Abstract
A simple assay for detection of compounds that bind to the active site in the transglycosylation domain of the essential bifunctional transglycosylase and transpeptidase penicillin-binding proteins (PBPs) is reported. The method is based on a competition with the specific transglycosylase inhibitor moenomycin. With moenomycin coupled to Affi-Gel beads, a simple filtration procedure allows the amount of labeled PBPs that bind to moenomycin beads in the presence of test substances to be determined. The PBPs can easily be labeled by the covalent binding of penicillin derivatives. Crude membrane extracts can be used as a source for the PBPs, and different kinds of labels for the penicillin-PBP complexes can be used. The assay can be adapted to high-throughput screens.
The β-lactam family of antibiotics is one of the most important and successful families of antibiotics for antibacterial chemotherapy for a number of different reasons. One reason is that they inhibit an enzymatic reaction, a dd-transpeptidation, which is essential for the mechanical strength of the bacterial exoskeleton, the murein (peptidoglycan) sacculus (19, 25). Another enzymatic reaction, murein transglycosylation, is as important for the formation of the murein sacculus, but until now no therapeutically useful antibiotics active against the formation of the murein sacculus have existed (11, 23). This report describes a simple assay that will allow screening for specific inhibitors of murein transglycosylases.
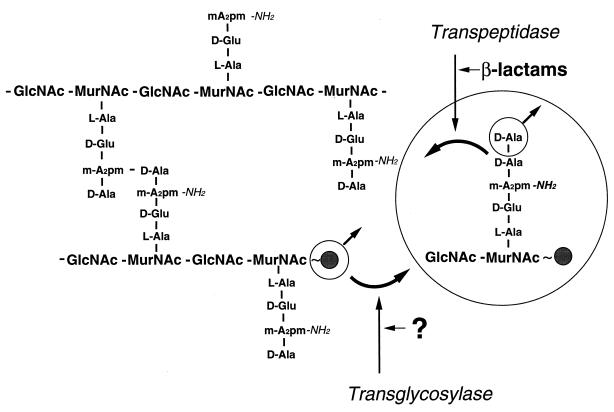
Murein is a cross-linked polymer that completely encloses the cell, thereby stabilizing it from rupture by the high level of intracellular turgor (14, 15). The murein netting is formed by polymerization of a peptidyl disaccharide subunit in two directions, resulting in a meshwork of glycan strands that are cross-linked by peptide bridges (11, 21). In the case of Escherichia coli or Bacillus cereus the murein precursor consists of N-acetylglucosaminyl-β-1,4-N-acetylmuramyl-l-alanyl-d-glutamyl-γ-meso-diaminopimelyl-d-alanyl-d-alanine (GlcNAc-MurNAc-l-Ala-d-Glu-γ-m-A2pm-d-Ala-d-Ala) that is linked via a pyrophosphate group to the C55 isoprenoid bactoprenol. Peptide-substituted polysaccharide strands are formed by transglycosylation of a lipid-linked nascent glycan strand to the lipid-linked murein precursor (Fig. 1) (23). They are then cross-linked by transpeptidation between the carboxyl end of the penultimate d-Ala of one peptide moiety and the ɛ amino group of the meso-diaminopimelyl (m-A2pm) of the peptide moiety that protrudes from a neighboring glycan strand (Fig. 1) (6, 11). Interestingly, a number of murein polymerases from different bacterial species are known. These murein polymerases are bifunctional proteins that combine a transpeptidase domain with a transglycosylase domain (7, 9, 12).
FIG. 1.
Polymerization of the murein subunit by transpeptidation and transglycosylation. A2pm, diaminopimelic acid; the solid ball represents the bactoprenol to which the subunit is linked via a pyrophosphate (∼).
Pencillins inhibit the cross-linking reaction by covalently binding to the transpeptidases because of a structural analogy with the d-alanyl-d-alanine terminus of the natural pentapeptide substrate (18, 27). Besides the dd-transpeptidases, dd-carboxypeptidases and dd-endopeptidases are also known to be penicillin-sensitive enzymes. The latter enzymes, however, are unlikely to be the major killing targets of penicillins (17, 19). Since some of the target enzymes of penicillin, which are referred to as penicillin-binding proteins (PBPs) (17), have been crystallized, details of the interaction of the penicillin molecule with the catalytic site of the enzymes are known (6, 10).
