Abstract
The occurrence of active efflux and cell wall modifications were studied in Salmonella enterica serovar Typhimurium mutants that were selected with enrofloxacin and whose phenotypes of resistance to fluoroquinolones could not be explained only by mutations in the genes coding for gyrase or topoisomerase IV. Mutant BN18/21 exhibited a decreased susceptibility to ciprofloxacin (MIC = 0.125 μg/ml) but did not have a mutation in the gyrA gene. Mutants BN18/41 and BN18/71 had the same substitution, Gly81Cys in GyrA, but exhibited different levels of resistance to ciprofloxacin (MICs = 2 and 8 μg/ml, respectively). None of the mutants had mutations in the parC gene. Evidence for active efflux was provided by a classical fluorimetric method, which revealed a three- to fourfold decrease in ciprofloxacin accumulation in the three mutants compared to that in the parent strain, which was annuled by addition of the efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone. In mutant BN18/71, a second fluorimetric method also showed a 50% reduction in the level of accumulation of ethidium bromide, a known efflux pump substrate. Immunoblotting and enzyme-linked immunosorbent assay experiments with an anti-AcrA antibody revealed that the resistance phenotype was strongly correlated with the expression level of the AcrAB efflux pump and suggested that decreased susceptibility to ciprofloxacin due to active efflux probably related to overproduction of this pump could occur before that due to gyrA mutations. Alterations were also found in the outer membrane protein and lipopolysaccharide profiles of the mutants, and these alterations were possibly responsible for the decrease in the permeability of the outer membrane that was observed in the mutants and that could act synergistically with active efflux to decrease the level of ciprofloxacin accumulation.
Fluoroquinolones are often the treatment of choice in the cases of life-threatening salmonellosis due to multidrug-resistant strains (4, 27). Salmonella sp. strains that exhibit treatment-compromising resistance to fluoroquinolones are uncommon, but the increasing incidence of strains with decreased susceptibility is a matter of concern (12, 28). In other gram-negative bacteria, such as Escherichia coli, Neisseria gonorrhoeae, or Klebsiella pneumoniae, high-level fluoroquinolone resistance is always associated with the presence of multiple mutations in the quinolone resistance-determining regions (QRDRs) of the genes that code for the intracellular targets of these antibiotics, gyrase (gyrA and gyrB) and topoisomerase IV (parC and parE) (2, 7, 11). For Salmonella enterica serovar Typhimurium, however, we showed in a previous study (9) that fluoroquinolone resistance is not well correlated with the presence of such mutations: highly fluoroquinolone-resistant mutants selected in vitro had no mutations in the genes that code for topoisomerase IV, and some had only one mutation in the gyrA gene, whereas E. coli isolates that exhibited the same level of resistance harbored at least two mutations in gyrA and one in parC (11).
In addition to this mechanism of target modification that is strictly specific to quinolones, gram-negative bacteria can face the presence of toxic compounds, including antibiotics, by excluding them from the cell. Changes in the cell envelope, including loss of outer membrane porins or alterations of the lipopolysaccharide (LPS), can be partially responsible for decreased susceptibility to a wide range of unrelated antibiotics (5, 26). Active efflux systems that act synergistically with the outer membrane could have a high level of participation in the intrinsic and the acquired antibiotic resistance of gram-negative bacteria (16, 22). Some of the multidrug efflux pumps, which belong to several families, exhibit a low specificity and thus confer decreased susceptibility or even clinically significant resistance to several classes of antibiotics when they are overexpressed. It was recently shown that an S. enterica serovar Typhimurium mutant that overproduces the AcrAB efflux pump (initially identified in E. coli as a close homolog of the MexAB efflux system of Pseudomonas aeruginosa) was more resistant than the parent strain to a wide variety of compounds such as fusidic acid, chloramphenicol, tetracycline, norfloxacin, and penicillin G (23).
The goal of the present study was to investigate if additional resistance mechanisms such as decreased cell envelope permeability or active efflux could explain the phenotype of S. enterica serovar Typhimurium mutants that were selected in vitro and that reached different levels of resistance to fluoroquinolone but that harbored not more than one gyrA mutation (9).
