Abstract
Syringomycin E is an antifungal cyclic lipodepsinonapeptide that inhibits the growth of Saccharomyces cerevisiae by interaction with the plasma membrane. A screen conducted to find the yeast genes necessary for its fungicidal action identified two novel syringomycin E response genes, SYR3 and SYR4. A syr3 mutant allele was complemented by ELO2 and ELO3. These genes encode enzymes that catalyze the elongation of sphingolipid very long chain fatty acids. Tetrad analysis showed that SYR3 was ELO2. Strains with deletions of SYR3/ELO2 and ELO3 were resistant to syringomycin E, and lipid analyses of both mutants revealed shortened fatty acid chains and lower levels of sphingolipids. SYR4 was identified by Tn5 inactivation of genomic library plasmids that complemented a syr4 mutant allele. SYR4 was found to be identical to IPT1, which encodes the terminal sphingolipid biosynthetic enzyme, mannosyl-diinositolphosphoryl-ceramide synthase. Deletion Δsyr4/ipt1 strains were viable, were resistant to syringomycin E, did not produce mannosyl-diinositolphosphoryl-ceramide, and accumulated mannosyl-inositolphosphoryl-ceramide. Accumulation of mannosyl-inositolphosphoryl-ceramide was not responsible for resistance since a temperature-sensitive secretory pathway mutant (sec14-3ts) accumulated this sphingolipid and was sensitive to syringomycin E. Finally, Δcsg1/sur1 and Δcsg2 strains defective in the transfer of mannose to inositolphosphoryl-ceramide were resistant to syringomycin E. These findings show that syringomycin E growth inhibition of yeast is promoted by the production of sphingolipids with fully elongated fatty acid chains and the mannosyl and terminal phosphorylinositol moieties of the polar head group.
Syringomycin E is a member of a family of small cyclic lipodepsinonapeptides (ca. 1,200 Da) produced by the plant bacterium Pseudomonas syringae pv. syringae (38). Other members include syringomycin A1 and G, the syringostatins, the syringotoxins, and the pseudomycins (2, 38). All possess a characteristic tetrapeptidyl sequence (dehydroaminobutanoic acid-hydroxyaspartic acid-chlorothreonine-serine) and a β-hydroxy fatty acid attached to the N-terminal serine. These metabolites are fungicidal to a broad range of fungi, including yeast and human pathogens (33), and they show relatively low levels of toxicity to plants (21) and cutaneous animal tissues (33). Syringomycin E was recently shown to be partly responsible for the biological control of fungal pathogens on postharvest citrus fruits by certain P. syringae pv. syringae strains (5). Syringomycin E interacts with the fungal plasma membrane, where it causes K+ efflux, Ca2+ influx, and changes in membrane potential by processes that are likely related to channel formation (14, 38).
Molecular genetic studies with yeast were initiated to more precisely define the antifungal mechanism of action of syringomycin E. Syringomycin E-resistant mutants of Saccharomyces cerevisiae were generated to permit identification of the mutated genes by complementation (39). Two genes, representing two of eight syringomycin E-resistant complementation groups, have been characterized. SYR1/ERG3 encodes the sterol C-5,6 desaturase for the biosynthesis of ergosterol, the primary sterol in the yeast plasma membrane (39). SYR2 is required for sphingoid base C-4 hydroxylation, a nonessential step in sphingolipid biosynthesis, revealing that C-4 OH-phytoceramide-based sphingolipids are required for syringomycin E action (7, 17). Thus, sterols and sphingolipids appear to be important factors for the susceptibility of yeast to syringomycin E.
Sphingolipids are involved in numerous cellular processes, such as protein anchoring, stress responses, and apoptosis (19, 20). In S. cerevisiae, the sphingolipids are predominantly located in the plasma membrane, and they constitute about 40% of the inositol-containing lipids in this membrane (10, 12). The three major species of S. cerevisiae sphingolipids differ by polar head group composition, and they are inositolphosphoryl-ceramide (IPC), mannosyl-inositolphosphoryl-ceramide (MIPC), and mannosyl-diinositolphosphoryl-ceramide [M(IP)2C] (20). Of the three, only IPC is essential for growth in standard laboratory growth media, and the specific functions of MIPC and M(IP)2C are not yet understood (10).
In addition to SYR2, several other yeast sphingolipid biosynthetic genes that are nonessential for growth have been identified (10). ELO2 and ELO3 are responsible for the conversion of C16 and C18 fatty acids to the very long chain (C20 to C26) fatty acids that are N-acylated to the ceramide moieties of sphingolipid molecules (27). Both genes provide the ability to make C20 and C24 acyl chains, but only ELO3 gives the ability to convert fatty acids from C24 to C26. ELO2 is identical to GNS1 and FEN1, and mutations of these genes confer resistance to the echinocandins, the sterol isomerase inhibitor SR31747, and fenpropimorph (13, 31). ELO3 is identical to SUR4, SRE1, and APA1; when ELO3 is mutated it confers resistance to SR31747 and causes decreased activity of the plasma membrane ATPase (9, 16, 31). Mutants with mutations in ELO2 and ELO3 have reduced levels of sphingolipids (27). IPT1 encodes the enzyme that catalyzes the terminal yeast sphingolipid biosynthetic step, which involves the transfer of phosphorylinositol from phosphatidylinositol to MIPC to form M(IP)2C and diacylglycerol (11, 23). Two genes, CSG1/SUR1 and CSG2, are necessary for mannosylation of IPC to MIPC (4). Although nonessential for growth, both are needed for growth in the presence of 50 mM Ca2+.
Here we describe findings that reveal the contributions of the biosynthetic genes described above to the susceptibility of yeast to syringomycin E. Two syringomycin E action genes (SYR3 and SYR4) are shown to be identical to ELO2 and IPT1, respectively, and mutants defective in these genes are shown to display resistance to syringomycin E. In addition, ELO3, CSG1/SUR1, and CSG2 are shown to promote susceptibility to syringomycin E. The findings reveal that production of sphingolipids with fully elongated very-long-chain fatty acids and with polar head groups that possess mannose and the terminal phosphorylinositol moieties promote the antifungal action of syringomycin E.