It is surprising that the second essential enzyme reaction needed for the formation of a mechanically strong murein, the transglycosylation reaction, has not yet been exploited for chemotherapeutically useful antibiotics. Mainly, only one antibiotic with a structure that specifically inhibits transglycosylation is known: moenomycin (Fig. 2) (8, 22, 26). Although they are quite effective in triggering lysis of both gram-positive and gram-negative bacteria, moenomycins have toxic side effects that prevent their clinical use.
FIG. 2.
Structure of moenomycin A. Ac, acetyl.
A major difficulty in any attempt to screen for transglycosylase inhibitors is the lack of a simple assay. The in vitro enzyme assay calls for the use of undecaprenyl-pyrophosphatidyl-disaccharidepentapeptide as a substrate (1, 11, 23). However, preparation of this lipid-linked murein precursor is quite cumbersome (1, 20). In order to establish a simple assay which could even be adopted as a high-throughput screening assay, we took advantage of the availability of the specific inhibitor, meonomycin. This allowed us to set up a competition assay that identifies compounds that interfere with the binding of moenomycin to the transglycosylase site of murein polymerases such as the bifunctional PBPs 1A and 1B of E. coli. The presence of a second binding site in the PBPs, the transpeptidation site, was used not only to specifically label the PBPs by using a radioactively labeled penicillin derivative but also to block this site from any unspecific (not transglycosylase directed) interaction of the test compounds with the target enzyme.
MATERIALS AND METHODS
Bacterial strains.
Wild-type strain E. coli MC1061 (3) or triple mutant D456, which lacks PBP 4, PBP 5, and PBP 6 (5), was used to prepare crude extracts of membrane proteins. For the preparation of extracts that lack PBP 1A or PBP 1B, strain SP1028 (ponA) or SP1026 (ponB), respectively, was used (28).
Preparation of crude membrane extracts.
Bacterial cells were grown in Luria-Bertani (LB) medium (10 g of casein hydrolysate per liter, 5 g of yeast extract per liter, 10 g of NaCl per liter) at 37°C with aeration. At an optical density (measured at 578 nm) of 0.6 to 0.8 the cells were cooled on ice for 10 min and were collected by centrifugation (7,000 × g, 10 min, 4°C). All the following steps were performed at 4°C. The pellet was resuspended in 10 mM Tris-maleate–10 mM MgCl2–0.02% NaN3 (pH 6.8). After the addition of 1 mM phenylmethylsulfonyl fluoride and 10 μg of DNase per ml, the cells were broken by passage through a French press at 16,000 lb/in2. The membranes were collected by ultracentrifugation (100,000 × g, 60 min) and were washed by stirring with 10 mM Tris-maleate–10 mM MgCl2–1 M NaCl–0.02% NaN3 (pH 6.8) for 1 h. Membrane proteins, including the high-molecular-weight PBPs, were solubilized by stirring overnight with 10 mM Tris-maleate–10 mM MgCl2–500 mM NaCl–0.02% NaN3–2% Triton X-100 (pH 6.8). The supernatant, after ultracentrifugation, was diluted with an equal volume of dialysis buffer and was dialyzed against 10 mM Tris-maleate–10 mM MgCl2–150 mM NaCl–0.02% NaN3 (pH 6.8). From 8 liters of cell culture, 48 ml of crude membrane extract containing 2.5 mg of protein per ml was prepared.
Labeling of PBPs.
The dialyzed membrane extract (48 ml) was incubated with 400 μl of [3H]benzylpenicillin (1 mCi/ml, 19 Ci/mmol; Amersham, Little Chalfont, England) at 37°C for 30 min. The reaction was stopped by the addition of 4.8 ml of 60 mg of penicillin G (Serva, Heidelberg, Germany) per ml. After a second incubation for 20 min at 37°C the extract was divided into 1.8-ml aliquots and was stored at −20°C. The extracts were stable for at least 1 month.
Immobilization of moenomycin.
Moenomycin A (kindly provided by W. Aretz, Hoechst-Marion-Roussel, Frankfurt, Germany) was coupled to activated agarose as described previously (24). Briefly, 10 ml of Affi-Gel 10 (Bio-Rad, Munich, Germany) was washed with methanol and was then added to a solution of 400 mg of moenomycin in 10 ml of methanol. The suspension was incubated for 96 h at 4°C with rotation of the tube head over head to avoid settling of the beads. The gel was washed on a filter: four times with 40 ml of methanol and two times with 40 ml of 10 mM Tris-maleate–10 mM MgCl2–150 mM NaCl–0.02% NaN3–0.2% Triton X-100 (pH 6.8) (TG buffer). To block the remaining coupling sites the material was incubated with TG buffer for 30 h at 4°C. The gel was washed three times with 40 ml of TG buffer and was resuspended to a final volume of 28 ml in TG buffer. This stock suspension of moenomycin–Affi-Gel beads could be stored at 4°C for 4 months with no loss in binding capacity for bifunctional PBPs.
