PURPOSE
Current surveillance imaging and tumor markers lack sensitivity for the early detection of recurrence in GI cancers. This study critically evaluates the current literature on the role of sequential measurement of circulating tumor DNA (ctDNA) before and after curative resection in informing recurrence.
METHODS
A systematic search using a predefined, registered protocol was conducted for studies published between January 2010 and May 2020. Included studies described patients with GI cancers treated with curative-intent surgical resection and measurement of ctDNA both before and after surgery. Patients were divided into three groups on the basis of the presence or absence of ctDNA at these time points. The primary outcome was recurrence-free survival (RFS).
RESULTS
The search yielded 3,873 articles; five met the inclusion criteria and collectively evaluated 57 patients. Pooled median RFS was 62 months (interquartile range 19 to not reached). Although median RFS was not reached in group 1 (– to –) or group 2 (+ to –), median RFS in group 3 (+ to +) was 15 months (interquartile range 9.6-60.4 months). Cox hazard ratio was 4.46 (95% CI, 1.17 to 16.99; P = .028) between group 1 and group 2, and 10.47 (95% CI, 2.91 to 37.74; P < .001) between group 2 and group 3.
CONCLUSION
Detectable ctDNA, either preoperatively or postoperatively, and its persistence after curative surgery are associated with a greater risk of recurrence and decreased RFS in GI cancers. Thus, perioperative measurement of ctDNA may be a useful postoperative risk stratification tool and guide additional therapies.
INTRODUCTION
GI malignancies are among the most common cancers, contributing to around 30% of the global cancer burden annually. These malignancies are aggressive and contribute to approximately 40% of global cancer deaths,1 with surgical resection remaining a cornerstone in clinical management. The postsurgical surveillance for these patients currently relies upon imaging modalities and/or serum tumor markers such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9). Despite advances in understanding molecular biology, therapeutic approaches, and post-treatment monitoring strategies, many patients relapse within 5 years of resection with curative-intent.2,3 Recurrence, both locoregional and distant, is associated with poor quality of life and dismal survival.4-6 Recurrence is also known to be dependent on various tumor-specific factors (type, stage, grade, and mutational status) and treatment-related factors such as completeness of resection; however, many drivers of this process remain unknown.7,8 Current imaging modalities and serum biomarkers used during post-treatment surveillance to detect recurrence after resection lack sensitivity and have not been shown to improve survival in randomized controlled trials.9
CONTEXT
Key Objective
What is the current role of perioperative circulating tumor DNA (ctDNA) evaluation in informing prognosis in patients undergoing curative-intent surgery for GI malignancies?
Knowledge Generated
This patient-level meta-analysis demonstrated that the perioperative evaluation of ctDNA levels is a useful risk stratification tool for GI cancers undergoing surgery. As compared to those with negative ctDNA, recurrence-free survival was significantly worse in patients with positive ctDNA (hazard ratio was 4.46 [95% CI, 1.17 to 16.99; P = .028] for those who became negative after surgery and 10.47 [95% CI, 2.91 to 37.74; P < .001] for those who stayed positive after surgery).
Relevance
Measurement of ctDNA appears to be promising in risk stratification of patients undergoing curative-intent surgery for GI cancers. Further research will elucidate whether this can be used to guide therapy decisions (such as neoadjuvant and adjuvant strategies) and in the early detection of recurrence.
Liquid biopsy is emerging as a promising tool in cancer diagnostics and has opened various avenues in the field of personalized medicine. Two types of liquid biopsies have been developed—circulating tumor cells (CTC) and circulating tumor DNA (ctDNA)—which have been exploited for their clinical validity for early cancer detection, improved staging, risk stratification, detection of new therapeutic targets, and monitoring response to therapy.10-13 One potential application of these biomarkers in early-stage cancers is the ability to predict disease recurrence after oncologic resection. This is based on the hypothesis that some patients with early-stage cancers have occult minimal residual disease after completing treatment, which is then responsible for relapse after initial therapy.12 Recent research suggests that the detection of CTCs, ctDNA, or cell-free DNA (cfDNA) after initial treatment in GI cancers can predict recurrence earlier than standard surveillance imaging and tumor markers, serving as a prognostic biomarker. Lead time in predicting disease may provide a window of opportunity that allows early application of salvage/alternative treatments. Whether such an approach will improve patient outcomes is being actively investigated. Most of the published studies are retrospective and have analyzed ctDNA in a small cohort of patients, thus providing limited evidence on the clinical validity of using ctDNA analysis as a biomarker to guide adjuvant therapy and predict therapeutic benefit.
We therefore performed a systematic review of the literature with patient-level meta-analysis to critically evaluate and quantitatively summarize the current evidence regarding the clinical validity of sequential cfDNA or ctDNA (herein referred to collectively as ctDNA) measurements before and after curative resection in informing the recurrence of GI cancers.
