Abstract
Accumulating data have brought the nonnucleoside reverse transcriptase (RT) inhibitors (NNRTIs) into the forefront of antiretroviral therapy. Among the emerging compounds in this class, a particularly attractive one is efavirenz (Sustiva), recently approved for clinical use by the U.S. Food and Drug Administration. In the present study, the equilibrium dissociation constants for efavirenz binding to the different catalytic forms of human immunodeficiency virus type 1 RT as well as the association and dissociation rates have been determined using a steady-state kinetic approach. In addition, the same enzymological analysis has been extended to the thio-substituted analog, sefavirenz, which showed comparable activity in vitro against RT. Both compounds have been found to act as purely uncompetitive inhibitors at low drug concentrations (5 to 50 nM) and as mixed noncompetitive inhibitors at higher doses (50 to 500 nM). This behavior can be interpreted in terms of the relative affinities for the different catalytic forms of the enzyme. Both efavirenz and sefavirenz showed increasing affinities for the different forms of RT in the following order: free enzyme < (i.e., bound with lower affinity) binary RT–template-primer (TP) complex < ternary RT-TP-deoxynucleoside triphosphate (dNTP) complex. The rate of binding of the two inhibitors to the different enzyme-substrate complexes was well below the diffusion limit (on the order of 104 M−1 s−1); however, both inhibitors, when bound to the ternary RT-TP-dNTP complex, showed very low dissociation rates, on the order of 10−4 s−1 for both compounds, typical of tightly binding inhibitors. Thus, efavirenz and its thio-substituted derivative sefavirenz appear to be peculiar in their mechanism of action, being selective tightly binding inhibitors of the ternary RT-TP-dNTP complex. Efavirenz is the first clinically approved NNRTI to show this property.
The virus-encoded human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is essential for the viral replication cycle and therefore represents a logical target for antiviral chemotherapy (11, 15). Recently, a class of inhibitors targeted to the viral RT, the so-called nonnucleoside RT inhibitors (NNRTIs), have gained a definitive place in the treatment of HIV-1 infections along with nucleoside RT inhibitors (NRTIs) and protease inhibitors (PIs) (5). These compounds, in spite of their different chemical structures, are highly specific for HIV-1 RT and bind to the enzyme at the same allosteric site, close to but distinct from the catalytic site, behaving as typically noncompetitive inhibitors with respect to the different substrates of the polymerization reaction (7, 28, 30). There are currently three NNRTIs approved for clinical use, nevirapine (Viramune), delavirdine (Rescriptor), and the most recently licensed, efavirenz (Sustiva). When used in monotherapy regimens, NNRTIs have rapidly selected for resistance (19, 20, 21, 22, 26, 27, 30), but the results of clinical trials with NNRTIs as components of highly effective antiretroviral therapy regimens in combination with NRTIs and/or PIs have been impressive (http://www.medscape.com/Medscape/HIV/TreatmentUpdate /1999/tu04/tu04-03html). In general, NNRTIs often show synergistic (or at least additive) effects in combination with NRTIs, as well as positive pharmacokinetic properties. In contrast to NRTIs or PIs, NNRTIs are characterized by less severe adverse effects for patients (4). There now exists a large amount of data justifying the use of NNRTIs plus NRTIs as initial therapy as well as in the treatment of individuals who have very advanced disease or who have already failed multiple NRTI or NRTI-PI combination therapies.
Among the emerging compounds in this class, a particularly attractive one is efavirenz. Efavirenz had very promising results in clinical trials aimed at evaluating its effect in association with NRTIs, NNRTIs, and PIs under a variety of clinical scenarios. It was particularly effective both in treatment-experienced individuals switched to the new therapy and in salvage regimens for patients not responding to standard NRTI-PI combinations. Like the other NNRTIs, however, efavirenz also selects for genotypic drug resistance, in particular, for the K103N mutation in the drug-binding site of HIV-1 RT (1, 9, 32, 33). This mutation was also the most frequently observed in samples from patients experiencing postvirological treatment failure and was already known to confer cross-resistance to other NNRTIs (30). These observations highlight the need for extended-spectrum efavirenz derivatives that may be active against the K103N mutant. A detailed understanding of the mechanism of action of efavirenz is an obligatory step in developing new molecules with a better profile of activity against drug-resistant mutants.
In the present study, the equilibrium dissociation constants for efavirenz binding to the different catalytic forms of HIV-1 RT as well as the association and dissociation rates have been determined using a steady-state kinetic approach. In order to evaluate how minor conformational changes in the structure of efavirenz could affect its binding to HIV-1 RT, a derivative bearing an oxocarbonyl-thiocarbonyl substitution has been synthesized and called sefavirenz. While sefavirenz displayed comparable activity in in vitro RT assays, the results indicated that the compounds bound with different affinities to the various catalytic forms of the enzyme-substrate complex.
MATERIALS AND METHODS
Chemicals.
[3H]dTTP (40 Ci/mmol) was from Amersham, and unlabeled deoxynucleoside triphosphates (dNTPs) were from Boehringer. Whatman was the supplier of the GF/C filters. All other reagents were of analytical grade and were purchased from Merck or Fluka.
Synthesis of compounds.
Melting points were measured using a Kofler hot-stage apparatus and are uncorrected. 1H and 13C nuclear magnetic resonance (NMR) spectra were recorded in CDCl3 at 300 and 75.46 MHz, respectively, using a Bruker ACE-300 spectrometer. 1H chemical shifts (δ) were reported with Me4Si (δ = 0.00 ppm) as an internal standard. 13C chemical shifts (δ) were reported with CDCl3 (central peak, δ = 77.00 ppm) as an internal standard. The following abbreviations are used: br, broad; s, singlet; d, doublet; dd, double doublet; and m, multiplet. Mass spectra were obtained on a Finnigan MAT 8222 spectrometer via the direct inlet. Electron ionization was performed at 70 eV and 0.5 mA with a source temperature of 250°C. Elemental analyses indicated by the symbols were within ±0.4% of the theoretical values and were performed on a Carlo Erba 1106 Elemental Analyzer. All reactions were monitored by thin-layer chromatography carried out on 0.25-mm Merck silica gel (60 F254) and visualized by UV light (δ = 264 or 365 nm); flash chromatography was performed with silica gel 60 (60 to 200 μm; Merck). High-pressure liquid chromatography (HPLC) analyses were run on a Merck-Hitachi L-7100 instrument equipped with an L-7400 UV detector and an L-7300 column oven. The column was an RP 18 Lichrospher 100 5-μm column (Merck). The conditions were as follows: eluent, CH3CN-H2O (65:35); flow rate, 1.1 ml/min; UV detector wavelength, 247 nm; and temperature, 25°C.