Assay performance.
The assay was performed in opaque 96-well, 0.45-μm-pore-size filter microtiter plates MultiScreen HV (Durapore membrane) with the MultiScreen filtration system (Millipore, Molsheim, Germany). To reduce the background signal, empty filters were preincubated for 6 h at 4°C with 150 μl of benzylpenicillin (30 mg/ml) in TG buffer. After filtration and washing with 200 μl of TG buffer the moenomycin–Affi-Gel bead suspension was added. Since only 2 μl of the stock suspension is needed per well, it was more accurate to prepare a dilution and pipet a larger volume. For example, if one plate with 96 wells was prepared, 250 μl of the moenomycin–Affi-Gel stock suspension was mixed with 2,250 μl of TG buffer. To each well except for the blank wells, 20 μl of the diluted suspension was added. By using a truncated tip with a wider opening, the pipetting of the suspension could be improved. In addition, it was important to mix the suspension from time to time to prevent settling of the beads during the filling of the wells. The beads were sedimented to the bottom of the wells by filtration with TG buffer. Afterward, TG buffer (20 μl) was added to each well to prevent the beads from drying out. Then, 50 μl of crude membrane extract containing tritium-labeled PBPs was put into each well. The compounds to be tested for their ability to inhibit the binding of the PBPs to moenomycin beads, e.g., moenomycin itself or its derivatives, were added as 5-μl volumes of solutions in dimethyl sulfoxide (DMSO) at various concentrations. Only DMSO was added to the control samples (no inhibition). The plate was sealed in plastic to prevent drying and was incubated overnight at 4°C. On the following day, the nonbound radioactivity was removed by filtration and the beads were extensively washed with TG buffer. The wells were rinsed with three times with 200 μl of TG buffer, followed by an incubation step with 200 μl of TG buffer for 10 min at 4°C. After filtration, two more washing steps with 200 μl of TG buffer were performed. Then, 30 μl of Opti Phase Supermix scintillation cocktail (Wallac Oy, Turku, Finland) was added and the radioactivity was measured with a MicroBeta Plus counter (Wallac Oy) according to the protocol of the manufacturer. Each compound as well as the controls and blanks was assayed in triplicate.
We also used a modified procedure in which nonlabeled extracts were incubated overnight with moenomycin–Affi-Gel beads. After the washing step (see above) the moenomycin-bound PBPs were labeled in the microtiter plate with an aliquot of 0.3 μl [3H]benzylpenicillin (0.3 μCi) by incubation at 37°C for 45 min. After another washing procedure, the radioactivity was measured as described above.
RESULTS
Determination of optimal assay conditions.
The principle of the assay is illustrated in Fig. 3 and is based on the competition of test compounds and moenomycin–Affi-Gel beads in binding to labeled PBPs. The assay is performed with the help of microtiter filter plates. It seemed important that, for the control samples, all the free enzyme was bound to the moenomycin beads. Therefore, the saturation value was determined. Saturation of binding was achieved with 1 μl of moenomycin beads. Since the pipetting of the beads was subject to some variations, we decided to routinely use a volume of 2 μl to ensure unambiguous saturating conditions.
FIG. 3.
Schematic representation of the competition assay for compounds that bind to the transglycosylase sites of PBPs. A membrane extract of a PBP 4, PBP 5, and PBP 6 deletion mutant was used for the example shown here. The arrows indicate PBPs that are bound to a soluble moenomycin antagonist (X) and that are therefore washed out. Thereby, the amount of PBPs retained by the moenomycin beads is decreased.
Binding of labeled PBPs to moenomycin beads depended on the incubation time. Rather long incubations of about 7.5 h were needed to obtain a stable signal that did not increase with further incubation. Probably, the diffusion rates of the different components, such as the beads and the enzyme-carrying detergent vesicles, are rather slow. Thus, we decided to use overnight incubations.
To reduce the background signal as much as possible it was important to preincubate the filters with unlabeled penicillin G. This step reduced any unspecific binding of free labeled penicillin as well as labeled penicilloyl enzyme via the penicillin moiety to the filters. Extensive washing after the incubation step also affected the background values. Besides washing by filtration, a short incubation with washing buffer was included in the procedure to effectively release low-molecular-weight label (free penicillin or degradation products) from the pores of the filters.