METHODS
The review protocol was registered with the International Prospective Register of Systematic Reviews (registration number: CRD42020186096). The report was prepared in accordance with the Meta-analysis Of Observational Studies in Epidemiology checklist.14
Search Strategy
Multiple iterations were used to develop a search strategy, which was applied to systematically search the PubMed database for articles measuring levels of CTCs or ctDNA before and after curative-intent surgery for GI cancers. Search terms are listed in the Data Supplement. Full details of the search strategy are available online.15
Study Selection
Two reviewers with extensive collective experience in molecular biology (P.J.H. and K.S.) independently screened titles and abstracts against a predefined inclusion and exclusion criteria. All available studies were screened. Manual reviews of the reference lists of the retrieved articles were performed to identify potentially useful articles. Full texts were then screened by two reviewers (P.J.H. and K.S.) independently, and disagreements were resolved by discussion with the senior author (K.K.T.). Studies were included on the basis of inclusion criteria defined as clinical trials, prospective cohort studies, and retrospective studies published in the English language between January 2010 and May 2020 involving adults age older than 18 years with GI cancer treated with curative-intent surgery. Subjects were required to have undergone perioperative measurement of CTCs, cfDNA, or ctDNA (both before and after surgery). Studies that did not involve both preoperative and postoperative measurement of either CTCs, cfDNA, or ctDNA were excluded. Primary gastric, pancreatic, ampullary, hepatobiliary, duodenal, jejunal, ileal, appendiceal, colorectal, anal, and mesothelial cancers were considered relevant to the scope of the review and included. Case reports, case series, nonhuman studies, and those not written in English were excluded.
Data Extraction and Quality Assessment
Data were extracted from the selected studies by two independent reviewers (P.J.H. and K.S.) using a predefined extraction form. The extraction form included the following data: the number of patients; demographics; type of primary cancer; liquid biopsy assay used (polymerase chain reaction [PCR], real-time PCR, next-generation sequencing [NGS], or flow cytometry), liquid biopsy type (CTCs, cfDNA, or ctDNA); time of sampling (preoperative or postoperative); presence and absence of CTCs, cfDNA, or ctDNA; change in the levels of CTCs, cfDNA, or ctDNA; time to recurrence; and survival data. The data about the race and ethnicity of patients could not be included because of nonavailability in primary studies. The primary outcome of interest was recurrence-free survival (RFS) using the median time to event as an effect measure with a 95% CI. Cox hazard ratios were calculated for between-group comparisons.
The quality of included studies was assessed using the Newcastle-Ottawa scale (NOS) for cohort and case-control studies as well as a predefined assessment form (Data Supplement).16 The NOS is a validated quality tool that scores from 0 to 9 with higher scores corresponding to a better quality of study. The predefined assessment form included assessment for the representativeness of patient population, sources of bias, description of ctDNA analysis methods and outcomes, and statistical quality and interpretation. In addition, scores were allotted for individual domains and a cumulative score was calculated. The scores ranged from 0 to 20, with higher scores corresponding to a better quality of the study.
Qualitative Synthesis
A descriptive qualitative synthesis of the results was undertaken for all the included studies in the systematic review. We assessed each patient individually, ensuring that all patients underwent surgery, with curative, rather than palliative, intent. Additionally, patients who had undergone curative interventions with modalities other than surgery such as radiofrequency ablation were excluded. Data extracted included ctDNA levels, RFS, and biomarker assessed (cfDNA v ctDNA v CTCs) as well as which scientific methods were used to measure the ctDNA levels.
Meta-Analysis
After relevant data extraction, patients were categorized into three groups on the basis of the status of ctDNA presurgery and postsurgery, as described below. Patients had either (1) negative ctDNA levels both before and after surgery, (2) positive ctDNA levels before surgery and negative after surgery, or (3) positive ctDNA levels both before and after surgery. We chose these groups on the basis of the most common patterns of ctDNA levels before and after surgery. This list is not exhaustive of all theoretical possibilities, such as having negative ctDNA levels before surgery and positive after surgery, but no patients in our included studies fit this category. In the real world, the biomarkers for such low-risk patients with postoperative recurrence might be urgently needed.
Because of the heterogeneity of the studies and our decision to categorize the patients into three groups, we performed a patient-level meta-analysis of the data. Statistical calculations were performed using RStudio software, version 1.1.414 (RStudio Inc, 2009-2018, Boston, MA). Recurrence data were approximated into recurrence rates using Kaplan-Meier nonparametric analyses.
RESULTS
Included Studies
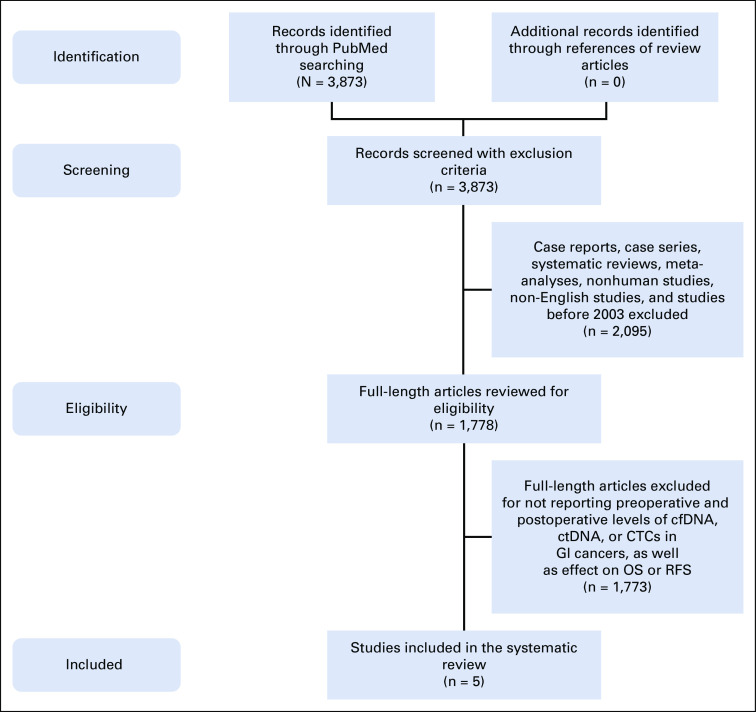
The search strategy retrieved 3,873 articles out of which 1,778 articles met inclusion criteria and were reviewed for eligibility. The large majority of studies were excluded because of the lack of both preoperative and postoperative ctDNA measurements. Five studies were ultimately included (Fig 1).