(i) 6-Chloro-4-cyclopropylethynyl-4-trifluoromethyl-1,4-dihydro-2H-3,1-benzoxazin-2-one (efavirenz).
The product was synthesized as described by Tan et al. (29); the mp was 133 to 136°C (hexane-toluene). HPLC analysis: retention time (RT), 6.57 min. 1H NMR (CDCl3): 0.85 (m, 2H), 0.94 (m, 2H), 1.40 (m, 1H), 6.81 (d, J = 8.5 Hz, 1H), 7.37 (dd, J = 2.5, 8.5 Hz, 1H), 7.49 (d, J = 2.5 Hz, 1H), 8.71 (br s, 1H). 13C NMR: 148.0, 133.2, 131.6, 129.0, 127.8, 127.7, 123.9, 120.1, 116.3, 115.7, 95.8, 77.3, 77.1, 76.9, 76.5, 55.0, 8.9, −0.7. Mass spectrometry: m/z (ra [relative abundance] %): 315 (M+, 30), 248 (23), 246 (100), 243 (33), 182 (13), 180 (36), 167 (12). Analysis calculated for C14H9NO2ClF3: C, 53.27; H, 2.87; N, 4.44. Found: C, 52.90; H, 2.92; N, 4.77.
(ii) 6-Chloro-4-cyclopropylethynyl-4-trifluoromethyl-1,4-dihydro-2H-3,1-benzoxazin-2-thione (Sefavirenz).
To a solution of efavirenz (506 mg, 1.602 mmol) in 15 ml of anhydrous toluene, Lawesson's reagent (324 mg, 0.801 mmol) was added, and the mixture was refluxed for 3.5 h. After the mixture was cooled to room temperature, the solid residue was removed by filtration. The solvent was distilled under reduced pressure, and the residue was purified by flash chromatography (hexane-ethyl acetate, 75:25). Crystallization from hexane gave white crystals; the mp was 147 to 148°C. HPLC analysis: RT, 7.29 min. 1H NMR (CDCl3): 0.85 (m, 2H), 0.94 (m, 2H), 1.39 (m, 1H), 6.80 (d, J = 8.5 Hz, 1H), 7.49 (dd, J = 2.5, 8.5 Hz, 1H), 7.52 (d, J = 2.5 Hz, 1H), 9.33 (br s, 1H). 13C NMR: 180.1, 131.8, 130.6, 130.5, 127.8, 127.4, 123.6, 118.7, 115.9, 115.3, 96.8, 79.6, 79.2, 78.6, 78.2, 55.5, 8.8, −0.7. Mass spectrometry: m/z (ra %): 331 (M+, 70), 296 (19), 262 (100), 243 (75), 234 (13), 224 (25), 167 (29), 87 (24). Analysis calculated for C14H9NOSClF3: C, 50.6; H, 2.73; N, 4.22. Found: C, 50.86; H, 2.77; N, 4.14.
Molecular modeling.
The calculations and simulation were performed on an O2 R10000 SGI workstation by using the software modules Discover and Builder of the Biosym/MSI software package. The structure of nevirapine was obtained from the atomic coordinates of the crystal structure of the HIV-1 RT–nevirapine complex (protein data bank file RVO). The structure of efavirenz was modeled using the Biosym/MSI software package, further subjected to the steepest descendent minimization for 1,000 steps, and then minimized with conjugate gradients for 10,000 steps. The superimposition of efavirenz and nevirapine gave a root mean square deviation value in the aligned position of 0.095.
Nucleic acid substrates.
The homopolymer poly(rA) (Pharmacia) was mixed at weight ratios (in nucleotides) of 10:1 with the oligomer oligo(dT)12–18 (Pharmacia) in 20 mM Tris-HCl (pH 8.0) containing 20 mM KCl and 1 mM EDTA; the mixture was heated at 65°C for 5 min and then slowly cooled at room temperature.
Expression and purification of recombinant HIV-1 RT forms.
Recombinant RT was expressed and purified to >95% purity as described previously (20). It had a specific activity on poly(rA)-oligo(dT) (see below) of 75,670 U/mg; 1 U of DNA polymerase activity corresponds to the incorporation of 1 nmol of dNMP into acid-precipitable material in 60 min at 37°C.
HIV-1 RT RNA-dependent DNA polymerase activity assay.
RNA-dependent DNA polymerase activity was assayed as follows. A final volume of 25 μl contained buffer A (50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol, 0.2 mg of bovine serum albumin per ml, 4% glycerol), 10 mM MgCl2, 0.5 μg of poly(rA)-oligo(dT) (10:1) (0.3 μM 3′-OH ends), 10 μM [3H]dTTP (1 Ci/mmol), and 2 to 4 nM RT. Reaction mixtures were incubated for 10 min at 37°C. Aliquots (20 μl) were then spotted on GF/C glass fiber filters, which were immediately immersed in 5% ice-cold trichloroacetic acid. Filters were washed twice in 5% ice-cold trichloroacetic acid and once in ethanol for 5 min and dried, and acid-precipitable radioactivity was quantitated by scintillation counting.
Inhibition assays.
Inhibition assay reactions were performed under the conditions described for the HIV-1 RT RNA-dependent DNA polymerase activity assay. Incorporation of radioactive dTTP into poly(rA)-oligo(dT) at different concentrations of DNA or dNTPs was monitored in the presence of increasing amounts of inhibitor. Data were then plotted according to Dixon (6).
Kinetics of inhibitor binding.
HIV-1 RT (20 to 40 nM) was incubated for 2 min at 37°C in a final volume of 4 μl in the presence of buffer A, 10 mM MgCl2, and 100 nM 3′-OH ends (for the formation of the RT–template-primer [TP] complex) or in the same mixture complemented with 10 μM unlabeled dTTP (for the formation of the RT-TP-dNTP complex). The inhibitor to be tested was then added to a final volume of 5 μl, at a concentration at which [EI]/[E0] = 1 − {1/[(1 + [I])/Ki]} > 0.9, where [E0] is free enzyme at the beginning of the reaction and [EI] is the enzyme-inhibitor complex. Then, 145 μl of a mixture containing buffer A, 10 mM MgCl2, and 10 μM [3H]dTTP (5 Ci/mmol) was added at different time points. After an additional 10 min of incubation at 37°C, 50-μl aliquots were spotted on GF/C filters, and acid-precipitable radioactivity was measured as described for the HIV-1 RT RNA-dependent DNA polymerase activity assay. The quantity (vt/v0) representing the normalized difference between the amount of dTTP incorporated at the zero time point and at the different time points was then plotted against time.
Kinetic parameter calculation.