Competition assay with moenomycin A as a test substance.
Two methods of labeling the PBPs have been compared with one another. In one method the membrane extracts were labeled with [3H]benzylpenicillin prior to the competition assay. This allowed the preparation of stocks that could be stored for at least 1 month. It had the advantage that one and the same PBP preparation can be used in several series of screenings (Fig. 4). In an alternative method only bound PBPs were labeled in the microtiter plates after the incubation of the membrane extract with the moenomycin beads and after a first washing step that removes unbound PBPs. This method (Fig. 5) yielded higher signals and a lower background (a 3% background compared to a 6% background by the method that used prelabeled membrane extracts). However, the latter method may not be the method of choice for screenings because it includes two additional washing steps and is prone to variations from experiment to experiment.
FIG. 4.
Competition of soluble moenomycin A with moenomycin beads for binding to [3H]benzylpenicillin-PBP complexes. Different amounts of moenomycin A were incubated, as described in detail in Materials and Methods, overnight at 6°C in the presence of moenomycin beads (2 μl) and [3H]benzylpenicillin-labeled membrane extracts (50 μl, 115 μg protein) from the indicated E. coli mutant. The radioactivity remaining on the filters was determined, and the background signal determined from incubation of labeled extract without moenomycin beads was substracted. The diagram shows the relative radioactivity compared to that from the incubation mixtures to which soluble moenomycin was not added. Rectangles, SP1028 (ponA); triangles, D456 (dacABC); circles, SP1026 (ponB).
FIG. 5.
Comparison of membrane extracts from different PBP mutants of E. coli. Membrane extracts (50 μl; 125 μg of protein) prepared from the indicated E. coli strains were incubated overnight at 6°C with moenomycin beads (2 μl) as described in Materials and Methods (grey columns). The background was determined by incubating the samples without moenomycin beads (black columns). After a washing step, [3H]benzylpenicillin (0.3 μl, 0.3 μCi) was added to the wells and the plates were incubated at 37°C for 45 min. After extensive washing the remaining radioactivity was determined. The average of two independent experiments is given. wt, wild type.
Under the established competition assay conditions, moenomycin A caused 50% inhibition of binding of the PBPs to the moenomycin beads at concentrations of 3 to 20 ng/ml (Fig. 4). Various derivatives of meonomycin obtained from Hoechst-Marion-Roussel did interfere with the binding to different extents. Mersacidin, which is known to bind to the murein lipid-bound precursors (2), and decaplanin, which also interferes with the lipid recycling process (12), had no inhibitory effect in the assay, even when they were added at rather high concentrations. The level of binding of labeled PBPs compared to that of the control was 111% in the presence of 0.2 mg of mersacidin per ml and 109% in the presence of 1 mg of decaplanin per ml.
Use of membrane extracts from a mutant lacking PBPs 4, 5, and 6.
The low-molecular-weight PBPs have dd-carboxypeptidase and/or dd-endopeptidase activity and thus bind to penicillin covalently (17). However, they lack a transglycosylase site and therefore do not contribute to the assay. Because they carry label they may interfere with the assay by increasing the background. Therefore, we tested whether the use of membrane proteins prepared from a triple mutant with dacA, dacB, and dacC deletions (5) would be of advantage. Figure 5 shows that, as expected, the use of a membrane extract from the mutant strain that lacked these low-molecular-weight PBPs decreased the background compared with that from the use of a membrane extract from wild-type cells.
Moenomycin competition assay with PBP 1A or PBP 1B.
Because of the availability of mutants that lack either PBP 1A or PBP 1B, we were able to perform the competition assay with one or the other major bifunctional transglycosylase and transpeptidase enzyme of E. coli. As shown in Fig. 4 and 5, there was a significant difference between the two PBPs in their binding to and competitive release from the moenomycin beads. The competition assay (Fig. 4) clearly reveals that the binding of PBP 1B to moenomycin is stronger than that of PBP 1A. A sevenfold higher concentration of free moenomycin (20 ng/ml) was needed to achieve a 50% reduction in binding of PBP 1B to moenomycin beads compared with that required to achieve a 50% reduction in binding of PBP 1A (3 ng of free moenomycin per ml).