FIG 1.
Flowchart depicting the search strategy used for selecting studies for the systematic review.17 cfDNA, cell-free DNA; CTC, circulating tumor cells; ctDNA, circulating tumor DNA; OS, overall survival; RFS, recurrence-free survival.
Quality Assessment and Descriptive Results
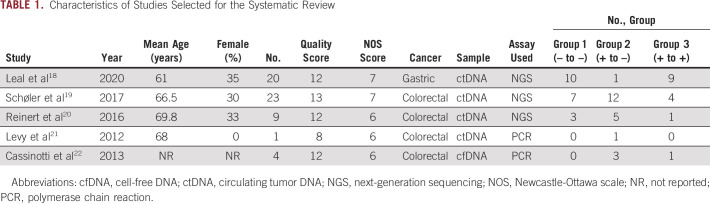
Included studies had NOS scores between six and seven, and quality scores ranging from 8 to 13 on the basis of our predefined quality assessment form (Table 1).18-22 The five studies comprised 57 patients who underwent measurement of ctDNA both before and after surgical resection of GI cancers (20 gastric and 37 colorectal). The median age for the entire patient population was 65 years (range, 30-82 years), and 32% of patients were female. Median follow-up time was 36 months (interquartile range, 19-44.5 months). Four studies measured levels of ctDNA and one study measured levels of cfDNA. No studies assessed CTC levels.
TABLE 1.
Characteristics of Studies Selected for the Systematic Review
Qualitative Synthesis
All the included studies were nonrandomized, observational studies. The characteristics of the selected studies are described in Table 1.18-22 Three of the studies (Reinert et al, Levy et al, and Schøler et al) included patients treated with surgery alone as well as those receiving both surgery and chemotherapy.19-21 Because we performed a patient-level meta-analysis, we were able to exclude patients treated with both surgery and chemotherapy from our study. The patient cohorts in Schøler et al and Reinert et al were from the similar study group; we carefully looked for the duplicates and excluded them from the final analysis. The study by Levy et al measured ctDNA levels of seven patients, but only one was treated with curative surgery alone who was then included in our patient-level meta-analysis. One study by Cassinotti et al measured cfDNA levels in 221 patients, but reported cfDNA levels of only four patients, without reporting cfDNA levels of the other 217.22 There was substantial heterogeneity among studies in terms of the techniques used to measure ctDNA levels. The three larger studies, encompassing 52 of 57 participants, used NGS to measure ctDNA levels,18-20 whereas the remaining two measured ctDNA using PCR and real-time PCR.21,22
Patient Categories
Patients were categorized into three groups (Table 1):
Group 1 (– to –): Negative ctDNA levels before and after surgery (n = 20, median age 68 years, 30% female)
Group 2 (+ to –): Positive ctDNA levels before surgery but negative levels after surgery (n = 22, median age 65 years, 26% female)
Group 3 (+ to +): Positive ctDNA levels both before and after surgery (n = 15, median age 62 years, 43% female).