MATERIALS AND METHODS
Bacterial strains and selection of fluoroquinolone-resistant mutants.
Fluoroquinolone-susceptible S. enterica serovar Typhimurium strain BN18 was isolated from the liver of a septicemic pigeon. Mutants BN18/21, BN18/41, and BN18/71 derived from this parent strain were obtained after, respectively, two, four, and seven selection steps on Mueller-Hinton agar plates supplemented with increasing concentrations of enrofloxacin (9). Our previous study showed that the QRDR sequences of gyrA, gyrB, parC, and parE of mutant BN18/21 were identical to those of parent strain BN18 (9). Mutants BN18/41 and BN18/71 were shown to have a single mutation in the QRDR of gyrA (Gly81Cys) but none in the QRDRs of gyrB, parC, and parE.
S. enterica serovar Typhimurium strain SH5014 and the corresponding multidrug-resistant mutant that overproduces the Acr pump, strain HN891, were used as controls for MIC determinations and in the immunoblotting experiments (23).
Susceptibility determinations.
MICs were determined by the standard agar doubling dilution method on Mueller-Hinton medium with inocula of 104 CFU per spot. MICs were determined after 18 h of incubation at 37°C. The antibiotics were purchased from the indicated manufacturers: nalidixic acid, penicillin G, carbenicillin, and cefoxitin, Sigma, Steinheim, Germany; ciprofloxacin, Bayer AG; and tetracycline and chloramphenicol, Boehringer Mannheim, Mannheim, Germany.
Accumulation studies. (i) Accumulation of ciprofloxacin.
Ciprofloxacin uptake was assayed by the method of Mortimer and Piddock (20), with some modifications. Bacteria were grown to the late logarithmic phase at 37°C in Luria-Bertani (LB) medium, harvested by centrifugation, washed in 50 mM sodium phosphate buffer (pH 7.0), and resuspended in the same buffer to an A600 of 12.0. The cells were equilibrated for 10 min at 37°C. Ciprofloxacin was then added to a final concentration of 10 μg/ml. After addition of ciprofloxacin, 0.5-ml samples were removed at different time intervals. Five minutes after addition of ciprofloxacin, the efflux pump inhibitor carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to the reaction mixture (final concentration, 100 μM). The samples were immediately diluted in 1 ml of ice-cold sodium phosphate buffer and were then centrifuged for 5 min at 5,600 × g. The pellet was washed once with 1 ml of ice-cold buffer and resuspended in 1 ml of 0.1 M glycine hydrochloride (pH 3.0) for at least 15 h at room temperature. The samples were then centrifuged at 5,600 × g for 10 min. The fluorescence of the supernatant was measured with a Quanta Master C-60 spectrofluorimeter (Photon Technology International, Monmouth Junction, N.J.) at excitation and emission wavelengths of 279 and 447 nm, respectively (3). The concentration of ciprofloxacin in the supernatant was calculated by comparison with a standard curve for ciprofloxacin (0.02 to 2.5 μg/ml) in 0.1 M glycine hydrochloride (pH 3.0). The results were expressed as nanograms of ciprofloxacin incorporated per milligram (dry weight) of bacteria. The experiments were performed at least three times to make sure of the reproducibility.
(ii) Accumulation of ethidium bromide.
A previously described fluorimetric method (1) was used, with slight modifications, to measure the accumulation of ethidium bromide. Briefly, cells were grown overnight, pelleted, and resuspended to an A600 of 0.2 in sodium phosphate buffer (pH 7.0). For demonstration of active efflux, ethidium bromide was added to the suspension at a final concentration of 2 μg/ml. Fluorescence was used as a measure of the amount of ethidium bromide incorporated by the cells and was recorded with a spectrofluorimeter at excitation and emission wavelengths of 530 and 600 nm, respectively. CCCP was added to the suspension (final concentration, 100 μM) after 400 s of incubation. The natural fluorescence of the cells was subtracted, and the fluorescence intensity was expressed in arbitrary units. For demonstration of differences in outer membrane permeability, cells were incubated with CCCP for 3 min before addition of ethidium bromide. Results were expressed as the fluorescence intensity of the cells incubated with ethidium bromide alone subtracted from that of the cells incubated with CCCP and ethidium bromide.
