In the title compound, the thiourea chromophore is planar to an r.m.s deviation of 0.032 Å with the thiolate sulfur atom being the most deviated. Bifurcated N—H⋯O intramolecular hydrogen bonds result in an S(6) supramolecular synthon. In the crystal, molecules are linked by N—H⋯O intermolecular hydrogen-bonding interactions and stabilized by C—H⋯π and π–π interactions.
Keywords: anthraquinone, thiourea, crystal structure, Hirshfeld surface, computational study
Abstract
The title compound, C18H14N2O3S, crystallizes in the orthorhombic crystal system and Pbca space group. The thiourea chromophore is planar to an r.m.s deviation of 0.032 Å with the thiolate sulfur atom being the most deviated. Bifurcated N—H⋯O intramolecular hydrogen bonds result in an S(6) supramolecular synthon. In the crystal, molecules are linked by N—H⋯O intermolecular hydrogen-bonding interactions and stabilized by C—H⋯π and π–π interactions. Hirshfeld surface analysis and fingerprint plot indicate the H⋯H intermolecular contacts as the highest contributor to the overall surface contacts (38%) and this is supported by the high dispersive and electrostatic interaction energies.
Chemical context
Anthraquinones, a group of tricyclic aromatic organic compounds, are the largest group of natural and synthetic quinones. A large number of them are well-known natural pigments found in plants, lichens, and fungi (Duval et al., 2016 ▸). These compounds exhibit important biological activities, including antitumor (Huang et al., 2007 ▸; Murdock et al., 1979 ▸, Shrestha et al., 2014 ▸, 2015 ▸; Chien et al., 2015 ▸), anti-inflammatory (Chien et al., 2015 ▸; Khan et al., 2011 ▸), diuretic (Chien et al., 2015 ▸), antiarthritic (Davis et al., 1986 ▸), antifungal (Wuthi-udomlert et al., 2010 ▸), antibacterial (Fosso et al., 2012 ▸), antimalarial (Winter et al., 1996 ▸), antioxidant (Dave & Ledwani, 2012 ▸), antileukemic (Chang & Lee, 1984 ▸; Ismail et al., 1997 ▸), antiviral and anti-HIV properties (Alves et al., 2004 ▸; Barnard et al., 1992 ▸; Schinazi et al., 1990 ▸; Schrader et al., 2000 ▸). Some aminoanthraquinone derivatives have also been reported to be good DNA intercalators (Hande, 2008 ▸; Schrader et al., 2000 ▸). The versatility of acyl thioureas stems from their ease of preparation and ability to introduce different functionalities, resulting in compounds with very interesting biological properties including antifungal (del Campo et al., 2002 ▸, 2004 ▸), antitumor (Sacht & Datt, 2000 ▸; Sacht et al., 2000 ▸; Hernández et al., 2005 ▸), antiviral, antibacterial, herbicidal, insecticidal and pharmacological activities (Binzet et al., 2006 ▸; Saeed et al., 2010 ▸). Recently, our research group reported the synthesis and crystal structures of a number of thiourea derivatives (Asegbeloyin et al., 2018 ▸, 2019 ▸; Okpareke et al., 2020 ▸; 2022 ▸; Oyeka et al., 2021 ▸). In a continuation of our series on thiourea derivatives, we present herein the crystal structure, Hirshfeld surface and computational study of a new potential biologically active thiourea derivative with an aminoanthraquinone moiety.

Structural commentary
The title compound crystallizes in the orthorhombic crystal system and Pbca space group. The molecular structure (Fig. 1 ▸) shows a central thiourea chromophore flanked on either side by methylene and anthraquinone units. The central thiourea moiety is essentially planar with an r.m.s deviation of 0.032 Å with the thiolate S atom being the most deviated out of the plane with a deviation of 0.044 (3) Å. The torsion angles between the thiourea and the adjourning methylene and anthraquinone moieties are −177.5 (2) and −140.8 (2)°, respectively, indicating that the anthraquinone moiety is slightly deviated from the thiourea plane, compared to the methylene moiety. The C1—N1—C5 bond angle of 126.09 (19)° subtended at the N1 atom is smaller than the less encumbered C2—N2—C1 angle [129.79 (19)°] subtended at N2 and larger than the central N1—C1—N2 [114.5 (2)°] bond angle subtended at the thiolate C1 carbon atom. The C1—N2 bond [1.395 (3) Å] is slightly longer than C1—N1 [1.364 (3) Å]. The thiourea carbonyl oxygen and imine groups are involved in a strong intramolecular N1—H1⋯O1 hydrogen bond (Table 1 ▸). The second amine nitrogen N2 is also involved in a hydrogen-bonding S(6) graph-set (Kansiz et al., 2022 ▸) interaction.
Figure 1.