MATERIALS AND METHODS
Strains and growth conditions.
The S. cervisiae strains used in the study are listed in Table 1. The strains were grown at 28 to 30°C with shaking in YPD (1% yeast extract, 2% peptone, 2% dextrose) medium or in a minimal medium, SC-leu, SC-his, or SC-ura, prepared as described by Kaiser et al. (22). Sporulation agar plates were prepared as described by Kaiser et al. (22). Escherichia coli MC1061 (ATCC 37535) carrying the Tn5 delivery vector pCHR81 was grown in Luria-Bertani (LB) medium that contained 10 μg of kanamycin per ml at 30°C to maintain the temperature-sensitive pCHR81 unless otherwise noted. E. coli DH5α was grown on LB medium at 37°C.
TABLE 1.
S. cerevisiae strains used in this study
| Strains | Genotype | Reference |
|---|---|---|
| W303-1A | MATa/MATα ade2/ade2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 | 7 |
| W303-2a | MATa/MATα ade2/ade2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 ELO2/elo2::HIS3 | This work |
| W303-3a | MATa/MATα ade2/ade2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 ELO3/elo3::HIS3 | This work |
| W303-4a | MATa/MATα ade2/ade2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 SYR4/syr4::URA3 | This work |
| SS33 | MATa his3 leu2 trp1 ura3 syr3 | 38 |
| SS56 | MATa his3 leu2 trp1 ura3 syr4(ipt1) | 38 |
| CTY1-1A | MATa ura3-52 Δhis3-200 lys2-801 sec14-3ts | 3 |
| CTY182 | MATa ura3-52 Δhis3-200 lys2-801 | 3 |
| W303H | ade2 his3 leu2 trp1 ura3 syr4 (ipt1)::URA3 (derived from W303-4a) | This work |
| W303I | ade2 his3 leu2 trp1 ura3 SYR4 (derived from W303-4a) | This work |
| W303C | MATα ade2 his3 leu2 trp1 ura3 (derived from W303-1A) | 7 |
| W303-Δelo2 | MATα ade2 his3 leu2 trp1 ura3 elo2::HIS3 (derived from W303-2a) | This work |
| W303-Δelo3 | MATα ade2 his3 leu2 trp1 ura3 elo3::HIS3 (derived from W303-3a) | This work |
| TDY2037 | MATα lys2 ura3-52 trp1Δleu2Δ) | 18 |
| TDY2038 | TDY2037 csg2::LEU2 | 18 |
| 39alt22A | MATα ura3 his4 leu2 trp1 | T. Dunn |
| 39alt22A Δcsg1 | 39alt22A csg1::LEU2 | T. Dunn |
Cloning of SYR3 and SYR4/IPT1.
S. cerevisiae SS33 (syr3) and SS56 (syr4) were transformed by electroporation with a centromeric plasmid pSB32-based yeast genomic library (gift from Philip Hieter, Johns Hopkins University School of Medicine). Transformants were replica plated as suspensions onto SC-ura agar medium with and without 0.5 μg of syringomycin E per ml for strain SS33 and 1 μg of syringomycin E per ml for strain SS56. The plates were incubated for 48 h, and colonies sensitive to syringomycin E were selected. Complementation and plasmid dependency were verified by isolating complementing plasmids and rescreening for the syringomycin E sensitivity phenotype. SYR3 was identified as ELO2 by sequencing multiple complementing plasmids with a primer directed toward pSB32, comparing overlapping sequences, and BLAST analysis of yeast genes catalogued in the Saccharomyces Genome Database (Stanford University).
Tn5 transposon disruption was used to localize and identify SYR4 within the complementing plasmid as described by Ohya (28), with slight modifications. A plasmid, p56-1A, that complemented the syr4 mutation in strain SS56 was used to transform E. coli MC1061 by electroporation. Fifty transformants, selected on 100 μg of ampicillin per ml and 10 μg of kanamycin per ml, were picked and used to inoculate 1-ml cultures in LB medium containing 200 μg of kanamycin per ml. This medium allowed selection for insertion of Tn5 into p56-1A since only strains carrying the transposon at high copy numbers grow in the presence of this level of kanamycin. The cultures were incubated at 37°C to prevent replication of the Tn5 source, pCHR81. Plasmids were isolated from 30 cultures that showed growth and were used to transform strain SS56. Transformants were screened on SC-ura with and without 1 μg of syringomycin E per ml. Colonies that carried p56-1A with a Tn5 disruption in SYR4 were identified by their resistance to syringomycin E.
SYR4 was identified by sequencing a Tn5-disrupted p56-1A that had lost its ability to complement the syringomycin E resistance phenotype. Sequencing was from two directions with a primer (GGTTCCGTTCAGGACGCTACTTGTGTATAAGAGTC) for the Tn5 transposon and a primer (ATCGACTACGCGATCATGGCCACCACACCCGTCCT) directed against pSB32. The sequence was then used for BLAST analyses of the Saccharomyces Genome Database (Stanford University) to identify the disrupted open reading frame.
Construction of ELO2 and ELO3 deletion strains.
ELO2 and ELO3 deletion strains W303-2a and W303-3a were constructed in the diploid strain W303-1A by using HIS3-disrupted plasmid constructs pCRelo2HIS and YEpelo3HIS provided by Charles Martin (Rutgers University), as described by Oh et al. (27). Transformants were selected on SC-his medium, sporulated, and dissected for tetrad analysis. Dissected tetrads were replica plated onto YPD agar medium with and without 1 μg of syringomycin E per ml and SC-his medium.
Construction of a strain with an SYR4 deletion.