All values were calculated by non-least-squares computer fitting of the experimental data to the appropriate rate equations. Kd values for the various reaction intermediates (Fig. 1A) were calculated according to the equations for uncompetitive inhibition and for mixed noncompetitive inhibition, respectively (6):
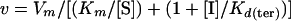 |
1 |
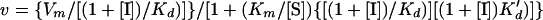 |
2 |
where Kd is Kd(bin) under the assay conditions specified in Fig. 1B, left panel, or Kd is Kd(E) under the conditions specified in Fig. 1B, right panel. Kd′ equalled Kd(ter) in both cases.
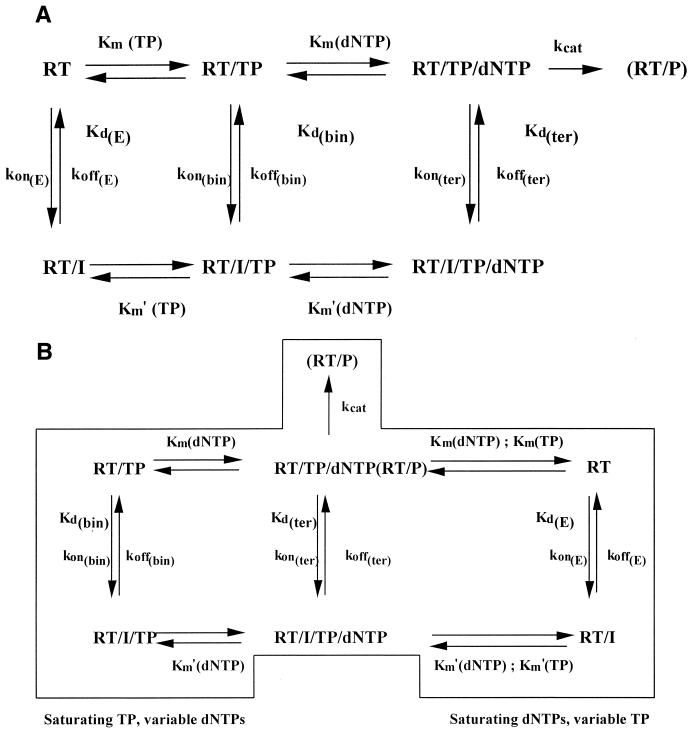
FIG. 1.
Simplified steady-state reaction scheme for interactions between HIV-1 RT and the inhibitors. (A) Steady-state reaction scheme for HIV-1 RT. I, inhibitor; Km, apparent Michaelis-Menten constant for the various enzyme-substrate complexes; Km′, apparent Michaelis-Menten constant for the various enzyme-substrate-inhibitor complexes; Kd(E), equilibrium dissociation constant for the RT-I complex; Kd(bin), equilibrium dissociation constant for the RT-I-TP complex; Kd(ter), equilibrium dissociation constant for the RT-I-TP-dNTP complex. kon and koff represent the binding and dissociation rates, respectively, for the corresponding equilibrium constants. (B) Simplified kinetic pathways for the inhibition of HIV-1 RT by efavirenz and sefavirenz, depending on various assay conditions. Abbreviations are as defined in panel A.
Apparent rate of binding (kapp) values were determined by fitting the experimental data to the single-exponent equation (vt/v0) = e−kappt, where t is time.
Determination of synergy.
Two approaches were used to determine synergy. The first was based on the median-effect method of Chou and Talalay as modified by Villahermosa et al. (31). Dose-response curves for the interaction between 3′-azido-2′, 3′-dideoxythymidine triphosphate (AZTTP) and efavirenz or sefavirenz were generated by fitting the experimental data to the equation
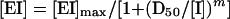 |
3 |
where [EI] is the fraction of inhibited enzyme, which was expressed as percent inhibition with respect to the control reaction without the inhibitor, and D50 was the concentration of inhibitor giving 50% inhibition. Accordingly, [EI]max was assumed to be 100% at infinite inhibitor concentrations. The parameter m is the sigmoidicity term.
The interaction index (I) was calculated according to the relationship I = (d1/D1) + (d2/D2), where d1 and d2 were the doses of the inhibitor giving 50% inhibition when tested in the combination (d1 + d2) and D1 and D2 are the D50 values of the corresponding inhibitor when tested alone. The I parameter is equivalent to the combin. index of Chou and Talalay. I of <1 indicates synergy, I of >1 indicates antagonism, and I of 1 indicates additivity, according to the mutually exclusive model for additivity of Chou and Talalay.
The second method was based on the Lowe additivity model as modified by Greco et al. (10). Dose-response curves for the interaction between AZTTP and efavirenz or sefavirenz were assumed to follow Hill's model and were generated by fitting the experimental data to the equation
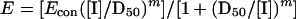 |
4 |
where E is the observed effect (percent activity), Econ is the control effect (activity in the absence of the inhibitor), and all the other parameters are as defined above. Effective inhibitor concentrations at different fractional inhibition levels were calculated from the parameters D50, [EI]con, and m according to the equation
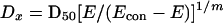 |
5 |
where Dx is the dose of drug giving a particular percent inhibition. I was then calculated according to Greco et al. (10) with the equation
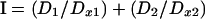 |
6 |
where D1 and D2 are the concentrations of the drugs in combination and Dx1 and Dx2 are the predicted inhibitory concentration of each drug giving the observed effect of the combination D1 + D2. I of <1 indicates synergy, I of >1 indicates antagonism, and I of 1 indicates additivity, according to the Lowe additivity model (10).
All the analyses were based on the results of three independent experiments for each drug combination, and the standard deviations (SD) for each parameter estimate are indicated.
RESULTS
Synthesis.
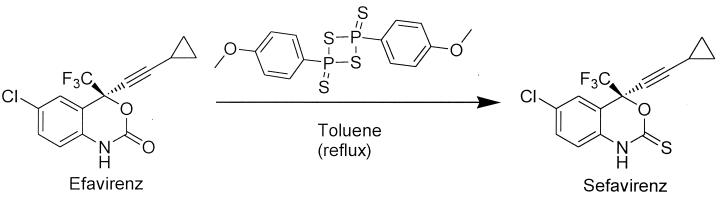
Efavirenz was synthesized as previously described (29). The thio-substituted analog sefavirenz was obtained by direct sulfuration (16) of efavirenz with Lawesson's reagent (Fig. 2). Although the sulfuration of carbamates has been reported (16), this is the first example of sulfuration of benzoxazin-2-ones with this reagent. The reaction yield is poor; however, after several recyclings of the unreacted product, the thio-substituted derivative can be obtained with a yield of >95%.
FIG. 2.
Drug structures.
Kinetic analysis.