View of the molecular structure of the title compound, with the atom labeling. Displacement ellipsoids are drawn at the 30% probability level. Intramolecular hydrogen bonds are shown as dashed lines.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯O1 | 0.86 | 1.98 | 2.685 (2) | 138 |
| N1—H1⋯O2 | 0.86 | 2.14 | 2.652 (2) | 117 |
| N2—H2⋯O3i | 0.86 | 2.19 | 3.038 (2) | 167 |
| C3—H3B⋯O2ii | 0.97 | 2.52 | 3.414 (3) | 153 |
| C15—H15⋯S1iii | 0.93 | 2.87 | 3.553 (2) | 131 |
| C17—H17⋯O2iv | 0.93 | 2.47 | 3.280 (3) | 145 |
Symmetry codes: (i)
 ; (ii)
; (ii)
 ; (iii)
; (iii)
 ; (iv)
; (iv)
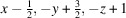 .
.
Supramolecular features
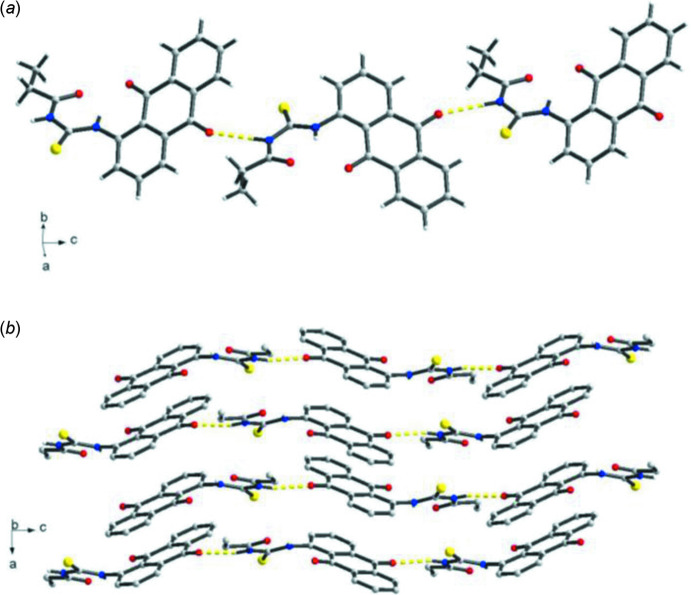
In the crystal, the molecules are linked by imine N—H⋯O (anthraquinone) hydrogen-bonding interactions, leading to supramolecular chains running along the c-axis direction (Fig. 2 ▸ a). Supramolecular layers are obtained from self-assembly of these chains via anthraquinone π–π stacking interactions along the ab plane with centroid–centroid distances of 3.916(3), 3.531(5), 3.701(2) and 3.705(2) Å (Fig. 2 ▸ b). These intermolecular interactions are balanced and stabilized by the phenyl C—H⋯O(carbonyl) and imine N—H⋯O(carbonyl) intramolecular S(6) synthon.
Figure 2.
(a) Supramolecular 1-D hydrogen-bonding interactions along c-axis direction of the title compound and (b) molecular aggregation structure of the crystal along the ab plane, showing repeating units of pairwise π–π stacking interactions.
Hirshfeld surface analysis and fingerprint plots
Hirshfeld surfaces (HS) and corresponding two-dimensional fingerprint plots (FPs) were calculated using the Crystal Explorer 17.5 software (Turner et al., 2017 ▸). The Hirshfeld surfaces mapped over d norm and shape-index were generated according to a procedure described by Tan et al. (2019 ▸) and used for further analysis of the intermolecular interactions. The HS mapped over d norm shows the most intense red regions around the thiourea N—H groups resulting from the amine-N—H⋯O (anthraquinone) hydrogen-bonding interactions (Fig. 3 ▸ a). Other intense red spots can be identified around the thiourea carbonyl oxygen and resulting from carbonyl C17—H17⋯O12 intermolecular interaction. Apart from the intense red spots, there are a number of other less intense red spots found around the alkyl C3 atom resulting from C3—H3B⋯O2 intermolecular interaction. Other intermolecular interactions in the Hirshfeld surface are the anthraquinone C—H⋯S(thiourea) and anthraquinone-C—H⋯H(alkyl) interactions shown respectively as pink and green dotted lines in Fig. 3 ▸ b. The anthraquinone π–π interactions can be seen in Fig. 3 ▸ c. The C⋯H/H⋯C contacts in the molecule are responsible for the molecular packing in the supramolecular structure and are the result of the C—H⋯π and π–π interactions (Tan & Tiekink, 2020 ▸) and are depicted by mapping the structure over the shape-index isosurface as shown in Fig. 3 ▸ d. The C—H⋯π interactions appear as hollow orange areas (π⋯H—C) and bulging blue areas (C—H⋯π) in the compound. The small blue regions surrounding a bright orange spot within the anthroquinone rings of the molecule indicate π–π stacking interactions.
Figure 3.
Hirshfeld surfaces mapped over (a), (b) and (c) d norm and (d) shape-index showing intermolecular atom-to-atom and π–π interactions in the crystal structure.