Recurrence-Free Survival
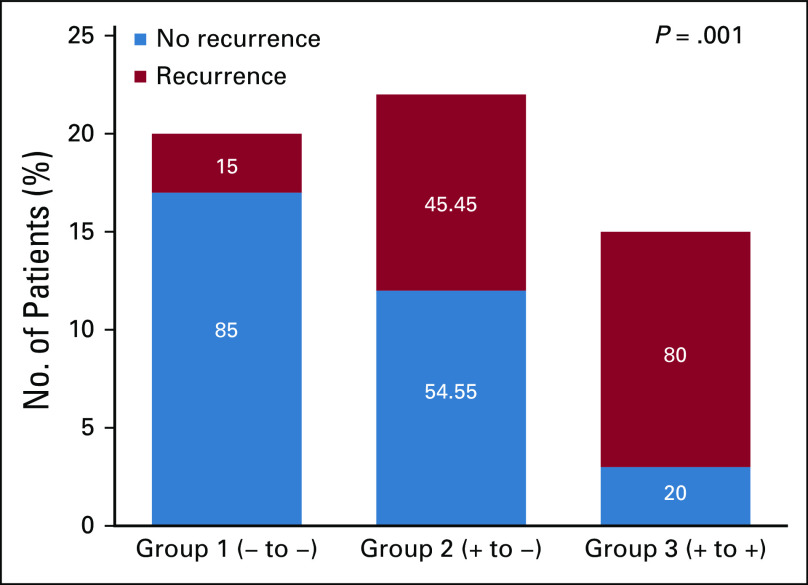
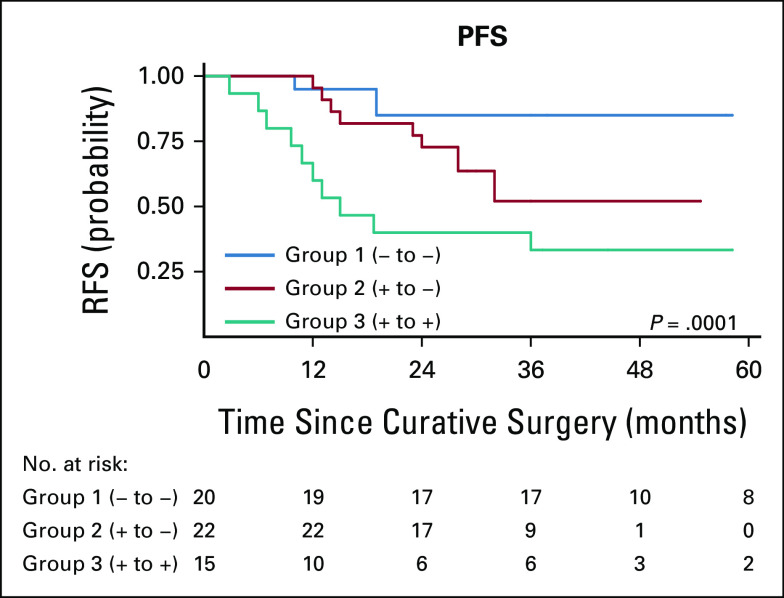
The median RFS was 62 months (25th quartile 19 months; 75th quartile was not reached because of censoring). Eighty-five percent (17/20) and 54.4% (12/22) of patients in groups 1 and 2, respectively, did not recur during the study period. By contrast, only 20% (3/15) of patients in group 3 had no recurrence throughout the length of the studies (Fig 2). Similarly, median RFS was not reached in group 1 and group 2 because of censoring (25th percentiles of 20 and 22 months, respectively), whereas median RFS in group 3 was 15 months (interquartile range, 9.6-60.4 months). Cox hazard ratio for recurrence between group 1 and group 2 was 4.46 (95% CI, 1.17 to 16.99; P = .028), and 10.47 (95% CI, 2.91 to 37.74; P < .001) between group 2 and group 3, demonstrating decreased RFS with increased ctDNA before or after surgery. Estimates of RFS were approximated into recurrence rates and used for plotting recurrence curves for different groups as shown in Figure 3. Group 1 had superior RFS relative to group 2, which was superior to group 3 (P = .001).
FIG 2.
Recurrence rates of cancer by patient group. Increased perioperative levels of circulating tumor DNA negatively correlate with recurrence-free survival (P = .001).
FIG 3.
Kaplan-Meier estimates for the RFS. Increased perioperative levels of circulating tumor DNA negatively correlate with RFS (log-rank P value = .0001). RFS, recurrence-free survival.
Subgroup Analysis
To determine whether cancer type affected RFS on the basis of grouping by ctDNA levels, a subgroup analysis by primary cancer type was performed, which showed similar trends in RFS in both patients with gastric (n = 20, log-rank P = .0023) and colorectal cancer (n = 37, log-rank P = .026; Fig 4).
FIG 4.
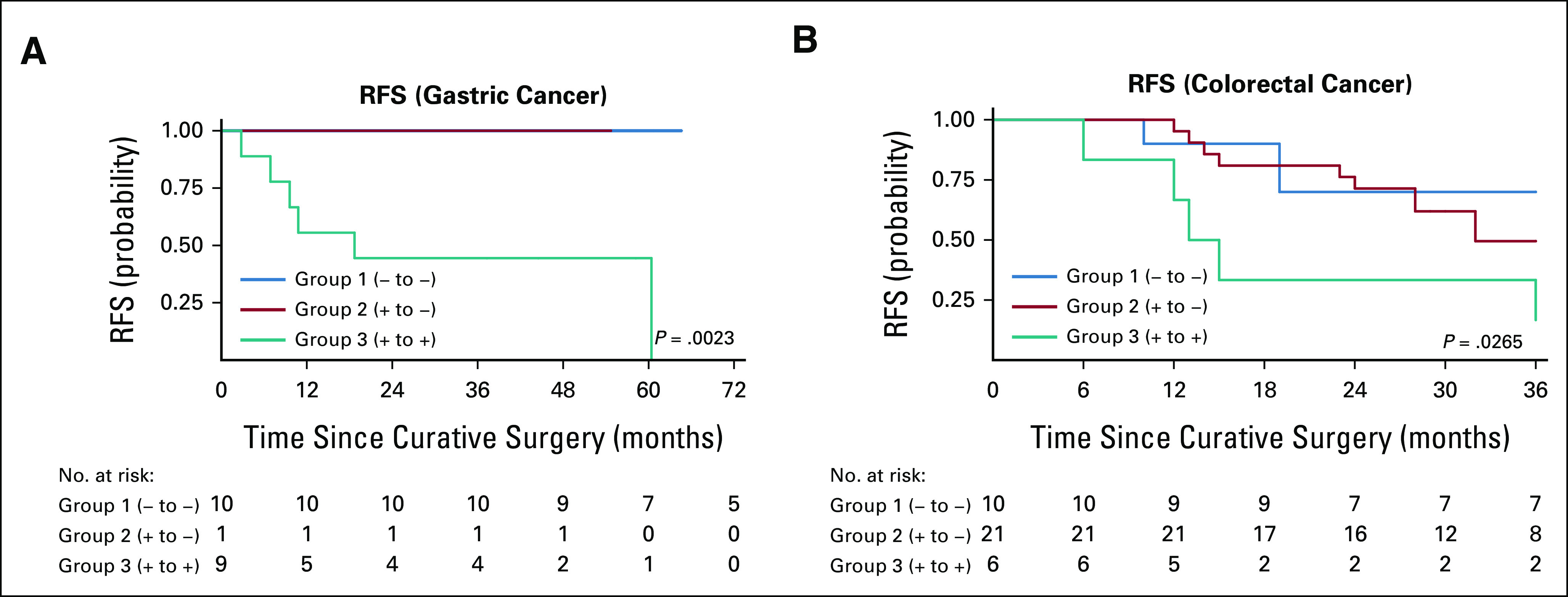
Kaplan-Meier estimates describing increased perioperative levels of cell-free DNA or circulating tumor DNA negatively correlate with RFS when patients with (A) gastric cancer (n = 20, log-rank P = .0023) are separated from patients with (B) colorectal cancer (n = 37, log-rank P = .0265). RFS, recurrence-free survival.
DISCUSSION
Detection of ctDNA is emerging as a potentially valuable method for post-treatment risk stratification, which may help guide subsequent clinical decisions and therapy. Several previous proof-of-principle studies have shown that detection of ctDNA after treatment (surgery or radical radiotherapy) is correlated with increased risk of relapse in multiple cancers, including gastric,23 colorectal,24,25 and pancreatic cancer.26 This systematic review and patient-level meta-analysis focuses on studies assessing the perioperative change in ctDNA levels for individual patients between samples collected preoperatively and postoperatively and further supports the evidence that perioperative ctDNA detection may predict early recurrence. Furthermore, a lack of clearance of detectable ctDNA after surgery may confer the highest risk of recurrence. Thus, similar to the usage of traditional biomarkers such as CEA and carbohydrate antigen 125 (CA-125), the serial measurement of ctDNA levels also has potential for pertinent clinical utility.