A 3.1-kb NsiI fragment was taken from p56-A1 and ligated into pRS415 (6) that had been digested with PstI. The resulting plasmid was digested with BamHI and XhoI, and the SYR4 fragment was purified and ligated into the psp72 vector (Promega, Madison, Wis.), which was also digested with BamHI and XhoI. A 1.1-kb deletion which resulted from PvuII digestion of the psp72::SYR4 construct was replaced with a 1.1-kb URA3 fragment obtained by digesting YCp50 with SmaI and PfmI and filling in the ends with the Klenow fragment. This final construct was linearized by digestion with ClaI and BstXI. All DNA working enzymes were from New England Biolabs (Beverly, Mass.). The linear fragment was used to transform diploid strain W303-1A by electroporation. Diploid transformants were selected on SC-ura medium, sporulated, and dissected for tetrad analysis (29). Dissected tetrads were replica plated onto YPD agar medium with and without 1 μg of syringomycin E per ml and SC-ura agar medium. SYR4 deletion was confirmed by Southern blot analysis (22).
Preparation of radiolabeled sphingolipids.
Yeast sphingolipids were labeled and extracted by the methods described by Smith and Lester (32), but with modifications. Twenty-milliliter cultures were grown at 30°C for 18 to 24 h in YPD medium containing 0.5 mCi of H332PO4. When appropriate, 0.2 mCi of [2-3H]myo-inositol was also added. Radiochemicals were from ICN Radiochemicals (Irvine, Calif.). Growth was terminated by addition of trichloroacetic acid to a final concentration of 5%. Cells were collected by centrifugation and washed once with H2O and were then resuspended in 1 ml of H2O. Lipids were extracted by addition of 1.4 ml of ethanol-ether-pyridine (1:0.33:0.067; vol per vol) and incubation at 57°C for 30 min. The debris was pelleted by centrifugation and supernatants were removed and placed into clean tubes. The samples were dried under N2 and stored at 4°C. The lipids were deacylated by two alternative methods. By method 1, dried lipid extracts were redissolved in 1 ml of solvent A (chloroform-methanol-water [16:16:5; vol per vol]). An equal volume of 0.2 N NaOH in methanol was added to each sample, and the mixture was incubated at 30°C for 45 min. To each sample, 1.1 ml of 0.5% (wt/vol) EDTA was added, and the mixtures were neutralized by addition of 0.2 ml of 1 N acetic acid. The nondeacylated lipids were extracted with 0.5 ml of chloroform, dried under N2, and resuspended in 0.5 ml of solvent A. By method 2, dried lipid extracts from 5 ml of culture were resuspended in 0.5 ml of methylamine reagent (25% methylamine in water [42.8%], methanol [45%], n-butanol [11.4%]). The samples were incubated at 53°C for 50 min before vacuum drying by centrifugation (SpeedVac; Savant Instruments). The dried samples were then suspended in 100 μl of solvent A, and 10 μl of each sample was used for thin-layer chromatographic analysis.
Thin-layer chromatography of radiolabeled lipids.
Labeled sphingolipids were separated by either one- or two-dimensional thin-layer chromatography as described by Steiner and Lester (35), with modifications. Samples were spotted onto 1-mm-thick silica G plates (Analtech Inc., Newark, Del.) that were dipped in 2.5% (wt/vol) EDTA, with the pH adjusted to 7.2 with NH4OH. The plates were dried at room temperature for 1 h and then overnight at 45°C. The plates were developed in the first dimension in CHCl3–CH3OH–4.2 N NH4OH (9:7:2; vol/vol). The second-dimension solvent was CHCl3-CH3OH-CH3COOH-H2O (15:6:4:1.6; vol/vol). The plates were dried for 30 min at room temperature and for 10 min at 50°C between the first- and second-dimension runs. Labeled lipids were detected by autoradiography. The identifications of IPC, MIPC, and M(IP)2C on the chromatograms were made by comparisons to the chromatographic positions of purified sphingolipids prepared by the methods of Smith and Lester (32). The identities of purified IPC, MIPC, and M(IP)2C were confirmed by electrospray ionization mass spectroscopy.
Fatty acid analysis.
Very long chain fatty acids were extracted from cells grown in 20 ml of SC-ura or SC-his media. Twenty A600 units of each strain was used for extraction. Reverse-phase high-pressure liquid chromatography (HPLC; column dimensions, 250 by 4.6 mm [Alltech Econosil C18 column]; particle size, 10 μm) was used to separate and quantify UV-absorbing phenacyl derivatives of the very-long-chain fatty acids. Saponification, derivatization, and HPLC analyses were performed as described by Lester et al. (25). Authentic fatty acid standards were obtained from Sigma Chemical Co.
RESULTS
S. cerevisiae SS33 and SS56 were used to clone the syringomycin E sensitivity genes SYR3 and SYR4, respectively. These are representative strains of two of eight distinct genetic complementation groups generated from a large collection of mutants resistant to growth inhibition by syringomycin E (39). SYR3 and SYR4 were cloned by gene complementation with yeast genomic libraries (29). Approximately 3,500 transformants of each strain were screened for recovery of growth sensitivity to syringomycin E at 1 μg per ml.
Cloning and identification of SYR3/ELO2 and ELO3.
Four syringomycin E-sensitive transformants of strain SS33 were isolated. The genomic library inserts of three complementing pSB32-based plasmids, 33-5, 33-6, and 33-15, were sequenced by using a primer directed toward pSB32. Sequence comparisons by BLAST analysis (1) with the Saccharomyces Genome Database (Stanford University) revealed that two of the plasmids, 33-5 and 33-6, contained the gene ELO2. ELO2 (which is the same as FEN1) encodes the enzyme that catalyzes the elongation of the sphingolipid very-long-chain fatty acids up to C24 (27). The genomic DNA insert of 33-15 contained sequences that corresponded to the gene ELO3. ELO3 (which is the same as SUR4) encodes an enzyme that catalyzes the elongation of very-long-chain fatty acids up to C26 and that can complement mutant defects of ELO2 (27).
Deletion of ELO2 and ELO3 and effect on susceptibility to syringomycin E.
To examine if ELO2 and ELO3 encoded functions that promoted growth inhibition by syringomycin E, we constructed strains with deletions of each of these genes by the one-step disruption method (30). Linearized HIS3 disruption plasmid constructs with Δelo2 or Δelo3 (pCRelo2HIS and YEpelo3HIS, respectively) (27) were used to transform diploid wild-type strain W303-1A. Transformants of each construct were sporulated and were subjected to tetrad analysis. Twenty-two tetrads from Δelo2/ELO2 diploids and seven tetrads from Δelo3/ELO3 diploids were dissected. Twenty-one of the former group of tetrads and all of the latter group of tetrads showed 2:2 cosegregation of the syringomycin E resistance phenotype and growth on SC-his medium.