Acr pump expression analysis by immunoblotting and ELISA.
Bacteria were grown overnight at 37°C in LB medium, harvested by centrifugation, and resuspended at an A600 of 10.0. Cells were diluted one-half in the sample buffer of Laemmli (15) and were heated for 10 min at 100°C. Whole-cell proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were transferred at 0.8 mA/cm2 to a nitrocellulose membrane. The membrane was washed three times with Tris-buffered saline (TBS; 0.15% NaCl, 10 mM Tris-HCl [pH 7.5]), saturated for 30 min at room temperature with TBS containing 1% skim milk, and incubated overnight at room temperature with an anti-AcrA polyclonal antibody (23) diluted 1/2,000 in TBS containing 0.05% Tween 20 (TBS-T) and 0.33% skim milk. After three washes in TBS-T, the membrane was incubated for 1 h with peroxidase conjugated to protein A diluted 1/1,000 in TBS-T (Sigma, St. Louis, Mo.). Finally, after three washes in TBS-T, the blot was developed by incubation at room temperature in a solution of TBS containing 0.06% 4-chloro-1-naphthol and 5 mM H2O2. The reaction was stopped by washes in distilled water.
For enzyme-linked immunosorbent assay (ELISA), bacteria resuspended at an A600 of 1.0 in phosphate-buffered saline (PBS; pH 7.2) were sonicated. A total of 100 μl of the sonicates was used to coat each well of 96-well polystyrene plates for 18 h at room temperature. The anti-AcrA antibody serially diluted from 1/500 to 1/256,000 in PBS containing 0.05% Tween 20 (PBS-T) was incubated on the plates for 2 h at 37°C. Binding of the anti-AcrA antibody was detected by a further incubation with the peroxidase-protein A conjugate diluted 1/1,000 in PBS-T. After incubation for 1 h at room temperature, the plates were filled with a substrate solution containing 4 mM H2O2 and 1 mM 2,2-azino-di-3-ethylbenzthiazoline-sulfonic acid. Excess reagents between the different incubations were removed by five washes in 0.9% NaCl solution containing 0.05% Tween 20. The plates were shaken for 1 h at room temperature, and optical density values were recorded with an automatic ELISA reader (Bio-Tek EL 312; Packard Instrument, Rungis, France). The levels of AcrA in the samples prepared from mutants were deduced from the differences in antibody titers between the mutants and the parent strain.
OMPs and LPS preparations.
Outer membrane proteins (OMPs) were extracted by the method of Jerse and Kaper (14), with slight modifications. Briefly, bacteria were grown overnight, harvested by centrifugation, washed once in 10 mM Tris-HCl (pH 8.0), and resuspended in the same buffer. Crude extracts prepared by sonication were centrifuged at 3,800 × g for 10 min. The supernatant was incubated with 1% Triton X-100 for 30 min at 37°C and ultracentrifuged at 100,000 × g for 1 h. The pellet was resuspended in 10 mM Tris (pH 8.0). Samples were prepared for SDS-PAGE by mixing them with an equal volume of the sample buffer of Laemmli (15) and heating the mixture for 10 min at 100°C. Protein gels were stained with Coomassie blue.
LPS was isolated by proteinase K treatment of whole cells as described by Hitchcock and Brown (13). LPS preparations tested by SDS-PAGE were stained with silver.
RESULTS
S. enterica serovar Typhimurium mutants BN18/21, BN18/41, and BN18/71, all derived from parent strain BN18, were chosen for this study because their phenotype of resistance to fluoroquinolones could not be totally explained only by the mutation identified in the gyrase (9). Mutant BN18/21 was resistant to nalidixic acid and exhibited decreased susceptibility to ciprofloxacin, but it had no mutations in the QRDRs of the genes gyrA, gyrB, parC, and parE. Mutant BN18/41 had a single mutation, Gly81Cys, in the gyrA gene but none in the other genes. Mutant BN18/71 had the same single mutation as BN18/41 but required fourfold more ciprofloxacin for inhibition. These observations led us to investigate the involvement of additional resistance mechanisms, that is, active efflux, decreased outer membrane permeability, and cell wall alterations.