A schematic reaction pathway for the inhibition of HIV-1 RT-catalyzed RNA-dependent DNA synthesis by efavirenz and/or sefavirenz is depicted in Fig. 1A. The data were analyzed according to the steady-state assumption that shortly after the initiation of the reaction, the enzyme-substrate complex is formed at the same rate as it dissociates. According to the ordered mechanism of the polymerization reaction, whereby TP binds first, followed by the addition of dNTP, the HIV-1 RT can be present in three different catalytic forms: as a free enzyme, in a binary complex with TP, and in a ternary complex with TP and dNTP (12, 17, 23, 24). Accordingly, it was assumed that the inhibitor could bind to any of these different forms and at the corresponding equilibria reported in Fig. 1A. The assay conditions used allowed processive synthesis by RT; thus, the complex of the enzyme with its products does not differ from the RT-TP complex, in the sense that the former shuttles back to the RT-TP state following incorporation and translocation along the template at a rate equal to the turnover number, kcat. The resulting rate equation for such a system is very complex and too impractical to be used. For these reasons, the general steady-state kinetic analysis was simplified by varying one of the substrates (either TP or dNTP) while the other was kept constant, as outlined in Fig. 1B. Because of the ordered mechanism of the two-substrate reaction catalyzed by HIV-1 RT, when the TP concentration was kept constant at a saturating level (50-fold over its Km) and inhibition was analyzed with various concentrations of dNTPs, at the steady state all the input RT was in the form of the RT-TP binary complex and only two forms of the enzyme (the binary complex and the ternary complex with dNTP) could react with the inhibitor, as shown in the left panel of Fig. 1B. Similarly, when the dNTP concentration was kept constant at a saturating level (fivefold over its Km) and inhibition was analyzed with various TP concentrations, RT was present either as a free enzyme or in the ternary complex with TP and dNTP, as shown in the right panel of Fig. 1B. Complex formation between RT and its substrates was assumed to occur with rapid equilibrium kinetics, so in the presence of saturating dNTP, conversion of the binary RT-TP complex into a ternary complex was assumed to occur at a much higher rate than inhibitor binding. Thus, in both cases, simple steady-state kinetic analysis could be used for the determination of the equilibrium dissociation constants of the different enzyme-inhibitor complexes.
Efavirenz and sefavirenz bind with different affinities to the binary RT-TP and ternary RT-TP-dNTP complexes.
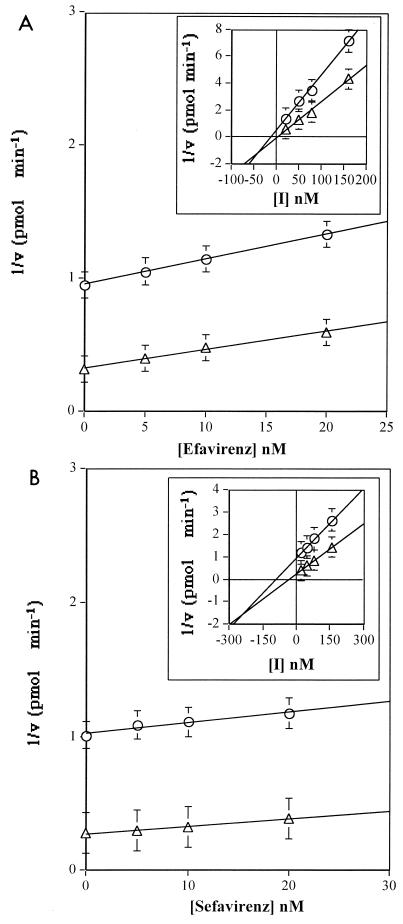
The effect of increasing concentrations of efavirenz on the RNA-dependent DNA synthesis catalyzed by HIV-1 RT on poly(rA)-oligo(dT) was tested with saturating TP and in the presence of two different dTTP concentrations. Under these conditions, only the binary RT-TP and the ternary RT-TP-dNTP complexes were available for inhibitor binding (Fig. 1B). The results are shown in Fig. 3A in the form of Dixon plots. When a range of concentrations of inhibitor from 5 to 180 nM was tested, the resulting inhibition displayed nonlinear kinetics, with a change in the slope of the curves. Inspection of the curves at low efavirenz concentrations (5 to 20 nM) showed a uncompetitive mechanism of inhibition. Fitting of the data to equation 1 for fully uncompetitive inhibition (see Materials and Methods) gave a value for the equilibrium dissociation constant for the ternary complex [Kd(ter)] of 4 nM, consistent with the reported Ki for efavirenz against HIV-1 RT. When the inhibitor was tested at higher concentrations, however, the resulting Dixon plot was diagnostic of a mixed noncompetitive mechanism of inhibition (Fig. 3A, inset). Fitting of the data to equation 2 gave a value for the equilibrium dissociation constant for the binary complex [Kd(bin)] of 30 nM. The two lines intersected below the x axis, according to the relationship Kd(bin) > Kd(ter). When sefavirenz was tested under the same conditions, a similar behavior was observed (Fig. 3B), with uncompetitive inhibition at low inhibitor concentrations and a calculated Kd(ter) value of 8 nM and mixed noncompetitive inhibition at higher concentrations (Fig. 3B, inset). The calculated Kd(bin) was 230 nM, indicating a reduced affinity of sefavirenz for the binary RT-TP complex with respect to efavirenz. Again, the two lines intersected below the x axis, according to the relationship Kd(bin) > Kd(ter). It should be noted that the observed mechanism is indicated with the term “uncompetitive” according to the standard nomenclature (6), in order to indicate the preferential binding of the inhibitor to the enzyme-substrate complex, a behavior which makes it distinct from other noncompetitive types of inhibition.
FIG. 3.
Inhibition of HIV-1 RT by efavirenz and sefavirenz, depending on variable dNTP concentrations. Reactions were carried out as described in Materials and Methods. (A) Dixon plot of RT inhibition by efavirenz in the presence of 2 μM dTTP (circles) or 10 μM dTTP (triangles) at inhibitor (I) concentrations of 5 to 25 nM or 25 to 160 nM (inset). (B) Dixon plot of RT inhibition by sefavirenz in the presence of 2 μM dTTP (circles) or 10 μM dTTP (triangles) at inhibitor concentrations of 5 to 20 nM or 20 to 160 nM (inset). Error bars show SD.
Efavirenz and sefavirenz show different affinities for binding to free RT.