To determine whether SYR3 was ELO2 or ELO3, tetrad analyses were performed with diploids produced by crossing haploid strain SS33 (syr3) with strains with disruptions in ELO2 or ELO3 (W303-Δelo2 or W303-Δelo3, respectively). All diploids were resistant to syringomycin at 1 μg per ml, and all meiotic segregants of nine tetrads derived from the SS33/W303-Δelo2 cross were also resistant. In contrast, diploids from the SS33/W303-Δelo3 cross sporulated poorly, and germination of meiotic segregants could not be achieved. The latter phenotypes are reminiscent of fen1/FEN1 and sur4/SUR4 (which is the same as elo2/ELO2 and elo3/ELO3) diploids (31), indicating that strain SS33 has a mutation in ELO2. Altogether, these results support the notion that SYR3 is ELO2, and like elo2 mutants, syr3 mutants can be complemented by ELO3.
Lipid analyses of strains W303-Δelo2 and W303-Δelo3.
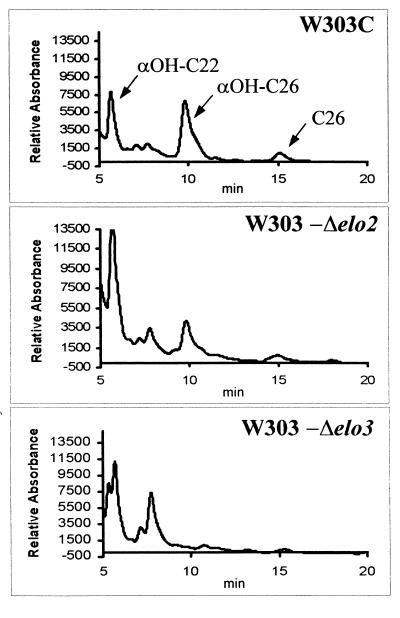
Long-chain fatty acid analysis of representative segregants from which genes were deleted (strains W303-Δelo2 and W303-Δelo3) showed defects in the synthesis of very long chain fatty acids (Fig. 1 and Table 2). Strain W303-Δelo2 produced elevated amounts of αOH-C22 very-long-chain fatty acids and reduced amounts of αOH-C:26 fatty acids. Strain W303-Δelo3 did not produce C26 very-long-chain fatty acids. Strain SS33 (syr3) showed the same fatty acid profile as strain W303-Δelo2. These fatty acid profiles are consistent with those reported by Oh et al. (27) for elo2 and elo3 mutants.
FIG. 1.
HPLC analysis of fatty acid derivatives from elo2 and elo3 syringomycin E-resistant mutants. Shown are the HPLC profiles of the phenacyl fatty acid derivatives of strains W303C (ELO2 ELO3), W303-Δelo2, and W303-Δelo3. Twenty microliters from each extract was analyzed as described in Materials and Methods.
TABLE 2.
Very-long-chain fatty acid compositions of Δelo2 and Δelo3 strains
| Strain | Mean ± SD % total peak areaa
|
|
|---|---|---|
| αOH:C22 | αOH:C26 | |
| W303C (wild type) | 0.612 ± 0.08 (n = 4) | 1.562 ± 0.76 (n = 4) |
| W303-Δelo2 | 0.707 ± 0.22 (n = 4) | 0.958 ± 0.56 (n = 4) |
| W303-Δelo3 | 0.648 ± 0.28 (n = 3) | NDb |
Determined from data presented in Fig. 1.
ND, not determined.
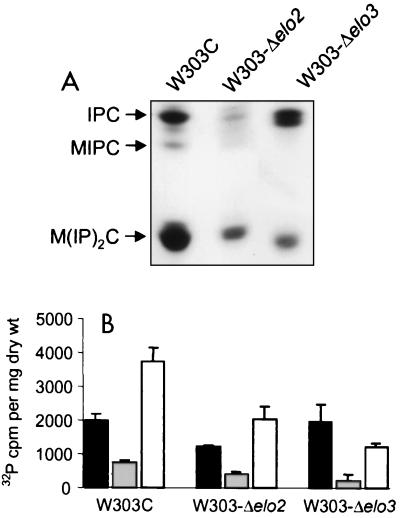
Oh et al. (27) showed that the chain lengths of the very-long-chain fatty acids influence the overall cellular sphingolipid composition. The sphingolipid levels in strains W303-Δelo2 and W303-Δelo3 were analyzed to determine if these effects occurred in the W303-1A genetic background. The sphingolipid profiles for strains W303-Δelo2 and W303-Δelo3 showed significant decreases in the levels of total sphingolipids as measured by the level of 32P incorporation (Fig. 2). Total sphingolipids of strains W303-Δelo2 and W303-Δelo3 had 32P counts of 3,654 ± 435 (standard deviation) (n = 3) and 3,426 ± 690 (n = 3) cpm per mg (dry weight) of cells, respectively, while the wild-type strain had 6,471 ± 501 (n = 3) cpm per mg (dry weight) of cells. Both mutant strains also showed significant decreases in M(IP)2C levels (Fig. 2). Although reduced in total quantities, the relative amounts of IPC, MIPC, and M(IP)2C in strain W303-Δelo2 resembled the relative amounts of these lipids in wild-type strain W303-1A, whereas strain W303-Δelo3 showed a relative increase in IPC levels (Fig. 2). The IPC species of the Δelo3 strain appeared to be heterogeneous, possibly due to production of very-long-chain fatty acids of various chain lengths.
FIG. 2.
Sphingolipid analysis of strains with Δelo2 and Δelo3 deletions. An autoradiogram of a thin-layer chromatographic plate with separated 32P-labeled sphingolipids from W303C (ELO2 ELO3), W303-Δelo2, and W303-Δelo3 is shown (A). The radiolabeled spots were scraped; and IPC (filled bar), MIPC (grey bar), and M(IP)2C (blank bar) were quantitated (B). Data are from three separate experiments, and the error bars represent standard deviations.