DISCUSSION
Since the binding of moenomycin to PBPs 1A and 1B is rather strong, only compounds with similar affinities may be detected in the assay unless high concentrations of the test compounds are used. To increase the sensitivity of the test for the identification of substances that display a lower affinity for the transglycosylase site compared to that of moenomycin A, the assay could be modified by using a moenomycin derivative as a ligand that has a lower binding constant.
The coupling of moenomycin is a rather slow reaction and takes 96 h in a cold room. That is not surprising because moenomycin A has no primary amino group (Fig. 2). It is still not clear where the coupling occurs. One possibility could be the hydroxyl group at the 1.4 substituted cyclopentenyl ring that is rather acid due to a keto-enol tautomery. Again, modifications of the method by using moenomycin derivatives with other functional groups may allow different and more efficient coupling procedures.
Importantly, the assay does not depend on purified enzymes, and crude fractions of solubilized membrane proteins can be used. If the interaction with specific PBPs is of interest, the respective deletion mutants can be used for the preparation of the membrane proteins. Purified PBPs may be used as well. The basic strategy can be modified in many ways to allow the application of quite different screening methods. Although the method uses crude membrane extracts, it exclusively measures the two essential bifunctional murein polymerases PBP 1A and PBP 1B (17, 19, 25). PBP 1C, a minor bifunctional PBP which has recently been cloned and characterized (16), does not bind to ampicillin or benzylpenicillin derivatives and thus is not detected in the assay.
The established assay identifies all compounds that interfere with the binding of moenomycin to either purified bifunctional PBPs or mixtures of bifunctional PBPs. For detection, the transpeptidase sites of the PBPs are covalently labeled with a penicillin derivative. In the study described in this report radioactively labeled penicillin was used. However, other, nonradioactive labels, such as fluorescence, could be used as well (4, 29). In the case of monofunctional transglycosylases that lack a PBP domain, the amount of transglycosylase that bound to the moenomycin beads could be determined with specific antibodies.
It is hoped that the strategy based on the specific binding of moenomycin in a competition assay sets the scene for a broader screening for transglycosylase-inhibitory substances. There is a good chance that in such screenings investigators can identify compounds that may turn out to be promising lead structures for novel antibacterial agents, which are so badly needed in our never ending battle against the increasing number of multidrug-resistant infections.
ACKNOWLEDGMENTS
We thank Uli Schwarz for interest and support and Vicky Kastner for critical reading of the manuscript.
REFERENCES
- 1.Anderson J S, Matsuhashi M, Haskin M A, Strominger J L. Biosynthesis of the peptidoglycan of bacterial cell walls. II. Phospholipid carriers in the reaction sequence. J Biol Chem. 1967;242:3180–3190. [PubMed] [Google Scholar]
- 2.Brötz H, Bierbaum G, Reynolds P E, Sahl H-G. The lantibiotic mersacidin inhibits peptidoglycan biosynthesis at the level of transglycosylation. Eur J Biochem. 1997;246:193–199. doi: 10.1111/j.1432-1033.1997.t01-1-00193.x. [DOI] [PubMed] [Google Scholar]
- 3.Casabadan M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli J. Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Dargis M, Malouin F. Use of biotinylated β-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria. Antimicrob Agents Chemother. 1994;38:973–980. doi: 10.1128/aac.38.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards D H, Donachie W D. Construction of a triple deletion of penicillin-binding proteins 4, 5 and 6 in Escherichia coli. In: de Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis. New York, N.Y: Plenum Press; 1993. pp. 369–374. [Google Scholar]
- 6.Ghuysen J-M. Biochemistry of the penicilloyl serine transferases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 103–129. [Google Scholar]
- 7.Goffin C, Ghuysen J-M. Multimodular penicillin binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huber G, Nesemann G. Moenomycin, an inhibitor of cell wall synthesis Biochem. Biophys Res Commun. 1968;30:7–13. doi: 10.1016/0006-291x(68)90704-3. [DOI] [PubMed] [Google Scholar]