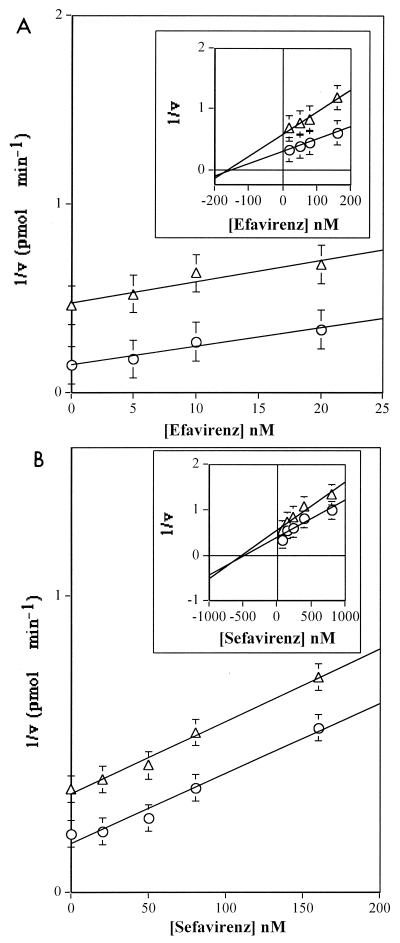
The effect of increasing concentrations of efavirenz on the RNA-dependent DNA synthesis catalyzed by HIV-1 RT with saturating dNTP was tested in the presence of two different TP (3′-OH primer ends) concentrations. According to Fig. 1B, right panel, the inhibitor could interact only with the free enzyme or the ternary RT-TP-dNTP complex. The results are shown in Fig. 4A. Dixon plots of the experimental data showed nonlinear kinetics, with uncompetitive inhibition at efavirenz concentrations of 5 to 20 nM (Fig. 4A) and mixed noncompetitive inhibition at higher concentrations (20 to 180 nM) (Fig. 4A, inset). The Kd(ter) value calculated according to the uncompetitive pathway was 4.5 nM, in good agreement with the value derived in the previous experiments (Fig. 3A). On the other hand, the equilibrium dissociation constant calculated for the free enzyme [Kd(E)] according to the mixed noncompetitive mechanism was 170 nM, indicating a poor affinity of efavirenz for the free enzyme. When sefavirenz was tested, a similar concentration dependence of the mechanism of inhibition was observed, with an uncompetitive Kd(ter) value of 7.5 nM (Fig. 4B). However, sefavirenz showed a significantly lower affinity for the free enzyme than efavirenz, with a Kd(E) derived according to the mixed noncompetitive mechanism of 750 nM. In both cases, the curves obtained at high concentrations of inhibitor intersected below the x axis, in accordance with the relationship Kd(E) > Kd(ter) (6).
FIG. 4.
Inhibition of HIV-1 RT by Efavirenz and Sefavirenz, depending on variable TP [poly(rA)-oligo(dT)] concentrations. Reactions were carried out as described in Materials and Methods. (A) Dixon plot of RT inhibition by efavirenz in the presence of 0.03 μM TP (as 3′-OH ends) (triangles) or 0.15 μM TP (circles) at inhibitor concentrations of 5 to 25 nM or 25 to 160 nM (inset). (B) Dixon plot of RT inhibition by sefavirenz in the presence of 0.03 μM TP (as 3′-OH ends) (triangles) or 0.15 μM TP (circles) at inhibitor concentrations of 20 to 160 nM or 80 to 800 nM (inset). Error bars show SD.
Determination of the rates for the formation and dissociation of the different RT-substrate-inhibitor complexes with efavirenz and sefavirenz.
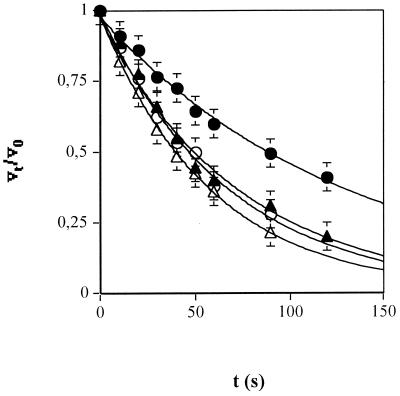
Both efavirenz and sefavirenz showed increasing affinities for the different catalytic forms of RT. However, given that Kd = koff/kon, the observed differences in the equilibrium dissociation constants could reflect various combinations of the association and dissociation rates kon and koff, respectively, for the different enzyme intermediates. In order to address this point, the apparent rate of binding (kapp) of the inhibitor to the binary RT-TP and ternary RT-TP-dNTP complexes was measured and koff an kon values were derived from the relationships kapp = kon(Kd + [I]) and Kd = koff/kon (20, 28). The experiment was done as described in Materials and Methods. Since the amount of dTTP incorporated at zero time (v0, uninhibited reaction) was proportional to the amount of RT-TP or RT-TP-dNTP present at the beginning of the reaction, v0 was also proportional to [E]0. The incorporation measured at subsequent time points (vt) was directly related to the amount of uninhibited enzyme ([E]t = [E]0 − [EI]t). Thus, since [E]0 decreased at a rate equal to the formation of the [EI] complex, the analysis of the dependence of vt/v0 on time allowed estimation of the kapp value.
Parallel reactions were run for 10 min at 37°C with enzyme, substrates, and inhibitors at the same concentrations as in the diluted mixture but without any preincubation. The incorporation observed in these control reactions was typically between 5 and 10% of v0 and was substracted as background. As shown in Fig. 5, the data points fitted the simple exponential relationship vt/v0 = e−kappt. The calculated kapp, kon, and koff values for efavirenz and sefavirenz are listed in Table 1, along with the corresponding Kd values. In both cases, formation of the RT-inhibitor-TP complex was characterized by lower association and higher dissociation rates with respect to the RT-inhibitor-TP-dNTP complex. Similar experiments were performed with nevirapine, another NNRTI included for comparison. Under the different experimental conditions tested, nevirapine, contrary to efavirenz and sefavirenz, showed a pure noncompetitive mechanism of action, as well as identical kon and koff values for binding to the different enzymatic forms of RT.
FIG. 5.
Determination of the apparent rate of inhibitor binding to the different enzyme-substrate complexes. Reactions were carried out as described in Materials and Methods. Computer fitting of the data to a simple exponential equation was used to generate progress curves for efavirenz (open symbols) or sefavirenz (filled symbols) binding to the RT-TP (circles) or RT-TP-dNTP (triangles) complexes. t, time. Error bars show SD.
TABLE 1.