Cloning and identification of SYR4/IPT1.
A single transformant of syringomycin E-resistant strain SS56 carried a plasmid, p56-1A, with an 8-kb genomic insert that restored the syringomycin E sensitivity phenotype. SYR4 was identified within this insert by disruption with a Tn5 transposable element by following the methods of Ohya (28). Thirty E. coli isolates carrying p56-1A plasmids with Tn5 insertions were isolated by their ability to grow on medium containing 200 μg of kanamycin per ml. The plasmids were isolated individually and were used to transform strain SS56. One plasmid unable to complement the SS56 syr4 mutation due to a Tn5 insertion in plasmid-borne SYR4 was recovered. Determination of the DNA sequence that flanked the Tn5 transposon followed by BLAST program comparisons with the Saccharomyces Genome Database revealed that SYR4 is identical to IPT1, the gene recently identified as the yeast phosphorylinositol transferase, M(IP)2C synthase (11).
Deletion of SYR4/IPT1 and effect on susceptibility to syringomycin E.
To verify that a mutation in SYR4/IPT1 was responsible for the syringomycin E resistance phenotype of SS56, a strain with a Δsyr4/ipt1 deletion was made by the one-step disruption method (30). A 1-kb fragment of the 5′ portion of SYR4/IPT1 was replaced with a 1.1-kb fragment containing the URA3 gene. This construct was linearized and was used to transform diploid strain W303-1A. The resulting transformants (e.g., strain W303-4a) were sporulated and were used for tetrad analysis. Four of the dissected tetrads were replica plated onto YPD agar medium with and without 1 μg of syringomycin E per ml and SC-ura agar medium. All four gave a 2:2 segregation pattern with cosegregation of syringomycin E resistance and growth on SC-ura medium, confirming that SYR4/IPT1 promoted growth inhibition by syringomycin E. Gene deletion was confirmed by Southern blot analysis of genomic DNA isolated from strain W303H (Δsyr4/ipt1) and strain W303I (SYR4/IPT1) by using SYR4/IPT1 as a probe (data not shown).
Effect of SYR4/IPT1 deletion on sphingolipid levels.
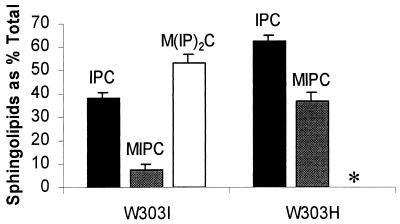
The sphingolipid compositions of strain W303H (Δsyr4/ipt1) and isogenic strain W303I (SYR4/IPT1) were determined by two-dimensional thin-layer chromatography after incorporation of [32P]H3PO4. Autoradiography of thin-layer chromatograms revealed the absence of M(IP)2C from strain W303H (Δsyr4/ipt1) (Fig. 3). The identity of M(IP)2C was confirmed by its ability to resist deacylation under mild alkaline conditions, colabeling with [3H]inositol, detection of mannose with orcinol staining, and negative-ion electrospray ionization mass spectral analysis. Dramatic increases in the amounts of MIPC (approximately fourfold above the wild-type levels) were observed with strain W303H (Δsyr4/ipt1) (Fig. 4). This is expected since MIPC is the substrate for Syr4p/Ipt1p (11). A smaller increase in IPC levels (approximately 33% above the wild-type levels) was also observed. MIPC represented 8% of the total inositol-containing sphingolipids in strain W303I (SYR4/IPT1), whereas it represented 37% in strain W303H (Δsyr4/ipt1), while the total radioactivity incorporated into sphingolipids was only slightly decreased (about 10%) in the mutant with the deletion.
FIG. 3.
Sphingolipid analysis of a strain with the Δsyr4/ipt1 deletion. The results of two-dimensional thin-layer chromatography of 32P-labeled lipids of W303H (Δsyr4/ipt1) (A) and W303I (SYR4/IPT1) (B) are shown.
FIG. 4.
Quantification of sphingolipids in a strain with the Δsyr4/ipt1 deletion. Strains W303I (SYR4/IPT1) and W303H (Δsyr4/ipt1) were labeled with [32P]H3PO4; and the sphingolipids IPC (filled bar), MIPC (grey bar), and M(IP)2C (open bar) were extracted and quantified as described in Materials and Methods. ∗, M(IP)2C was not detected in extracts from strain W303H (Δsyr4/ipt1). Data were obtained from three separate experiments. Error bars represent standard deviations.
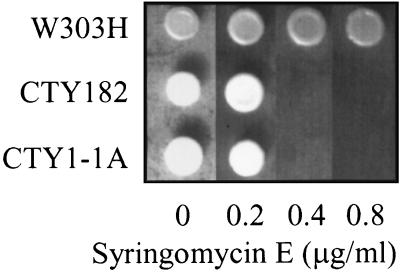
To determine if the increased MIPC levels in syr4/ipt1 mutants caused less susceptibility to syringomycin E, the relationship between syringomycin E sensitivity and MIPC accumulation was investigated. S. cerevisiae CTY1-1A contains a temperature-sensitive allele of SEC14, which encodes a phosphatidylinositol transfer protein required for secretion (3). This strain accumulates twofold higher levels of MIPC than its parental wild-type strain, CTY182, at permissive temperatures (36). In the present study, strains CTY1-1A and CTY182 were similarly sensitive to syringomycin E (Fig. 5). This result indicates that accumulation of MIPC was not the cause of the resistance of the strain W303H (Δsyr4/ipt1) to syringomycin E.
FIG. 5.
Syringomycin E sensitivity of MIPC-accumulating strain CTY1-1A (sec14-3ts). Strains CTY1-1A (sec14-3ts), CTY-182 (SEC14), and W303H (Δsyr4/ipt1) were plated onto YPD agar medium containing 0, 0.2, 0.4, or 0.8 μg of syringomycin E per ml and were grown at a permissive temperature (28°C).