- 9.Ishino F, Mitsui K, Tamaki S, Matsuhashi M. Dual enzyme activities of cell wall peptidoglycan synthesis, peptidoglycan transglycosylase and penicillin-sensitive transpeptidase, in purified preparations of Escherichia coli penicillin-binding protein 1A. Biochem Biophys Res Commun. 1980;97:287–293. doi: 10.1016/s0006-291x(80)80166-5. [DOI] [PubMed] [Google Scholar]
- 10.Kelly J A, Moews P C, Knox J R, Frère J M, Ghuysen J M. Penicillin target enzyme and the antibiotic binding site. Science. 1982;218:479–481. doi: 10.1126/science.7123246. [DOI] [PubMed] [Google Scholar]
- 11.Matsuhashi M. Utilization of lipid-linked precursors and the formation of peptidoglycan in the proces of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 55–71. [Google Scholar]
- 12.Nakagawa J, Tamaki S, Matsuhashi M. Purified penicillin binding proteins 1Bs from Escherichia coli membrane showing activities of both peptidoglycan polymerase and peptidoglycan crosslinking enzyme. Agric Biol Chem. 1979;43:1379–1380. [Google Scholar]
- 13.Neu H C, Chin N X, Niu W W. In vitro activity of the new glycopeptide decaplanin. Eur J Clin Microbiol Infect Dis. 1992;11:458–462. doi: 10.1007/BF01961864. [DOI] [PubMed] [Google Scholar]
- 14.Park J T. The murein sacculus. In: Neidhardt F C, editor. Escherichia coli and Salmonella. Washington, D.C.: American Society for Microbiology; 1996. pp. 48–57. [Google Scholar]
- 15.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, United Kingdom: Chapman & Hall; 1980. [Google Scholar]
- 16.Schiffer G, Höltje J-V. Cloning and characterization of PBP 1C, a third member of the multimodular class A of penicillin-binding proteins of Escherichia coli J. Biol Chem. 1999;274:32031–32039. doi: 10.1074/jbc.274.45.32031. [DOI] [PubMed] [Google Scholar]
- 17.Spratt B G. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tipper D J, Strominger J L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-d-alanyl-d-alanine. Proc Natl Acad Sci USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomasz A. Penicillin-binding proteins and the antibacterial effectiveness of beta-lactam antibiotics. Rev Infect Dis. 1986;8(Suppl. 3):260–378. doi: 10.1093/clinids/8.supplement_3.s260. [DOI] [PubMed] [Google Scholar]
- 20.Umbreit J N, Strominger J L. Isolation of the lipid intermediate in peptidoglycan biosynthesis from Escherichia coli. J Bacteriol. 1972;112:1306–1309. doi: 10.1128/jb.112.3.1306-1309.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Heijenoort J. Biosynthesis of the bacterial peptidoglycan unit. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 39–54. [Google Scholar]
- 22.van Heijenoort Y, Derrien M, van Heijenoort J. Polymerization by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coli K12 and its inhibition by antibiotics. FEBS Lett. 1978;89:141–144. doi: 10.1016/0014-5793(78)80540-7. [DOI] [PubMed] [Google Scholar]
- 23.van Heijenoort Y, van Heijenoort J. Biosynthesis of the peptidoglycan of Escherichia coli K12—properties of the in vitro polymerization by transglycosylation. FEBS Lett. 1980;110:241–244. doi: 10.1016/0014-5793(80)80082-2. [DOI] [PubMed] [Google Scholar]
- 24.Vollmer W, von Rechenberg M, Höltje J-V. Demonstration of molecular interactions between the murein polymerase PBP1B, the lytic transglycosylase MltA, and the scaffolding protein MipA of Escherichia coli. J Biol Chem. 1999;274:6726–6734. doi: 10.1074/jbc.274.10.6726. [DOI] [PubMed] [Google Scholar]
- 25.Waxman D J, Strominger J L. Penicillin. Annu Rev Biochem. 1983;52:825–869. doi: 10.1146/annurev.bi.52.070183.004141. [DOI] [PubMed] [Google Scholar]
- 26.Welzel P, Kunisch F, Kruggel F, Stein H, Scherkenbeck J, Hiltmann A, Dudeck H, Müller D, Maggio J E, Fehlhaber H-W, Seibert G, van Heijenort Y, van Heijenoort J. Moenomycin A: minimum structural requirements for biological activity. Tetrahedron. 1987;43:585–598. [Google Scholar]
- 27.Wise E M, Jr, Park J T. Penicillin: its basic site of action as an inhibitor of a peptide cross-linking reaction in cell wall mucopeptide synthesis. Proc Natl Acad Sci USA. 1965;54:75–81. doi: 10.1073/pnas.54.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yousif S Y, Broome-Smith J K, Spratt B G. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
- 29.Zhao G, Meier T I, Kahl S D, Gee K R, Blaszczak L C. Bocillin FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob Agents Chemother. 1999;43:1124–1128. doi: 10.1128/aac.43.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]