Kinetic parameters for binding of efavirenz, sefavirenz, and nevirapine to different catalytic forms of HIV-1 RT
| Drug | Catalytic form | Mean ± SD
|
|||
|---|---|---|---|---|---|
| Kd (nM) | kapp (s−1) | kon (s−1 M−1)a | koff (s−1)b | ||
| Efavirenz | RT | 170 ± 5 | NAc | NA | NA |
| RT-TPd | 30 ± 2 | (6 ± 1) × 10−3 | (1.3 ± 0.2) × 104 | (4 ± 1) × 10−4 | |
| RT-TP-dNTPe | 4 ± 0.5 | (7 ± 1) × 10−3 | (3.4 ± 0.2) × 104 | (1.6 ± 0.5 × 10−4 | |
| Sefavirenz | RT | 750 ± 15 | NA | NA | NA |
| RT-TPd | 230 ± 20 | (4 ± 1) × 10−3 | (0.5 ± 0.1) × 104 | (1 ± 0.5) × 10−3 | |
| RT-TP-dNTPe | 8 ± 0.5 | (6 ± 1) × 10−3 | (5.5 ± 0.5) × 104 | (4 ± 1) × 10−4 | |
| Nevirapine | RT | 400 ± 15 | NA | NA | NA |
| RT-TPd | 400 ± 20 | (5.7 ± 1) × 10−3 | (0.15 ± 0.03) × 104 | (6 ± 0.5) × 10−4 | |
| RT-TP-dNTPe | 450 ± 25 | (6 ± 1) × 10−3 | (0.2 ± 0.05) × 104 | (6 ± 1) × 10−4 | |
Calculated from kapp = kon (Ki + [I]).
Calculated from Kd = koff/kon.
NA, not applicable.
Efavirenz and sefavirenz show synergistic inhibition of HIV-1 RT in combination with AZTTP.
The effect of various combinations of efavirenz and AZTTP on the RNA-dependent DNA synthetic activity of HIV-1 RT was tested as described by Villahermosa et al. (31) and Greco et al. (10) (see Materials and Methods). Briefly, dose-response curves for each inhibitor alone were obtained within a wide range of concentrations and compared to inhibition curves obtained with combinations of the inhibitors at a fixed molar ratio. This ratio was determined according to the different potencies of the compounds, ensuring in this way that both inhibitors significantly contributed to the inhibition observed. Data were fitted to the corresponding equations (see Materials and Methods), and the calculated parameters for both efavirenz and sefavirenz combinations with AZTTP are listed in Table 2. Efavirenz was found to be significantly synergistic with AZTTP in its inhibition of HIV-1 RT at fractional inhibition levels of 20 to 90%. This result was in agreement with previous observations indicating a synergistic action of efavirenz in combination with NRTIs in in vitro RT inhibition assays. The thio-substituted analog sefavirenz displayed a similar behavior, but its synergistic effect was slightly reduced, as reflected by higher indexes estimated for the different fractional inhibition levels. The reduced I for sefavirenz can be explained in terms of the observed reduced affinity of the thio-substituted analog for the RT-TP-dNTP complex (Table 1).
TABLE 2.
Calculated interaction parameters for combinations of AZTTP with efavirenz or sefavirenza
| Drug(s) | Reference | Mean ± SD
|
||||||
|---|---|---|---|---|---|---|---|---|
| D50 (nM) | d1 (nM) | d2 (nM) | d1 + d2 (nM) | I
|
||||
| 50 | 20 | 90 | ||||||
| Efavirenz (d1) | 31 | 11 ± 2 | NA | NA | NA | NA | ||
| 10 | 13 ± 2 | NA | NA | NA | NA | NA | NA | |
| Sefavirenz (d1) | 31 | 25 ± 1 | NA | NA | NA | NA | ||
| 10 | 30 ± 1 | NA | NA | NA | NA | NA | NA | |
| AZTTP (d2) | 31 | 19 ± 2 | NA | NA | NA | NA | ||
| 10 | 21 ± 2 | NA | NA | NA | NA | NA | NA | |
| Efavirenz + AZTTP | 31 | NA | 5.9 ± 0.3 | 1.4 ± 0.2 | 7.3 ± 0.3 | 0.6 ± 0.2 | ||
| 10 | NA | 5.6 ± 0.3 | 1.5 ± 0.2 | 7.1 ± 0.3 | 0.5 ± 0.2 | 0.58 ± 0.05 | 0.4 ± 0.1 | |
| Sefavirenz + AZTTP | 31 | NA | 11.2 ± 0.2 | 2.8 ± 0.3 | 14 ± 1 | 0.68 ± 0.05 | ||
| 10 | NA | 13.6 ± 0.2 | 3.4 ± 0.3 | 17 ± 1 | 0.61 ± 0.05 | 0.64 ± 0.04 | 0.55 ± 0.1 | |
See the text for details. NA, not applicable. I50, I20, and I90, Berenbaum's I for 50, 20, and 90% inhibition, respectively.
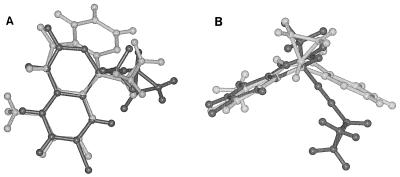
A modeled tridimensional structure of efavirenz shows analogy to nevirapine. As shown in Fig. 6, the energy-minimized tridimensional structure of efavirenz showed a conformation similar to the one assumed by the NNRTI nevirapine in the RT-nevirapine complex, as revealed by crystal structure determination. In particular, the oxygen substituent at position C-2 of efavirenz is perfectly superimposable and coplanar with the oxygen at position C-6 of nevirapine. For nevirapine, this position has been proposed to make important contacts with different residues of the NNRTI-binding pocket, including Phe 227 and Val 106. It is conceivable that similar contacts are also important for the stabilization of efavirenz. Thus, substitution of the oxygen with a sulfur atom at this position in sefavirenz could impair some of these interactions.
FIG. 6.
Superimposition of efavirenz and nevirapine. Ball and stick models are shown for efavirenz (dark grey) and nevirapine (light grey). Drawing was performed with the program Insight II. (A) Top view. (B) Front view.