Although the use of ctDNA for cancer risk stratification is still in its early stages, there is evidence that it may help inform the initiation of targeted therapy options. As a proof of concept, PCR-based methods analyzing RAS mutational status in patients with colorectal cancer have shown high concordance with tissue mutational status in the metastatic setting and are US Food and Drug Administration–approved to determine the eligibility of antiepidermal growth factor receptor therapy.27,28 Similarly, for other cancers, analyzing the mutational and epigenetic profiles of circulating DNA may inform tumor sensitivity or resistance to available therapies. Analysis of changes in ctDNA during therapy may also guide the adoption of alternative therapeutic options. Perhaps most importantly, early detection of changes in ctDNA, especially ctDNA during post-treatment surveillance, may indicate recurrence of radiographically occult micrometastatic disease and may potentially guide early treatment, with an intention of improving survival outcomes. In some instances, such as patients with high risk of peritoneal disease, such detection may justify further diagnostic testing such as a laparoscopy. However, until the therapeutic implications of early detection of disease are clarified via clinical trials, the use of ctDNA in management decisions needs to be carefully assessed by the physician. The psychologic impact of positive tests is not inconsequential for the patient and their caregivers. False-positive results of ctDNA can also substantially affect the well-being of patients. Active (ClinicalTrials.gov identifier: NCT03803553, NCT04680260, NCT04920032) and future clinical trial results will hopefully shed light on these remaining questions.
Currently, ctDNA assays are not widely accepted into routine clinical practice. A 2018 review by ASCO and the College of American Pathologists concluded that several questions must first be answered to validate their clinical utility.11 First, a prospective registry study should be conducted in a patient cohort matching the intended-use population, as all studies before this point have retrospective design. Moreover, studies must conduct imaging upon detection of increased levels of these liquid biopsy markers. Currently, the BESPOKE study (ClinicalTrials.gov identifier: NCT04264702) has begun enrolling patients to prospectively study the role of ctDNA in determining the rate of recurrence of patients with stage II and III colorectal cancer. Additional pragmatic challenges include establishing the most clinically relevant method to standardize the quantitative assessment of ctDNA among laboratories to enable direct comparability.
ctDNA is most often detected using NGS or PCR to identify a known set of genetic mutations. The sensitivity and specificity of these techniques are limited, as mutations are infrequent in a given tumor and are not disease-specific,11,29 which has led to interest in the utility of epigenetic modifications in detecting tumor-specific mutations in the circulating DNA. Notably, recent studies have demonstrated distinct signatures of 5-hydroxymethylcytosine in several GI cancers.29-32 Detection of these signatures provides several theoretical advantages over traditional techniques identifying gene mutations. Epigenetic changes may be present more frequently than gene mutations and therefore allow greater sensitivity. Epigenetic signatures may also be informative of the site of metastasis, response to treatment, and the risk of recurrence. Moreover, it is possible that tumor growth may damage surrounding normal tissue, causing release of normal tissue cfDNA. As techniques mapping epigenetic profiles improve, it may be possible to detect abnormalities in levels of both ctDNA or normal tissue cfDNA indicating early malignancy (Fig 5).