The overall two-dimensional fingerprint plot (Spackman & McKinnon, 2002 ▸; Tan & Tiekink, 2020 ▸) and those delineated into H⋯H, H⋯O/O⋯H, H⋯C/C⋯H, C⋯C, S⋯H/H⋯S and C⋯O/O⋯C interactions are illustrated in Fig. 4 ▸, and their percentage contributions are presented in Table 2 ▸. The overall fingerprint plot comprises all intermolecular contacts in the molecule and exhibits a shield-like profile with two symmetric spikes on each side of a triangular protrusion. These spikes are also observed in the fingerprint plots for the O⋯H/H⋯O contacts, which make a 19.5% contribution to the overall surface contact, but not in the other surface contacts. These spikes are due to the C—H⋯O and N2—H2⋯O3 hydrogen-bonding interactions in the crystal structure of the title compound. H⋯H contacts are the single highest contributor to the overall surface with a 38.0% contribution and and result from C—H⋯H and H⋯H dispersion interactions. The other major surface contacts are C⋯H/H⋯C (13.0%) S⋯H/H⋯S (10.8%), and C⋯C (11.2%), showing that C⋯H and π intermolecular contacts contribute significantly to the overall stability of the supramolecular architecture in the crystal structure (Ekowo et al., 2020 ▸; Izuogu et al., 2020 ▸).
Figure 4.
The overall and individual two-dimensional fingerprint plots for intermolecular contacts in the crystal structure.
Table 2. Percentage contributions of intermolecular contacts to the Hirshfeld surface.
| Contact | Percentage contribution |
|---|---|
| H⋯H | 38.0 |
| H⋯O/O⋯H | 19.5 |
| C⋯H/H⋯C | 13.0 |
| C⋯C | 26.3 |
| H⋯H | 11.2 |
| S⋯H/H⋯S | 10.8 |
| C⋯O/O⋯C | 2.7 |
| N⋯H/H⋯N | 1.4 |
| C⋯O/O⋯C | 1.3 |
Interaction energy calculations
The interaction energies between pairs of molecules within the crystal of the title compound were calculated by adding up the four energy components, viz. electrostatic (E ele), polarization (E pol), dispersion (E dis), and exchange repulsion (E rep) (Tan et al., 2019 ▸; Ayiya & Okpareke, 2021 ▸). The energies were obtained by calculating the wave function of each pair of molecules or atoms at the B3LYP/6-31G(d,p) level of theory (Ayiya & Okpareke, 2021 ▸; Izuogu et al., 2020 ▸). Quantitative estimations of the strength and nature of the intermolecular interactions in title compound crystal with individual energy components (E ele, E pol, E dis, and E rep) as well as the sum of the energy components E tot are presented in Table 3 ▸. This shows that the dispersive component of the energy makes the most significant contribution to the total interaction energy profile in the crystal structure, probably due to the intermolecular dispersive π interactions resulting from the π–π stacking of adjacent anthraquinone ring systems in the crystal. The electrostatic component is the second highest contributor to the total interaction energy and probably results from the C⋯H, H⋯H and van der Waals interactions. A graphical representation of the magnitude of the interaction energies is presented in Fig, 5a–d in the form of energy frameworks to show the supramolecular architecture using cylindrical poles joining the centroids of molecular pairs. The red, green, and blue color-coded frameworks in Fig. 5 ▸ a, 5b, and 5c, respectively, represent the E ele, E dis, and E tot, energy components for intermolecular interactions in crystal of the title compound, while Fig. 5 ▸ d shows the annotated E tot energy. The magnitude of the cylindrical pipes indicates the significance of the E ele energy component to the total interaction energy and the molecular packing in the crystal.
Table 3. A summary of the calculated interaction energies for the title compound (kJ mol−1).
Please define N and R
| N | Symop | R | E_ele | E_pol | E_dis | E_rep | E_tot |
|---|---|---|---|---|---|---|---|
| 1 |
x, −y +
 , z +
, z +

|
14.92 | 0.6 | −0.2 | −2.7 | 0.4 | −1.6 |
| 0 | -x, −y, −z | 6.11 | −24.1 | −4.8 | −85.9 | 77.8 | −55.8 |
| 0 | -x +
 , −y, z +
, −y, z +

|
11.23 | −33.2 | −7.5 | −17.8 | 38.4 | −32.3 |
| 1 | -x +
 , −y, −z +
, −y, −z +

|
7.82 | −17.7 | −6.2 | −44.9 | 42.1 | −36.4 |
| 0 | -x +
 , y +
, y +
 , z
, z
|
9.48 | −0.7 | −1.1 | −13.3 | 8.2 | −8.0 |
| 0 |
x +
 , −y +
, −y +
 , −z
, −z
|
8.88 | −10.8 | −3.0 | −17.6 | 14.2 | −20.1 |
| 0 |
x, −y +
 , z +
, z +

|
13.01 | −0.0 | −0.5 | −9.9 | 3.6 | −6.8 |
| 1 | -x, y +
 , −z +
, −z +

|
12.22 | −0.1 | −0.7 | −10.2 | 8.5 | −4.2 |
| 0 | -x, −y, −z | 5.85 | −11.3 | −1.1 | −69.5 | 42.1 | −47.3 |
Figure 5.