Susceptibility of Δcsg1/sur1 and Δcsg2 deletion mutants to syringomycin E.
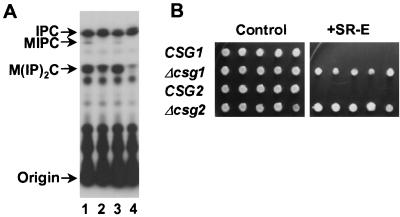
Because the steps preceding and following the addition of mannose in the sphingolipid biosynthetic pathway lead to syringomycin E resistance, it was of interest to study the importance of the mannosylation step itself for susceptibility to syringomycin E. Strains that had Δcsg1/sur1 or Δcsg2 deletions and that were defective for mannosylation were less susceptible to syringomycin E than isogenic wild-type strains (Fig. 6). Mutants with both types of deletion displayed no MIPC and had increased levels of IPC compared to the levels in isogenic wild-type strains, in agreement with previous reports (4, 8, 42) (Fig. 6). However, although reduced in amounts, M(IP)2C was still detected in lipid extracts of the strains with Δcsg1/sur1 and Δcsg2 deletions, with the latter showing barely detectable levels.
FIG. 6.
Effects of CSG1 and CSG2 deletions on sphingolipid composition and syringomycin E sensitivity. (A) Thin-layer chromatograph of sphingolipids of strains with Δcsg1 and Δcsg2 deletions. Yeast strains were grown and labeled in YPD medium containing 10 μCi of [32P]phosphoric acid per ml for 16 h. Lipids were extracted, deacylated with methylamine reagent, and analyzed by thin-layer chromatography. Lane 1, 39alt22A (CSG1 CSG2); lane 2, 39alt22A Δcsg1 (csg1::LEU2 CSG2); lane 3, TDY2037 (CSG1 CSG2); lane 4, TDY2038 (CSG1 csg2::LEU2). (B) Syringomycin E sensitivity of csg1 and csg2 strains. Five colonies of each strain were plated onto YPD medium with 0.5 μg of syringomycin E per ml. The strains shown are as follows: CSG1, strain 39alt22A; Δcsg1, strain 39alt22AΔcsg1; CSG2, strain TDY2037; Δcsg2, strain TDY2038.
DISCUSSION
The findings presented above identify five sphingolipid biosynthetic genes that promote the susceptibility of S. cerevisiae to syringomycin E. SYR3/ELO2 and ELO3, SYR4/IPT1, and CSG1/SUR1 and CSG2 encode enzymes responsible for elongation of the N-acylated very-long-chain fatty acids, attachment of the terminal phosphorylinositol group, and mannosylation, respectively, of the yeast sphingolipids. Although they facilitate sensitivity to syringomycin E, these genes are not required for growth in standard laboratory growth media. Mutations of these genes confer resistance to other antifungal cyclic lipodepsinonapeptide analogs of P. syringae pv. syringae. The degree of resistance, however, appears to vary according to the gene and analog. Cross-resistance to syringotoxin and syringostatin A is displayed in syringomycin E-resistant mutants of eight different gene complementation groups corresponding to genes that include the sphingoid base C-4 hydroxylase gene, SYR2, SYR3/ELO2, and SYR4/IPT1 (39). By comparison, yeast mutants defective in SYR2 are strongly resistant to pseudomycin B, whereas mutants defective in SYR3/ELO2, ELO3, SYR4/IPT1, CSG1/SUR1, and CSG2 are only slightly resistant to this analog (at concentrations that are twofold higher than those needed to inhibit parental wild-type strains) (D. Young and J. Radding, unpublished data).
How the functions of these genes allow sensitivity to syringomycin E is not clear. One possibility is that the structural modifications that they impart are necessary for proper binding of syringomycin E to the plasma membrane that leads to channel formation (14). M(IP)2C and MIPC (produced by Syr4p/Ipt1p and by Csg1p/Sur1p and Csg2p, respectively) are abundant in the plasma membrane (24) and will significantly increase the negative charge density on the outer surface of this membrane. The cationic cyclic peptidyl portion of syringomycin E should interact favorably with these sphingolipid polar head groups. Similarly, the full-length very-long-chain fatty acids produced by Syr3p/Elo2p and Elo3p may be needed for optimal positioning of the sphingolipids in the plasma membrane to promote syringomycin E binding and channel formation. A second possibility is that these functions modulate the relative amounts of IPC, MIPC, and M(IP)2C to either promote or prevent the ability of syringomycin E to interact with the plasma membrane. A Δsyr3/elo2 mutant produced small amounts of IPC, MIPC, and M(IP)2C, consistent with a previous report (27), and a Δelo3 mutant produced significantly less MIPC and M(IP)2C but normal amounts of IPC (Fig. 2). As expected, a Δsyr4/ipt1 mutant lacked M(IP)2C and accumulated MIPC (Fig. 4). Syringomycin E resistance did not appear to result from MIPC accumulation since a secretory pathway mutant (sec14-3ts) accumulated MIPC and was sensitive to syringomycin E (Fig. 5). Likewise, the Δcsg1/sur1 and Δcsg2 mutants lacked MIPC and accumulated IPC (Fig. 6). It is not known if IPC accumulation results in resistance to syringomycin E. Finally, it is possible that M(IP)2C with fully elongated very long chain fatty acids may be the source of growth-inhibitory phytoceramide. Thus, syringomycin E may activate M(IP)2C hydrolysis to generate phytoceramide analogous to hydrolytic generation of ceramide from sphingomyelin in mammalian cells (19, 20). Addition of cell-permeative (short-chain) analogs of phytoceramide to yeast are reported to inhibit growth (26), and a yeast M(IP)2C hydrolase activity has been measured (41).
Both SYR3/ELO2 and ELO3 fully restored the sensitivity of strain SS33 to syringomycin E, but the mutation in this strain was in SYR3/ELO2 rather than ELO3. Oh et al. (27) showed that overexpression of either ELO2 or ELO3 fully complemented elo2 mutants with production of sphingolipids with all of the very-long-chain fatty acids (C22 to C26). The inverse complementation of elo3 mutants by ELO2, however, was not complete since it did not allow production of sphingolipids with C26 very-long-chain fatty acids. Also, in the present work, crosses between strains SS33 and W303-Δelo3 produced diploids that sporulated poorly and that prevented germination of haploids consistent with previously described phenotypes of analogous elo2/ELO2 and elo3/ELO3 diploids (31).