FIG 5.
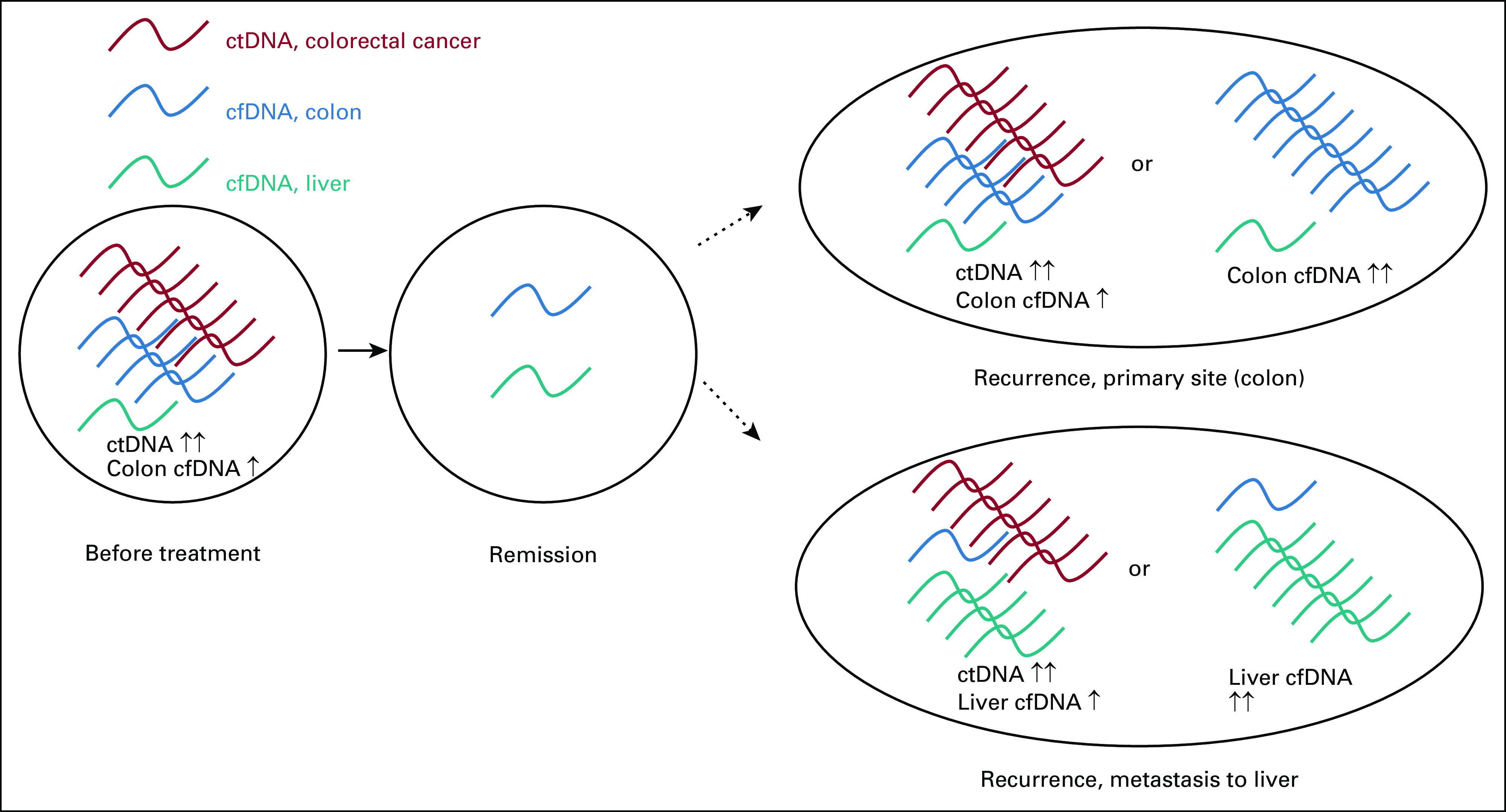
Hypothesized model of utility of epigenetic signatures to detect recurrence of cancers, that is, colorectal cancer. Before treatment, colorectal cancer ctDNA (red) is detectable in the blood. Elevated levels of normal tissue cfDNA from the colon (blue) may also be detectable because of damage from malignancy. After treatment, ctDNA is no longer detectable, and normal tissue cfDNA levels return to normal. Cancer recurrence at the primary site (colon) may be marked by increased colorectal cancer ctDNA and/or increased normal colon tissue cfDNA. Cancer recurrence at distant metastatic sites, such as the liver, may be marked by increased colorectal cancer ctDNA and/or increased normal liver tissue cfDNA (teal). cfDNA, cell-free DNA; ctDNA, circulating tumor DNA.