Perspective views of the energy frameworks of the title compound showing (a) electrostatic, (b) dispersion, (c) total energy and (d) annotated total energy. The cylindrical radius is proportional to the relative strength of the corresponding energies and they were adjusted to the same scale factor of 100 with a cut-off value of 5 kJmol−1 within 2 x 2 x 2 unit cells.
Database survey
Anthraquinones derivatives with thiourea unit are scarce and our search for the basic architecture of the compound in the Cambridge Structural Database (CSD, version 5.42, update of May 2021; Groom et al., 2016 ▸) did not reveal any structure similar to the title compound.
Synthesis and crystallization
A solution of propionyl chloride (1.85 g, 0.02 mol) dissolved in 40 mL acetone was mixed with 30 mL of an acetone solution of potassium thiocyanate (1.94 g, 0.02 mol). The reaction mixture was refluxed for 30 min to give a suspension of propionylisothiocyanate, which was left to cool to room temperature. 1-Aminoanthraquinone (4.47 g, 0.02 mol) was dissolved in 40 mL of acetone and the resulting solution was mixed with the suspension of propionylisothiocyanate, and the mixture was stirred for 2 h. The resultant reddish suspension was filtered, and left at room temperature for 96 h to obtain a reddish crystalline solid of the title compound.
Refinement
Crystal data, collection and structure refinement details are summarized in Table 4 ▸. The carbon-bound H atoms were placed in calculated positions and were included in the refinement using the riding-model approximation with U iso(H) set to 1.2U eq(C). The nitrogen-bound H atoms were located in the difference-Fourier maps and refined freely with appropriate RIGU restraints placed on the bonds.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C18H14N2O3S |
| M r | 338.37 |
| Crystal system, space group | Orthorhombic, P b c a |
| Temperature (K) | 100 |
| a, b, c (Å) | 7.3003 (1), 18.9557 (3), 21.9045 (3) |
| V (Å3) | 3031.19 (8) |
| Z | 8 |
| Radiation type | Cu Kα |
| μ (mm−1) | 2.07 |
| Crystal size (mm) | 0.18 × 0.12 × 0.08 |
| Data collection | |
| Diffractometer | XtaLAB Synergy, Dualflex, Pilatus 200K |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2018 ▸) |
| T min, T max | 0.869, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18022, 3013, 2816 |
| R int | 0.034 |
| (sin θ/λ)max (Å−1) | 0.624 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.048, 0.144, 1.13 |
| No. of reflections | 3013 |
| No. of parameters | 218 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.67, −0.64 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022003127/zn2016sup1.cif
Supporting information file. DOI: 10.1107/S2056989022003127/zn2016Isup2.cml
CCDC reference: 2161135
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are thankful to the University of Nigeria Nsukka for research funding and the School of Chemical Sciences of the University of Auckland for the use of their X-ray diffractometer.
supplementary crystallographic information
Crystal data
| C18H14N2O3S | Dx = 1.483 Mg m−3 |
| Mr = 338.37 | Cu Kα radiation, λ = 1.54184 Å |
| Orthorhombic, Pbca | Cell parameters from 10712 reflections |
| a = 7.3003 (1) Å | θ = 4.0–74.2° |
| b = 18.9557 (3) Å | µ = 2.07 mm−1 |
| c = 21.9045 (3) Å | T = 100 K |
| V = 3031.19 (8) Å3 | Block, clear colourless |