Both Δcsg1/sur1 and Δcsg2 deletion strains still produced M(IP)2C, albeit at lower levels compared to the levels produced by isogenic wild-type strains. This observation suggests that these two genes are functionally redundant for production of M(IP)2C or that other routes for its synthesis exist. Similar low levels of M(IP)2C production by these mutants were observed by Daum et al. (8). However, Beeler et al. (4) and Zhao et al. (42) observed that Δcsg1/sur1 and Δcsg2 mutants did not produce M(IP)2C. Conceivably, because of the relatively high polarity of M(IP)2C and the use of different lipid extraction solvents, these discrepancies are due to differences in extraction efficiencies of this sphingolipid.
Because mutants defective in SYR3/ELO2, ELO3, SYR4/IPT1, CSG1/SUR1, and CSG2 were viable, it may be speculated that natural resistance to the pseudomonad cyclic lipodepsipeptides may be common among the yeasts and fungi found in nature. However, determination of the nonessentiality of these genes has been confined to studies with nutrient-rich laboratory growth media (e.g., YPD medium) and under favorable growth conditions (e.g., high-level oxygen tension). These genes may conceivably have more critical contributions to growth and survival under the typically more stringent conditions of natural environments. Thus, the prevalence and propensity for development of fungal resistance to these cyclic lipodepsipeptides in natural environments remain to be determined.
In summary, the results show that sphingolipids play major roles in the susceptibility of yeast to the plant bacterial and antifungal metabolite syringomycin E. When combined with the previous finding that sphingoid base C-4 hydroxylation is needed for syringomycin E action (17), a general picture emerges in which structural modifications that occur during sphingolipid biosynthesis promote susceptibility to this antifungal agent. Sterols also influence the ability of syringomycin E to inhibit yeasts (37, 40) as well as formation of ion channels in planar lipid bilayers (15). It is of interest to determine (i) if these two lipid classes work independently or synergistically to promote syringomycin E action and (ii) by what mechanisms these lipids may facilitate ion channel formation in the yeast plasma membrane.
ACKNOWLEDGMENTS
This research was supported by the Lilly Research Laboratories, the Utah Agricultural Experiment Station (project 607), and the National Science Foundation (grant 9003398).
We thank Teresa Dunn for providing strains TDY2037, TDY2038, 39alt22A, and 39alt22AΔcsg1 and Charles Martin for providing plasmids pCRelo2HIS and YEpelo3HIS. We acknowledge the assistance of S. Sonobe and R. Miyaoka (Hiroshima University).
Footnotes
Utah Agricultural Experiment Station paper 7221.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ballio A, Bossa F, Di Giorgio D, Ferranti P, Paci M, Scaloni A, Segre A, Strobel G. Novel bioactive lipodepsipeptides from Pseudomonas syringae: the pseudomycins. FEBS Lett. 1994;355:96–100. doi: 10.1016/0014-5793(94)01179-6. [DOI] [PubMed] [Google Scholar]
- 3.Bankaitis V A, Malehorn D E, Emr S D, Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beeler T J, Fu D, Rivera J, Monaghan E, Gable K, Dunn T M. SUR1 (CSG1/BCL21), a gene necessary for growth of Saccharomyces cerevisiae in the presence of high Ca2+ concentrations at 37°C, is required for mannosylation of inositolphosphorylceramide. Mol Gen Genet. 1997;255:570–579. doi: 10.1007/s004380050530. [DOI] [PubMed] [Google Scholar]
- 5.Bull C T, Wadsworth M L, Sorensen K N, Takemoto J Y, Austin R K, Smilanick J L. Syringomycin E produced by biological control agents controls green mold on lemons. Biol Control. 1998;12:89–95. [Google Scholar]
- 6.Christianson T, Sikorski R, Dante M, Shero J, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 7.Cliften P, Wang Y, Mochizuki D, Miyakawa T, Wangspa R, Hughes J, Takemoto J Y. SYR2, a gene necessary for syringomycin growth inhibition of Saccharomyces cerevisiae. Microbiology. 1996;142:477–484. doi: 10.1099/13500872-142-3-477. [DOI] [PubMed] [Google Scholar]
- 8.Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, Kelly D E, Kelly S, Schweizer E, Schuller H J, Hojad U, Greiner E, Finger K. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 9.Desfarges L, Durrens P, Juguelin H, Cassagne C, Bonneu M, Aigle M. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast. 1993;9:267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- 10.Dickson R C, Lester R L. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426:347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- 11.Dickson R C, Nagiec E E, Wells G B, Nagiec M M, Lester R L. Synthesis of mannose-(inositol-P)2-ceramide, the major sphingolipid in Saccharomyces cerevisiae, requires the IPT1 (YDR072c) gene. J Biol Chem. 1997;272:29620–29625. doi: 10.1074/jbc.272.47.29620. [DOI] [PubMed] [Google Scholar]
- 12.Dickson R C, Wells G B, Schmidt A, Lester R L. Isolation of mutant Saccharomyces cerevisiae strains that survive without sphingolipids. Mol Cell Biol. 1990;10:2176–2181. doi: 10.1128/mcb.10.5.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.el-Sherbeini M, Clemas J A. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-beta-glucan in vitro. J Bacteriol. 1995;177:3227–3234. doi: 10.1128/jb.177.11.3227-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feigin A M, Takemoto J Y, Wangspa R, Teeter J H, Brand J G. Properties of voltage-gated ion channels formed by syringomycin E in planar lipid bilayers. J Membr Biol. 1996;149:41–47. doi: 10.1007/s002329900005. [DOI] [PubMed] [Google Scholar]