DISCUSSION
A detailed understanding of the mechanism of action of efavirenz is an obligatory step in the development of new derivatives with a better activity profile against drug-resistant mutants. In the present study, the equilibrium dissociation constants for efavirenz binding to the different catalytic forms of HIV-1 RT as well as the association and dissociation rates have been determined using a steady-state kinetic approach. In addition, the same enzymological analysis has been extended to its thio-substituted analog, sefavirenz, which showed comparable activity in vitro against RT. Both compounds displayed nonlinear kinetics of inhibition (Fig. 3 and 4). They acted as purely uncompetitive inhibitors at low drug concentrations (5 to 50 nM) and as mixed noncompetitive inhibitors at higher doses (50 to 500 nM). According to the reaction scheme illustrated in Fig. 1A and to the equilibrium dissociation constants for inhibitor binding listed in Table 1, this behavior can be interpreted in terms of the relative affinities for the different catalytic forms of the enzyme. Both efavirenz and sefavirenz showed increasing affinities for the different forms of RT in the following order: free enzyme < (i.e., bound with lower affinity) binary RT-TP complex < ternary RT-TP-dNTP complex. Thus, when only the binary RT-TP and the ternary RT-TP-dNTP complexes were available for inhibitor binding (Fig. 1B, left panel), at low drug concentrations the inhibitor interacted only with the ternary complex, the affinity for the binary complex being too low. In such a situation, the inhibition followed a fully uncompetitive mechanism according to equation 1 (see Materials and Methods). When the inhibitor concentrations were raised, a ternary complex with the inhibitor (RT-inhibitor-TP) could also be formed; thus, the inhibition was governed by equation 2, describing a mixed noncompetitive mechanism. The same was true for the example illustrated in Fig. 1B, right panel. Under these conditions, at low efavirenz and sefavirenz concentrations, only the quaternary complex with the inhibitor (RT-inhibitor-TP-dNTP) was formed, revealing purely uncompetitive inhibition. At higher drug doses, an RT-inhibitor complex started to be formed, resulting in mixed noncompetitive inhibition.
Both efavirenz and sefavirenz displayed the lowest affinity toward the free enzyme, but with a different selectivity ratio, Kd(E)/Kd(ter), which was 42.5 for efavirenz but which was increased to 93.7 for Sefavirenz. The greatest difference between efavirenz and sefavirenz was seen in the selectivity for the ternary complex versus the binary complex, since the Kd(bin)/Kd(ter) value of 7.5 for efavirenz was increased to 28.5 for sefavirenz.
Determination of the binding (kon) and dissociation (koff) rates for the interaction of the two inhibitors with the different catalytic forms of HIV-1 RT showed that the increases in the koff value for the conversion of the RT-inhibitor-TP-dNTP complex into the RT-inhibitor-TP complex were similar for both inhibitors (2- and 2.5-fold, respectively), whereas the kon value for drug binding to the RT-inhibitor-TP complex was reduced 10-fold for sefavirenz but only 2.5-fold for efavirenz. The rates of binding of the two inhibitors to the different enzyme-substrate complexes were well below the diffusion limit (on the order of 104 M−1 s−1) but substantially higher than those for nevirapine (another clinically used NNRTI) (20). In comparison, the estimated rate for RT-TP complex formation was shown to be on the order of 106 M−1 s−1, thus justifying the assumption of faster equilibrium kinetics for binding of the enzyme to its substrates than to the inhibitor (12, 23, 24). The quaternary complex RT-inhibitor-TP-dNTP showed very low koff values, on the order of 10−4 s−1 for both compounds (Tables 1 and 2), typical of tightly binding inhibitors.
Both efavirenz and sefavirenz showed significant synergy in combination with AZTTP. Comparison of the observed D50 values with the corresponding d1 and d2 values (Table 2) showed an apparent 6- to 12-fold increase in the potency of AZTTP when combined with efavirenz or sefavirenz. Since these inhibitors have a greater affinity for the RT-TP-dNTP complex (Table 1), it is unlikely that this increase was due to an effect on the kon for complex formation between the RT-inhibitor-TP intermediate and AZTTP. The observed synergy was most likely due to the accumulation of the chain-terminated complex RT-TP-AZTMP, which in turn could be converted to a dead-end RT-inhibitor-TP-AZTMP quaternary complex with a very low dissociation rate (koff), thus accounting for the observed effect on the equilibrium constant for AZTTP inhibition.
Other classes of NNRTIs have been reported to follow complex kinetics of inhibition (2, 3, 8). For example, carboxanilide UC38 interacted specifically either with the binary RT-TP complex or with the ternary RT-TP-dNTP complex; however, the equilibrium dissociation constants were 100- to 1,000-fold higher than the corresponding constants for efavirenz, thus suggesting much lower binding and/or higher dissociation rates. The thiocarboxanilide derivative UC781 showed a preference for the different catalytic forms of RT similar to that of efavirenz, but the dissociation rate for the RT-inhibitor-TP-dNTP complex was 10-fold higher than the corresponding rate for efavirenz, resulting an even more tightly binding inhibitor. Thus, efavirenz and its thio-substituted derivative sefavirenz appear to be unique in their mechanism of action, being selective tightly binding inhibitors of the ternary RT-TP-dNTP complex. Efavirenz is thus the first clinically approved NNRTI to show this property. Preliminary in vitro characterization of RT inhibition by efavirenz showed either noncompetitive or mixed but not uncompetitive inhibition. This difference could be attributable either to the TP utilized or to the range of inhibitor concentrations used for the analysis.
We found that there was a reduced affinity of sefavirenz for both free RT and the RT-TP complex. This selectivity for the ternary complex can be explained by the crystallographic structures of different RT-substrate-inhibitor complexes (13, 14, 18, 25). The specific effect seen for the substitution of a sulfur atom in place of an oxygen atom in reducing the affinity of sefavirenz for both free RT and the RT-TP binary complex could reflect the different structures of unbound RT with respect to the binary or ternary complexes with its substrates. In fact, in the ternary complex, there are large-scale structural differences as well as local conformational changes in comparison with both the free enzyme and the RT-TP complex. Thus, it is possible that the structure of the ternary complex displayed the optimal side-chain conformation for inhibitor binding and that the thio substitution at position C-6 of sefavirenz made this compound more sensitive to the structural differences between the different catalytic forms of the enzyme-substrate complexes.
ACKNOWLEDGMENTS
This work was supported by an ISS-AIDS fellowship (to G.M.); by the CNR Target Project on Biotechnology (to S.S.); by the ISS II AIDS Research National Program, project 2.1.3, research proposal 133 (to S.S.); and by TMR grant ERBMRXCT 970125 (to S.S.). FAR 1999 (Università degli Studi, Pavia, Italy) is gratefully acknowledged.