| Z = 8 | 0.18 × 0.12 × 0.08 mm |
| F(000) = 1408 |
Data collection
| XtaLAB Synergy, Dualflex, Pilatus 200K diffractometer | 3013 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, PhotonJet (Cu) X-ray Source | 2816 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.034 |
| ω scans | θmax = 74.3°, θmin = 4.0° |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2018) | h = −8→8 |
| Tmin = 0.869, Tmax = 1.000 | k = −23→22 |
| 18022 measured reflections | l = −26→26 |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.048 | H-atom parameters constrained |
| wR(F2) = 0.144 | w = 1/[σ2(Fo2) + (0.0724P)2 + 3.6939P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.13 | (Δ/σ)max < 0.001 |
| 3013 reflections | Δρmax = 0.67 e Å−3 |
| 218 parameters | Δρmin = −0.64 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.72292 (8) | 0.44246 (3) | 0.25812 (3) | 0.02439 (19) | |
| O2 | 0.7597 (2) | 0.64440 (8) | 0.41206 (7) | 0.0228 (4) | |
| O3 | 0.6789 (2) | 0.46534 (8) | 0.60029 (7) | 0.0244 (4) | |
| O1 | 0.9337 (3) | 0.66694 (9) | 0.29076 (7) | 0.0287 (4) | |
| N1 | 0.8521 (3) | 0.53993 (9) | 0.33766 (8) | 0.0195 (4) | |
| H1 | 0.889018 | 0.582673 | 0.342345 | 0.023* | |
| N2 | 0.8371 (3) | 0.57086 (10) | 0.23530 (8) | 0.0223 (4) | |
| H2 | 0.815839 | 0.557602 | 0.198428 | 0.027* | |
| C12 | 0.6591 (3) | 0.62664 (11) | 0.51256 (9) | 0.0171 (4) | |
| C5 | 0.8416 (3) | 0.49780 (11) | 0.39044 (9) | 0.0172 (4) | |
| C13 | 0.6390 (3) | 0.58121 (11) | 0.56248 (9) | 0.0169 (4) | |
| C10 | 0.7788 (3) | 0.52594 (11) | 0.44627 (9) | 0.0154 (4) | |
| C11 | 0.7332 (3) | 0.60171 (11) | 0.45330 (9) | 0.0163 (4) | |
| C9 | 0.7647 (3) | 0.48093 (11) | 0.49745 (9) | 0.0159 (4) | |
| C6 | 0.9019 (3) | 0.42781 (11) | 0.38807 (10) | 0.0200 (5) | |
| H6 | 0.951456 | 0.410016 | 0.352119 | 0.024* | |
| C7 | 0.8881 (3) | 0.38493 (11) | 0.43898 (10) | 0.0208 (5) | |
| H7 | 0.927247 | 0.338318 | 0.436683 | 0.025* | |
| C15 | 0.5707 (3) | 0.60672 (12) | 0.61753 (10) | 0.0220 (5) | |
| H15 | 0.556732 | 0.576471 | 0.650632 | 0.026* | |
| C8 | 0.8170 (3) | 0.41032 (11) | 0.49330 (10) | 0.0179 (4) | |
| H8 | 0.804179 | 0.380524 | 0.526757 | 0.022* | |
| C14 | 0.6936 (3) | 0.50620 (11) | 0.55722 (9) | 0.0172 (4) | |
| C18 | 0.6119 (3) | 0.69770 (11) | 0.51866 (10) | 0.0223 (5) | |
| H18 | 0.625588 | 0.728232 | 0.485743 | 0.027* | |
| C2 | 0.8937 (3) | 0.64023 (12) | 0.24157 (10) | 0.0230 (5) | |
| C17 | 0.5446 (3) | 0.72278 (13) | 0.57380 (11) | 0.0262 (5) | |
| H17 | 0.513700 | 0.770149 | 0.577811 | 0.031* | |
| C1 | 0.8093 (3) | 0.51926 (12) | 0.27974 (10) | 0.0203 (5) | |
| C16 | 0.5234 (3) | 0.67731 (13) | 0.62298 (11) | 0.0254 (5) | |
| H16 | 0.477295 | 0.694251 | 0.659739 | 0.031* | |
| C3 | 0.8997 (4) | 0.68035 (13) | 0.18245 (11) | 0.0298 (6) | |
| H3A | 0.777627 | 0.681545 | 0.164961 | 0.036* | |
| H3B | 0.978880 | 0.655647 | 0.154039 | 0.036* | |
| C4 | 0.9688 (5) | 0.75570 (14) | 0.19012 (13) | 0.0380 (6) | |
| H4A | 0.965684 | 0.779397 | 0.151405 | 0.057* | |
| H4B | 1.092260 | 0.754860 | 0.205179 | 0.057* | |
| H4C | 0.891940 | 0.780289 | 0.218617 | 0.057* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0304 (3) | 0.0229 (3) | 0.0198 (3) | −0.0057 (2) | 0.0008 (2) | −0.0018 (2) |
| O2 | 0.0358 (9) | 0.0158 (7) | 0.0170 (7) | −0.0024 (6) | 0.0019 (7) | 0.0034 (6) |