- 15.Feigin A M, Schagina L V, Takemoto J Y, Teeter J H, Brand J G. The effect of sterols on the sensitivity of membranes to the channel-forming antifungal antibiotic, syringomycin E. Biochim Biophys Acta. 1997;1324:102–110. doi: 10.1016/s0005-2736(96)00214-3. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Arranz M, Maldonado A M, Mazon M J, Portillo F. Transcriptional control of yeast plasma membrane H(+)-ATPase by glucose. Cloning and characterization of a new gene involved in this regulation. J Biol Chem. 1994;269:18076–18082. [PubMed] [Google Scholar]
- 17.Grilley M M, Stock S D, Dickson R C, Lester R L, Takemoto J Y. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273:11062–11068. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- 18.Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sur2p and Scs7p. J Biol Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- 19.Hannun Y A. Functions of ceramide in coordinating cellular responses to stress. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 20.Hannun Y A, Obeid L M. Ceramide: an intracellular signal for apoptosis. Trends Biochem Sci. 1995;20:73–77. doi: 10.1016/s0968-0004(00)88961-6. [DOI] [PubMed] [Google Scholar]
- 21.Iacobellis N S, Lavermicocca P, Grgurina I, Simmaco M, Ballio A. Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol Mol Plant Pathol. 1992;40:107–116. [Google Scholar]
- 22.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 23.Leber A, Fischer P, Schneiter R, Kohlwein S D, Daum G. The yeast mic2 mutant is defective in the formation of mannosyl-diinositolphosphorylceramide. FEBS Lett. 1997;411:211–214. doi: 10.1016/s0014-5793(97)00692-3. [DOI] [PubMed] [Google Scholar]
- 24.Lester R L, Dickson R C. Sphingolipids with inositolphosphate-containing head groups. Adv Lipid Res. 1993;26:253–274. [PubMed] [Google Scholar]
- 25.Lester R L, Wells G B, Oxford G, Dickson R C. Mutant strains of Saccharomyces cerevisiae lacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J Biol Chem. 1993;268:845–856. [PubMed] [Google Scholar]
- 26.Nickels J T, Broach J R. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 27.Oh C S, Toke D A, Mandala S, Martin C E. ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J Biol Chem. 1997;272:17376–17384. doi: 10.1074/jbc.272.28.17376. [DOI] [PubMed] [Google Scholar]
- 28.Ohya Y. LA-PCR-based quick method for the identification of genes responsible for the complementation of Saccharomyces cerevisiae mutations. BioTechniques. 1996;20:777–789. doi: 10.2144/96205bm07. [DOI] [PubMed] [Google Scholar]
- 29.Rose M D, Winston F, Heiter P. Methods in yeast genetics; a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 30.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–221. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 31.Silve S, Leplatois P, Josse A, Dupuy P H, Lanau C, Kaghad M, Dhers C, Picard C, Rahier A, Taton M, Le Fur G, Caput D, Ferrara P, Loison G. The immunosuppressant SR 31747 blocks cell proliferation by inhibiting a steroid isomerase in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2719–2727. doi: 10.1128/mcb.16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith S, Lester R L. Inositol phosphorylceramide, a novel substance and the chief member of a major group of yeast sphingolipids containing a single inositol phosphate. J Biol Chem. 1974;249:3395–3405. [PubMed] [Google Scholar]
- 33.Sorensen K N, Kim K H, Takemoto J Y. In vitro antifungal and fungicidal activities and erythrocyte toxicities of cyclic lipodepsinonapeptides produced by Pseudomonas syringae pv. syringae. Antimicrob Agents Chemother. 1996;40:2710–2713. doi: 10.1128/aac.40.12.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorensen K N, Wanstrom A A, Allen S D, Takemoto J Y. Efficacy of syringomycin E in a murine model of vaginal candidiasis. J Antibiot. 1998;51:743–749. doi: 10.7164/antibiotics.51.743. [DOI] [PubMed] [Google Scholar]
- 35.Steiner S, Lester R L. Studies on the diversity of inositol-containing yeast phospholipids: incorporation of 2-deoxyglucose into lipid. J Bacteriol. 1972;109:81–88. doi: 10.1128/jb.109.1.81-88.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stock S D, Hama H, DeWald D B, Takemoto J Y. SEC14-dependent secretion in Saccharomyes cerevisiae. Non-dependence on sphingolipid synthesis-coupled diacylglycerol production. J Biol Chem. 1999;274:12979–12983. doi: 10.1074/jbc.274.19.12979. [DOI] [PubMed] [Google Scholar]
- 37.Taguchi N, Takano Y, Julmanop C, Wang Y, Stock S, Takemoto J, Miyakawa T. Identification and analyses of the Saccharomyces cerevisiae SYR1 reveals that ergosterol is involved in the action of syringomycin. Microbiology. 1994;140:353–359. doi: 10.1099/13500872-140-2-353. [DOI] [PubMed] [Google Scholar]
- 38.Takemoto J Y. Bacterial phytotoxin syringomycin and its interaction with host membranes. In: Verma D P S, editor. Molecular signals in plant-microbe communications. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 247–260. [Google Scholar]
- 39.Takemoto J Y, Yu Y, Stock S D, Miyakawa T. Yeast genes involved in growth inhibition by Pseudomonas syringae pv. syringae syringomycin family lipodepsipeptides. FEMS Microbiol Lett. 1993;114:339–342. doi: 10.1111/j.1574-6968.1993.tb06595.x. [DOI] [PubMed] [Google Scholar]
- 40.Wangspa R, Takemoto J Y. Role of ergosterol in growth inhibition of Saccharomyces cerevisiae by syringomycin E. FEMS Microbiol Lett. 1998;167:215–220. doi: 10.1111/j.1574-6968.1998.tb13231.x. [DOI] [PubMed] [Google Scholar]
- 41.Wells G B, Dickson R C, Lester R L. Heat-induced elevation of ceramide in Saccharomyces cerevisiae via de novo synthesis. J Biol Chem. 1998;273:7235–7243. doi: 10.1074/jbc.273.13.7235. [DOI] [PubMed] [Google Scholar]
- 42.Zhao C, Beeler T, Dunn T D. Suppressors of the Ca2+-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J Biol Chem. 1994;269:21480–21488. [PubMed] [Google Scholar]