., Zhu, S.-R. & Liu, L. (2019). Chem. Commun. 55, 12072–12075. [DOI] [PubMed]
- Nalli, M., Armijos Rivera, J. I., Masci, D., Coluccia, A., Badia, R., Riveira-Muñoz, E., Brambilla, A., Cinquina, E., Turriziani, O., Falasca, F., Catalano, M., Limatola, C., Esté, J. A., Maga, G., Silvestri, R., Crespan, E. & La Regina, G. (2020). Eur. J. Med. Chem. 208, 112696–112718. [DOI] [PubMed]
- Nosenko, Y., Wiosna-Sałyga, G., Kunitski, M., Petkova, I., Singh, A., Buma, W. J., Thummel, R. P., Brutschy, B. & Waluk, J. (2008). Angew. Chem. Int. Ed. 47, 6037–6040. [DOI] [PubMed]
- Rigaku (2019). CrystalStructure. Rigaku Corporation, Tokyo, Japan.
- Rigaku OD (2018). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2020). Acta Cryst. E76, 1–11. [DOI] [PMC free article] [PubMed]
- Suresh, D., Ferreira, B., Lopes, P. S., Gomes, C. S. B., Krishnamoorthy, P., Charas, A., Vila-Viçosa, D., Morgado, J., Calhorda, M. J., Maçanita, A. L. & Gomes, P. T. (2016). Dalton Trans. 45, 15603–15620. [DOI] [PubMed]
- Tian, Y., Tian, L., Li, C., Jia, X. & Li, J. (2016). Org. Lett. 18, 840–843. [DOI] [PubMed]
- Wang, Z., Li, T., Xing, S. & Zhu, B. (2018). Org. Biomol. Chem. 16, 5021–5026. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989022002973/yk2166sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989022002973/yk2166Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989022002973/yk2166Isup3.cml
CCDC reference: 2159620
Additional supporting information: crystallographic information; 3D view; checkCIF report