| O3 | 0.0347 (10) | 0.0221 (8) | 0.0163 (8) | 0.0002 (7) | 0.0017 (6) | 0.0062 (6) |
| O1 | 0.0417 (11) | 0.0264 (9) | 0.0180 (8) | −0.0020 (7) | −0.0021 (7) | −0.0015 (6) |
| N1 | 0.0294 (10) | 0.0152 (8) | 0.0140 (9) | −0.0018 (7) | 0.0012 (7) | 0.0009 (7) |
| N2 | 0.0289 (10) | 0.0232 (10) | 0.0148 (9) | −0.0010 (8) | −0.0003 (8) | −0.0001 (7) |
| C12 | 0.0153 (10) | 0.0190 (10) | 0.0168 (10) | −0.0003 (8) | −0.0012 (8) | 0.0007 (8) |
| C5 | 0.0192 (11) | 0.0175 (10) | 0.0149 (10) | −0.0025 (8) | −0.0018 (8) | 0.0025 (8) |
| C13 | 0.0142 (10) | 0.0199 (10) | 0.0165 (10) | −0.0004 (8) | −0.0019 (8) | 0.0005 (8) |
| C10 | 0.0151 (10) | 0.0146 (10) | 0.0165 (10) | −0.0016 (7) | −0.0031 (8) | 0.0013 (8) |
| C11 | 0.0168 (10) | 0.0166 (10) | 0.0156 (10) | −0.0022 (8) | −0.0032 (8) | 0.0005 (8) |
| C9 | 0.0138 (10) | 0.0178 (10) | 0.0162 (10) | −0.0028 (8) | −0.0021 (8) | 0.0014 (8) |
| C6 | 0.0224 (11) | 0.0189 (10) | 0.0187 (10) | 0.0014 (8) | 0.0004 (8) | −0.0010 (8) |
| C7 | 0.0240 (11) | 0.0146 (10) | 0.0239 (11) | 0.0015 (9) | −0.0020 (9) | 0.0013 (8) |
| C15 | 0.0208 (11) | 0.0289 (12) | 0.0164 (10) | 0.0010 (9) | 0.0004 (8) | 0.0016 (9) |
| C8 | 0.0183 (10) | 0.0164 (10) | 0.0190 (10) | −0.0011 (8) | −0.0024 (8) | 0.0042 (8) |
| C14 | 0.0158 (10) | 0.0202 (10) | 0.0156 (10) | −0.0026 (8) | −0.0033 (8) | 0.0022 (8) |
| C18 | 0.0267 (11) | 0.0174 (10) | 0.0228 (11) | 0.0025 (9) | 0.0006 (9) | 0.0015 (8) |
| C2 | 0.0235 (11) | 0.0230 (11) | 0.0226 (11) | 0.0006 (9) | 0.0019 (9) | 0.0008 (9) |
| C17 | 0.0277 (12) | 0.0211 (11) | 0.0298 (12) | 0.0043 (9) | 0.0021 (10) | −0.0033 (9) |
| C1 | 0.0205 (11) | 0.0218 (11) | 0.0185 (10) | 0.0013 (8) | 0.0006 (8) | 0.0017 (8) |
| C16 | 0.0258 (12) | 0.0293 (12) | 0.0212 (11) | 0.0037 (9) | 0.0028 (9) | −0.0048 (9) |
| C3 | 0.0398 (14) | 0.0298 (13) | 0.0199 (11) | −0.0015 (11) | 0.0037 (10) | 0.0016 (9) |
| C4 | 0.0559 (18) | 0.0280 (13) | 0.0302 (13) | −0.0085 (12) | 0.0012 (12) | 0.0085 (10) |
Geometric parameters (Å, º)
| S1—C1 | 1.656 (2) | C9—C14 | 1.487 (3) |
| O2—C11 | 1.228 (3) | C6—H6 | 0.9300 |
| O3—C14 | 1.226 (3) | C6—C7 | 1.384 (3) |
| O1—C2 | 1.226 (3) | C7—H7 | 0.9300 |
| N1—H1 | 0.8600 | C7—C8 | 1.384 (3) |
| N1—C5 | 1.407 (3) | C15—H15 | 0.9300 |
| N1—C1 | 1.364 (3) | C15—C16 | 1.387 (3) |
| N2—H2 | 0.8600 | C8—H8 | 0.9300 |
| N2—C2 | 1.385 (3) | C18—H18 | 0.9300 |
| N2—C1 | 1.395 (3) | C18—C17 | 1.388 (3) |
| C12—C13 | 1.400 (3) | C2—C3 | 1.503 (3) |
| C12—C11 | 1.484 (3) | C17—H17 | 0.9300 |
| C12—C18 | 1.397 (3) | C17—C16 | 1.388 (3) |
| C5—C10 | 1.411 (3) | C16—H16 | 0.9300 |
| C5—C6 | 1.399 (3) | C3—H3A | 0.9700 |
| C13—C15 | 1.392 (3) | C3—H3B | 0.9700 |
| C13—C14 | 1.481 (3) | C3—C4 | 1.524 (4) |
| C10—C11 | 1.482 (3) | C4—H4A | 0.9600 |
| C10—C9 | 1.412 (3) | C4—H4B | 0.9600 |
| C9—C8 | 1.395 (3) | C4—H4C | 0.9600 |
| C5—N1—H1 | 117.0 | C9—C8—H8 | 120.3 |
| C1—N1—H1 | 117.0 | C7—C8—C9 | 119.49 (19) |
| C1—N1—C5 | 126.06 (19) | C7—C8—H8 | 120.3 |
| C2—N2—H2 | 115.1 | O3—C14—C13 | 121.5 (2) |
| C2—N2—C1 | 129.79 (19) | O3—C14—C9 | 120.32 (19) |
| C1—N2—H2 | 115.1 | C13—C14—C9 | 118.13 (18) |
| C13—C12—C11 | 121.73 (19) | C12—C18—H18 | 120.0 |
| C18—C12—C13 | 119.5 (2) | C17—C18—C12 | 120.1 (2) |
| C18—C12—C11 | 118.74 (19) | C17—C18—H18 | 120.0 |
| N1—C5—C10 | 121.02 (18) | O1—C2—N2 | 123.4 (2) |