The major limitation of the present meta-analysis is the heterogeneity of the included studies. Three of the studies included patients treated with surgery alone as well as patients treated with both surgery and chemotherapy. Also, techniques of assessment of ctDNA differed. The data regarding genes used for the ctDNA analysis could not be compared. Furthermore, the combined sample size is small to ascertain the utility of ctDNA as a predictive biomarker. Additionally, the diagnostic performance of ctDNA was not compared directly to the existing tumor markers. Also, the clinical stage was not available for all patients, and the pathologic stage available was heterogeneous and missing for 8.7% of patients, making stage-wise comparisons difficult. Finally, the observational nature of all included studies limits the strength of their findings. Validation of these findings should be confirmed with a multi-institutional, prospective registry study.
In conclusion, detectable ctDNA, either preoperatively or postoperatively, and persistence after curative resection are associated with a greater risk of recurrence and decreased RFS. Thus, perioperative evaluation of ctDNA levels may be useful as a postoperative risk stratification tool. Future clinical trials assessing the utility of postresection ctDNA measurements to guide additional treatment decisions for GI malignancies should be encouraged.
Chuan He
Stock and Other Ownership Interests: Accent Therapeutics, Shanghai Epican Genetech
Consulting or Advisory Role: Accent Therapeutics
Patents, Royalties, Other Intellectual Property: Wisegene licensed TAB-seq from the University of Chicago
Oliver S. Eng
Stock and Other Ownership Interests: Iovance Biotherapeutics, Zymeworks, Alpine Immune Sciences, Aileron Therapeutics
Daniel V. T. Catenacci
Honoraria: Genentech/Roche, Lilly, Amgen, Foundation Medicine, Taiho Pharmaceutical, Guardant Health, Merck, Bristol Myers Squibb, Gritstone Oncology, Five Prime Therapeutics, Astellas Pharma, Seattle Genetics, Tempus, Pieris Pharmaceuticals, Daiichi Sankyo/UCB Japan, Zymeworks, QED Therapeutics, Natera, Archer, Novartis
Consulting or Advisory Role: Genentech/Roche, Amgen, Merck, Lilly, Taiho Pharmaceutical, Bristol Myers Squibb, Astellas Pharma, Seattle Genetics, Daiichi Sankyo/UCB Japan, Zymeworks, Guardant Health
Speakers' Bureau: Guardant Health, Genentech, Lilly, Merck, Tempus, Daiichi Sankyo/Astra Zeneca
No other potential conflicts of interest were reported.
SUPPORT
P.J.H. is supported by NIH Medical Scientist National Research Service Award (T32GM007281). A.D. and H.D.D.W. are supported by the Irving Harris Foundation Grant.
AUTHOR CONTRIBUTIONS
Conception and design: Ankit Dhiman, Hunter D. D. Witmer, Chuan He, Daniel V. T. Catenacci, Kiran K. Turaga
Administrative support: Kiran Turaga
Provision of study materials or patients: Kiran Turaga
Collection and assembly of data: Phillip J. Hsu, Khushboo Singh, Ankit Dhiman, Daniel V. T. Catenacci, Kiran K. Turaga
Data analysis and interpretation: Phillip J. Hsu, Ankit Dhiman, Hunter D. D. Witmer, Oliver S. Eng, Daniel V. T. Catenacci, Mitchell C. Posner
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Chuan He
Stock and Other Ownership Interests: Accent Therapeutics, Shanghai Epican Genetech
Consulting or Advisory Role: Accent Therapeutics
Patents, Royalties, Other Intellectual Property: Wisegene licensed TAB-seq from the University of Chicago
Oliver S. Eng
Stock and Other Ownership Interests: Iovance Biotherapeutics, Zymeworks, Alpine Immune Sciences, Aileron Therapeutics
Daniel V. T. Catenacci
Honoraria: Genentech/Roche, Lilly, Amgen, Foundation Medicine, Taiho Pharmaceutical, Guardant Health, Merck, Bristol Myers Squibb, Gritstone Oncology, Five Prime Therapeutics, Astellas Pharma, Seattle Genetics, Tempus, Pieris Pharmaceuticals, Daiichi Sankyo/UCB Japan, Zymeworks, QED Therapeutics, Natera, Archer, Novartis
Consulting or Advisory Role: Genentech/Roche, Amgen, Merck, Lilly, Taiho Pharmaceutical, Bristol Myers Squibb, Astellas Pharma, Seattle Genetics, Daiichi Sankyo/UCB Japan, Zymeworks, Guardant Health
Speakers' Bureau: Guardant Health, Genentech, Lilly, Merck, Tempus, Daiichi Sankyo/Astra Zeneca
No other potential conflicts of interest were reported.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. : Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209-249, 2021 [DOI] [PubMed] [Google Scholar]
- 2.Guraya SY: Pattern, stage, and time of recurrent colorectal cancer after curative surgery. Clin Colorectal Cancer 18:e223-e228, 2019 [DOI] [PubMed] [Google Scholar]
- 3.Spolverato G, Ejaz A, Kim Y, et al. : Rates and patterns of recurrence after curative intent resection for gastric cancer: A United States multi-institutional analysis. J Am Coll Surg 219:664-675, 2014 [DOI] [PubMed] [Google Scholar]
- 4.Shariat-Madar B, Jayakrishnan TT, Gamblin TC, et al. : Surgical management of bowel obstruction in patients with peritoneal carcinomatosis. J Surg Oncol 110:666-669, 2014 [DOI] [PubMed] [Google Scholar]