REFERENCES
- 1.Adkins J C, Noble S. Efavirenz. Drugs. 1998;56:1055–1066. doi: 10.2165/00003495-199856060-00014. [DOI] [PubMed] [Google Scholar]
- 2.Barnard J, Borkow G, Parniak M. The thiocarboxanilide nonnucleoside UC781 is a tight-binding inhibitor of HIV-1 reverse transcriptase. Biochemistry. 1997;36:7786–7792. doi: 10.1021/bi970140u. [DOI] [PubMed] [Google Scholar]
- 3.Debyser Z, Vandamme A-M, Pauwels R, Baba M, Desmyter J, Clercq E D. Kinetics of inhibition of endogenous human immunodeficiency virus type 1 reverse transcription by 2′,3′-dideoxynucleoside 5′-triphosphate, tetrahydroimidazo[4,5,1-jk]-[1,4]-benzodiazepin-2-(1H)-thione and 1-[(2-hydroxyethoxy)methyl]-6-(phenylthio)thymine derivatives. J Biol Chem. 1992;267:11769–11776. [PubMed] [Google Scholar]
- 4.De Clercq E. Perspectives of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Farmaco. 1999;54:26–45. doi: 10.1016/s0014-827x(98)00103-7. [DOI] [PubMed] [Google Scholar]
- 5.De Clercq E. HIV-1-specific reverse transcriptase inhibitors: highly selective inhibitors of human immunodeficiency virus type 1 that are specifically targeted at the viral reverse transcriptase. Med Res Rev. 1993;13:229–258. doi: 10.1002/med.2610130303. [DOI] [PubMed] [Google Scholar]
- 6.Dixon M, Webb E C. Enzymes. London, England: Longman; 1979. [Google Scholar]
- 7.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher R S, Syed K, Mithani S, Dimitrienko G I, Parniak M A. Carboxanilide derivative non-nucleoside inhibitors of HIV-1 reverse transcriptase interact with different mechanistic forms of the enzyme. Biochemistry. 1995;34:4346–4353. doi: 10.1021/bi00013a025. [DOI] [PubMed] [Google Scholar]
- 9.Graul A, Rabasseda J, Castañer J. Efavirenz. Drugs Future. 1998;23:133–141. [Google Scholar]
- 10.Greco W R, Bravo G, Parsons J C. The search for synergy: a critical review from a response surface perspective. Pharm Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 11.Hottiger M, Hübscher U. Human immunodeficiency virus type 1 reverse transcriptase. Biol Chem Hoppe-Seyler. 1996;377:97–120. [PubMed] [Google Scholar]
- 12.Hsieh J-C, Zinnen S, Modrich P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J Biol Chem. 1993;268:24607–24613. [PubMed] [Google Scholar]
- 13.Hsiou Y, Ding J, Das K, Clark A D, Jr, Hughes S H, Arnold E. Structure of unliganded HIV-1 reverse transcriptase at 2.7 Å resolution: implications of conformational changes for polymerization and inhibition mechanisms. Structure. 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1672. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 15.Hübscher U, Spadari S. DNA replication and chemotherapy. Physiol Rev. 1994;74:259–304. doi: 10.1152/physrev.1994.74.2.259. [DOI] [PubMed] [Google Scholar]
- 16.Joergensen K A, Ghattas A B, Lawesson S O. The ⩵C⩵S → ⩵C⩵O transformation using the soft NO(+)-species. Tetrahedron. 1982;38:1163–1168. [Google Scholar]
- 17.Kati W M, Johnson K A, Jerva L F, Anderson K S. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992;267:25988–25997. [PubMed] [Google Scholar]
- 18.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 19.Larder B A. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J Gen Virol. 1994;75:951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- 20.Maga G, Amacker M, Ruel N, Hübscher U, Spadari S. Resistance to nevirapine of HIV-1 reverse transcriptase mutants: loss of stabilizing interactions and thermodynamic or steric barriers are induced by different single amino acid substitutions. J Mol Biol. 1997;274:738–747. doi: 10.1006/jmbi.1997.1427. [DOI] [PubMed] [Google Scholar]
- 21.Mellors J W, Dutschman G E, Im G J, Tramontano E, Winkler S R, Cheng Y C. In vitro selection and molecular characterization of human immunodeficiency virus-1 resistant to non-nucleoside inhibitors of reverse transcriptase. Mol Pharmacol. 1992;41:446–451. [PubMed] [Google Scholar]
- 22.Oxford J S, Al-Jabri A A, Stein C A, Levantis P. Analysis of resistance mutants of viral polymerases. Methods Enzymol. 1996;275:555–601. doi: 10.1016/s0076-6879(96)75031-0. [DOI] [PubMed] [Google Scholar]
- 23.Reardon J E. Human immunodeficiency virus reverse transcriptase: a kinetic analysis of RNA-dependent and DNA-dependent DNA polymerization. J Biol Chem. 1993;268:8743–8751. [PubMed] [Google Scholar]
- 24.Reardon J E. Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry. 1992;31:4473–4479. doi: 10.1021/bi00133a013. [DOI] [PubMed] [Google Scholar]
- 25.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, Stammers D. High resolution structures of HIV-1 RT from four RT- inhibitor complexes. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 26.Richman D, Shih C K, Lowy I, Prodanovich P, Goff S, Griffin J. Human immunodeficiency virus type 1 mutants resistant to nonnucleoside inhibitors of reverse transcriptase arise in tissue culture. Proc Natl Acad Sci USA. 1991;88:11241–11245. doi: 10.1073/pnas.88.24.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richman D D, Havlir D, Corbeil J, Looney D, Ignacio C, Spector S A, Sullivan J, Cheeseman S, Barringer K, Pauletti D, Shih C-K, Myers M, Griffin J. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J Virol. 1994;68:1660–1666. doi: 10.1128/jvi.68.3.1660-1666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spence R A, Kati W M, Anderson K S, Johnson K A. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan L, Chen C-Y, Tyller R D, Grabowski E J J, Reider P J. A novel, highly enantioselective ketone alkynylation reaction mediated by chiral zinc aminoalkoxides. Angew Chem Int Ed Engl. 1999;38:711–713. doi: 10.1002/(SICI)1521-3773(19990301)38:5<711::AID-ANIE711>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Tantillo C, Ding J, Jacobo-Molina A, Nanni R G, Boyer P L, Hughes S H, Pauwels R, Andries K, Janssen P A, Arnold E. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. J Mol Biol. 1994;243:369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 31.Villahermosa M L, Martinez-Irujo J J, Cabodevilla F, Santiago E. Synergistic inhibition of HIV-1 reverse transcriptase by combinations of chain-terminating nucleotides. Biochemistry. 1997;36:13223–13231. doi: 10.1021/bi970852k. [DOI] [PubMed] [Google Scholar]
- 32.Young S D, Britcher S F, Tran L O, Payne L S, Lumma W C, Lyle T A, Huff J R, Anderson P A, Olsen D B, Carroll S S, Pettibone D J, O'Brien J A, Ball R G, Balani S K, Lin J H, Chen I-W, Schleif W A, Sardana V V, Long W J, Byrnes V W, Emini E A. L-743,726 (DMP-266): a novel, highly potent nonnucleoside inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39:2602–2605. doi: 10.1128/aac.39.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young S D, Payne L S, Britcher S F, Tran L O, Lumma W C J. Benzoxazinones as inhibitors of HIV reverse transcriptase. Merck & Co., Inc. U.S. patent 5,663,169. September 1997.. [Google Scholar]