| C6—C5—N1 | 119.38 (19) | O1—C2—C3 | 122.8 (2) |
| C6—C5—C10 | 119.54 (19) | N2—C2—C3 | 113.80 (19) |
| C12—C13—C14 | 120.11 (19) | C18—C17—H17 | 119.9 |
| C15—C13—C12 | 120.1 (2) | C18—C17—C16 | 120.1 (2) |
| C15—C13—C14 | 119.83 (19) | C16—C17—H17 | 119.9 |
| C5—C10—C11 | 121.97 (18) | N1—C1—S1 | 127.30 (17) |
| C5—C10—C9 | 118.90 (19) | N1—C1—N2 | 114.5 (2) |
| C9—C10—C11 | 119.10 (19) | N2—C1—S1 | 118.19 (16) |
| O2—C11—C12 | 119.42 (19) | C15—C16—C17 | 120.3 (2) |
| O2—C11—C10 | 121.78 (19) | C15—C16—H16 | 119.9 |
| C10—C11—C12 | 118.78 (18) | C17—C16—H16 | 119.9 |
| C10—C9—C14 | 121.97 (18) | C2—C3—H3A | 109.0 |
| C8—C9—C10 | 120.52 (19) | C2—C3—H3B | 109.0 |
| C8—C9—C14 | 117.50 (18) | C2—C3—C4 | 112.9 (2) |
| C5—C6—H6 | 119.9 | H3A—C3—H3B | 107.8 |
| C7—C6—C5 | 120.3 (2) | C4—C3—H3A | 109.0 |
| C7—C6—H6 | 119.9 | C4—C3—H3B | 109.0 |
| C6—C7—H7 | 119.5 | C3—C4—H4A | 109.5 |
| C6—C7—C8 | 121.0 (2) | C3—C4—H4B | 109.5 |
| C8—C7—H7 | 119.5 | C3—C4—H4C | 109.5 |
| C13—C15—H15 | 120.0 | H4A—C4—H4B | 109.5 |
| C16—C15—C13 | 119.9 (2) | H4A—C4—H4C | 109.5 |
| C16—C15—H15 | 120.0 | H4B—C4—H4C | 109.5 |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···O1 | 0.86 | 1.98 | 2.685 (2) | 138 |
| N1—H1···O2 | 0.86 | 2.14 | 2.652 (2) | 117 |
| N2—H2···O3i | 0.86 | 2.19 | 3.038 (2) | 167 |
| C3—H3B···O2ii | 0.97 | 2.52 | 3.414 (3) | 153 |
| C15—H15···S1iii | 0.93 | 2.87 | 3.553 (2) | 131 |
| C17—H17···O2iv | 0.93 | 2.47 | 3.280 (3) | 145 |
Symmetry codes: (i) −x+3/2, −y+1, z−1/2; (ii) x+1/2, y, −z+1/2; (iii) −x+3/2, −y+1, z+1/2; (iv) x−1/2, −y+3/2, −z+1.
References
- Alves, D. S., Pérez-Fons, L., Estepa, A. & Micol, V. (2004). Biochem. Pharmacol. 68, 549–561. [DOI] [PubMed]
- Asegbeloyin, J. N., Ifeanyieze, K. J., Okpareke, O. C., Oyeka, E. E. & Groutso, T. V. (2019). Acta Cryst. E75, 1297–1300. [DOI] [PMC free article] [PubMed]
- Asegbeloyin, J. N., Oyeka, E. E., Okpareke, O. & Ibezim, A. (2018). J. Mol. Struct. 1153, 69–77.
- Ayiya, B. B. & Okpareke, O. C. (2021). J. Chem. Crystallogr., https://doi.org/10.1007/s10870-021-00902-4.
- Barnard, D. L., Huffman, J. H., Morris, J. L., Wood, S. G., Hughes, B. G. & Sidwell, R. W. (1992). Antiviral Res. 17, 63–77. [DOI] [PubMed]
- Binzet, G., Arslan, H., Flörke, U., Külcü, N. & Duran, N. (2006). J. Coord. Chem. 59, 1395–1406.
- Campo, R. del, Criado, J. J., García, E., Hermosa, M. R., Jiménez-Sánchez, A., Manzano, J. L., Monte, E., Rodríguez-Fernández, E. & Sanz, F. (2002). J. Inorg. Biochem. 89, 74–82. [DOI] [PubMed]
- Campo, R. del, Criado, J. J., Gheorghe, R., González, F. J., Hermosa, M., Sanz, F., Manzano, J. L., Monte, E. & Rodríguez-Fernández, E. (2004). J. Inorg. Biochem. 98, 1307–1314. [DOI] [PubMed]
- Chang, P. & Lee, K. H. (1984). Phytochemistry, 23, 1733–1736.
- Chien, S. C., Wu, Y.-C., Chen, Z.-W. & Yang, W. C. (2015). Evid. Based Complementary Altern. Med. pp. 1–14.
- Dave, H. & Ledwani, L. (2012). Indian J. Nat. Prod. Resour. 3, 291–319.
- Davis, R. H., Agnew, P. S. & Shapiro, E. (1986). J. Am. Podiatric Med. Assoc. 76, 1–8. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Duval, J., Pecher, V., Poujol, M. & Lesellier, E. (2016). Ind. Crops Prod. 94, 812–833.
- Ekowo, L. C., Eze, S. I., Ezeorah, J. C., Groutso, T., Atiga, S., Lane, J. R., Okafor, S., Akpomie, K. G. & Okparaeke, O. C. (2020). J. Mol. Struct. 1210, 127994.