- 5.Olson TJP, Pinkerton C, Brasel KJ, et al. : Palliative surgery for malignant bowel obstruction from carcinomatosis a systematic review. JAMA Surg 149:383-392, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klaver YLB, Lemmens VEPP, Creemers GJ, et al. : Population-based survival of patients with peritoneal carcinomatosis from colorectal origin in the era of increasing use of palliative chemotherapy. Ann Oncol 22:2250-2256, 2011 [DOI] [PubMed] [Google Scholar]
- 7.Elias D, Honoré C, Dumont F, et al. : Results of systematic second-look surgery plus hipec in asymptomatic patients presenting a high risk of developing colorectal peritoneal carcinomatosis. Ann Surg 254:289-293, 2011 [DOI] [PubMed] [Google Scholar]
- 8.Enblad M, Graf W, Birgisson H: Risk factors for appendiceal and colorectal peritoneal metastases. Eur J Surg Oncol 44:997-1005, 2018 [DOI] [PubMed] [Google Scholar]
- 9.Goéré D, Malka D, Tzanis D, et al. : Is there a possibility of a cure in patients with colorectal peritoneal carcinomatosis amenable to complete cytoreductive surgery and intraperitoneal chemotherapy? Ann Surg 257:1065-1071, 2013 [DOI] [PubMed] [Google Scholar]
- 10.Merker JD, Oxnard GR, Compton C, et al. : Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol 36:1631-1641, 2018 [DOI] [PubMed] [Google Scholar]
- 11.Weiser DA, West-Szymanski DC, Fraint E, et al. : Progress toward liquid biopsies in pediatric solid tumors. Cancer Metastasis Rev 38:553-571, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pantel K, Alix-Panabières C: Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat Rev Clin Oncol 16:409-424, 2019 [DOI] [PubMed] [Google Scholar]
- 13.Donaldson J, Park BH: Circulating tumor DNA: Measurement and clinical utility. Annu Rev Med 69:223-234, 2018 [DOI] [PubMed] [Google Scholar]
- 14.Stroup DF, Berlin JA, Morton SC, et al. : Meta-analysis of observational studies in epidemiology. JAMA 283:2008-2012, 2000 [DOI] [PubMed] [Google Scholar]
- 15.Hsu P, Turaga K, Singh K: Utility of circulating tumor cells, circulating tumor DNA, and cell free DNA in informing prognosis of gastrointestinal cancers. PROSPERO 2020 CRD42020186096. https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=186096
- 16.Deeks JJ, Dinnes J, D'Amico R, et al. : Evaluating non-randomised intervention studies. Health Technol Assess 7:1-173, 2003 [DOI] [PubMed] [Google Scholar]
- 17.Moher D, Liberati A, Tetzlaff J, et al. : Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med 6:e1000097, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leal A, van Grieken NCT, Palsgrove DN, et al. : White blood cell and cell-free DNA analyses for detection of residual disease in gastric cancer. Nat Commun 11:525, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schøler LV, Reinert T, Ørntoft MBW, et al. : Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res 23:5437-5445, 2017 [DOI] [PubMed] [Google Scholar]
- 20.Reinert T, Schøler LV, Thomsen R, et al. : Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 65:625-634, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Levy M, Benesova L, Lipska L, et al. : Utility of cell-free tumour DNA for post-surgical follow-up of colorectal cancer patients. Anticancer Res 32:1621-1626, 2012 [PubMed] [Google Scholar]
- 22.Cassinotti E, Boni L, Segato S, et al. : Free circulating DNA as a biomarker of colorectal cancer. Int J Surg 11:S54-S57, 2013. (suppl 1) [DOI] [PubMed] [Google Scholar]
- 23.Maron SB, Chase LM, Lomnicki S, et al. : Circulating tumor DNA sequencing analysis of gastroesophageal adenocarcinoma. Clin Cancer Res 25:7098-7112, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diehl F, Schmidt K, Choti MA, et al. : Circulating mutant DNA to assess tumor dynamics. Nat Med 14:985-990, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tie J, Wang Y, Tomasetti C, et al. : Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8:346ra92, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietrasz D, Pécuchet N, Garlan F, et al. : Plasma circulating tumor DNA in pancreatic cancer patients is a prognostic marker. Clin Cancer Res 23:116-123, 2017 [DOI] [PubMed] [Google Scholar]
- 27.Vidal J, Muinelo L, Dalmases A, et al. : Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 28:1325-1332, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmiegel W, Scott RJ, Dooley S, et al. : Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: Concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol 11:208-219, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian X, Sun B, Chen C, et al. : Circulating tumor DNA 5-hydroxymethylcytosine as a novel diagnostic biomarker for esophageal cancer. Cell Res 28:597-600, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Han X, Gao C, et al. : 5-Hydroxymethylome in circulating cell-free DNA as a potential biomarker for non-small-cell lung cancer. Genomics Proteomics Bioinformatics 16:187-199, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song CX, Yin S, Ma L, et al. : 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res 27:1231-1242, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Zhang X, Lu X, et al. : 5-Hydroxymethylcytosine signatures in circulating cell-free DNA as diagnostic biomarkers for human cancers. Cell Res 27:1243-1257, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]