- Fosso, M. Y., Chan, K. Y., Gregory, R. & Chang, C. T. (2012). ACS Comb. Sci. 14, 231–235. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hande, K. R. (2008). Update on Cancer Therapeutics, 3, 13–26.
- Hernández, W., Spodine, E., Beyer, L., Schröder, U., Richter, R., Ferreira, J. & Pavani, M. (2005). Bioinorg. Chem. Appl. 3, 299–316. [DOI] [PMC free article] [PubMed]
- Huang, Q., Lu, G., Shen, H. M., Chung, M. C. & Ong, C. N. (2007). Med. Res. Rev. 27, 609–630. [DOI] [PubMed]
- Ismail, N. H., Ali, A. M., Aimi, N., Kitajima, M., Takayama, H. & Lajis, N. H. (1997). Phytochemistry, 45, 1723–1725.
- Izuogu, D. C., Asegbeloyin, J. N., Jotani, M. M. & Tiekink, E. R. T. (2020). Acta Cryst. E76, 697–702. [DOI] [PMC free article] [PubMed]
- Kansiz, S., Yesilbag, S., Dege, N., Saif, E. & Agar, E. (2022). Acta Cryst. E78, 84–87. [DOI] [PMC free article] [PubMed]
- Khan, K., Karodi, R., Siddiqui, A., Thube, S. & Rub, R. (2011). Int. J. Appl. Res. Nat. Prod. 4, 28–36.
- Murdock, K., Child, R., Fabio, P., Angier, R. D., Wallace, R. E., Durr, F. E. & Citarella, R. (1979). J. Med. Chem. 22, 1024–1030. [DOI] [PubMed]
- Okpareke, O. C., Henderson, W., Akkoç, S. & Coban, B. (2022). Inorg. Chim. Acta, 531, 120707.
- Okpareke, O. C., Henderson, W., Lane, J. R. & Okafor, S. N. (2020). J. Mol. Struct. 1203, 127360.
- Oyeka, E. E., Babahan, I., Eboma, B., Ifeanyieze, K. J., Okpareke, O. C., Coban, E. P., Özmen, A., Coban, B., Aksel, M., Özdemir, N., Groutso, T. V., Ayogu, J. I., Yildiz, U., Bilgin, M. D., Biyik, H. H., Schrage, B. R., Ziegler, C. J. & Asegbeloyin, J. N. (2021). Inorg. Chim. Acta, 528, 120590.
- Rigaku OD (2018). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Sacht, C. & Datt, M. (2000). Polyhedron, 19, 1347–1354.
- Sacht, C., Datt, M. S., Otto, S. & Roodt, A. (2000). J. Chem. Soc. Dalton Trans. pp. 727–733.
- Saeed, S., Rashid, N., Ali, M., Hussain, R. & Jones, P. G. (2010). Eur. J. Chem. 1, 221–227. [DOI] [PubMed]
- Schinazi, R. F., Chu, C. K., Babu, J. R., Oswald, B. J., Saalmann, V., Cannon, D. L., Eriksson, B. F. & Nasr, M. (1990). Antiviral Res. 13, 265–272. [DOI] [PubMed]
- Schrader, K. K., Dayan, F. E., Allen, S. N., de Regt, M. Q., Tucker, C. S. & Paul, R. N. Jr (2000). Int. J. Plant Sci. 161, 265–270. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shrestha, J. P., Fosso, M. Y., Bearss, J. & Chang, C. T. (2014). Eur. J. Med. Chem. 77, 96–102. [DOI] [PubMed]
- Shrestha, J. P., Subedi, Y. P., Chen, L. & Chang, C. T. (2015). Med. Chem. Commun. 6, 2012–2022.
- Spackman, M. A. & McKinnon, J. J. (2002). CrystEngComm, 4, 378–392.
- Tan, S. L., Jotani, M. M. & Tiekink, E. R. T. (2019). Acta Cryst. E75, 308–318. [DOI] [PMC free article] [PubMed]
- Tan, S. L. & Tiekink, E. R. T. (2020). Acta Cryst. E76, 102–110. [DOI] [PMC free article] [PubMed]
- Turner, M., McKinnon, J., Wolff, S., Grimwood, D., Spackman, P., Jayatilaka, D. & Spackman, M. (2017). Crystal Explorer 17.5. University of Western Australia.
- Winter, R., Cornell, K. A., Johnson, L. L., Ignatushchenko, M., Hinrichs, D. J. & Riscoe, M. K. (1996). Antimicrob. Agents Chemother. 40, 1408–1411. [DOI] [PMC free article] [PubMed]
- Wuthi-udomlert, M., Kupittayanant, P. & Gritsanapan, W. (2010). J. Health Res. 24, 117–122.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989022003127/zn2016sup1.cif
Supporting information file. DOI: 10.1107/S2056989022003127/zn2016Isup2.cml
CCDC reference: 2161135
Additional supporting information: crystallographic information; 3D view; checkCIF report