Susceptibilities to unrelated antibiotics.
The mutants selected with enrofloxacin exhibited higher levels of resistance than the parent strain not only to antibiotics of the same family (i.e., quinolones) but also to structurally unrelated compounds (Table 1). The MICs of tetracyclines and chloramphenicol were increased fourfold and eightfold in mutants BN18/21 and BN18/71, respectively. The MICs of β-lactams were increased fourfold (penicillin G and carbenicillin) and eightfold (cefoxitin) for mutant BN18/21. For mutants BN18/41 and BN18/71, the MICs of the three β-lactams were increased 8-fold for penicillin G and carbenicillin and 16-fold for cefoxitin. These results strongly suggested that the serial passages on fluoroquinolone had resulted in the selection of an MDR (multiple drug resistance) phenotype, classically characterized by an enhanced activity of a broad-specificity drug efflux pump (22).
TABLE 1.
Susceptibilities to quinolones and structurally unrelated antibiotics, GyrA substitutions, and AcrA expression levels of control strains, parent strain BN18, and mutants derived from BN18
| Strain | MIC (μg/ml)a
|
GyrA substitution | AcrA production levelb | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Nal | Enr | Cip | Cm | Tc | Pen | Cb | Fox | |||
| SH5014 | 4 | NDc | 0.03 | 4 | 1 | 16 | 8 | 4 | ND | ND |
| HN891 | 8 | ND | 0.06 | 8 | 2 | 64 | 32 | 16 | ND | ND |
| BN18 | 8 | 0.06 | 0.03 | 1 | 4 | 8 | 4 | 1 | None | 100 |
| BN18/21 | 32 | 1 | 0.125 | 4 | 16 | 32 | 16 | 8 | None | 350 |
| BN18/41 | >1,024 | 8 | 2 | 8 | 16 | 64 | 32 | 16 | Gly81Cys | 700 |
| BN18/71 | >1,024 | 32 | 8 | 8 | 32 | 64 | 32 | 16 | Gly81Cys | 1,000 |
Nal, nalidixic acid; Enr, enrofloxacin; Cip, ciprofloxacin; Cm, chloramphenicol; Tc, tetracycline; Pen, penicillin G; Cb, carbenicillin; Fox, cefoxitin.
Expressed as a percentage of the production level of strain BN18.
ND, not determined.
Evidence for active efflux. (i) Ciprofloxacin accumulation.
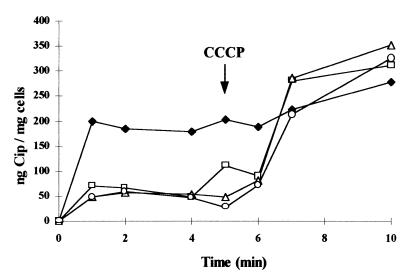
Ciprofloxacin uptake appeared to be drastically reduced in the three mutants compared to that in the parent strain (Fig. 1). At steady state, reached within 1 min following addition of ciprofloxacin, all three mutants accumulated about three- to fourfold less ciprofloxacin than the parent strain. The lack of correlation between the MICs of the antibiotics for the mutants and the level of accumulation of ciprofloxacin could be due to the technical limits of this fluorometric method. Addition of CCCP, a proton motive force uncoupler, induced a very rapid and significant increase in cell-associated ciprofloxacin in the three mutants. As a consequence, at steady state following addition of CCCP, the accumulation of ciprofloxacin was nearly identical in all the strains. These results indicated that an active efflux process limits the accumulation of ciprofloxacin by the cells in a much more efficient way in the mutants than in the parent strain.
FIG. 1.
Accumulation of ciprofloxacin (Cip) by parent strain BN18 (⧫) and mutants BN18/21 (□), BN18/41 (▵), and BN18/71 (○). CCCP (100 μM) was added at the time indicated by the arrow.
(ii) Ethidium bromide accumulation.
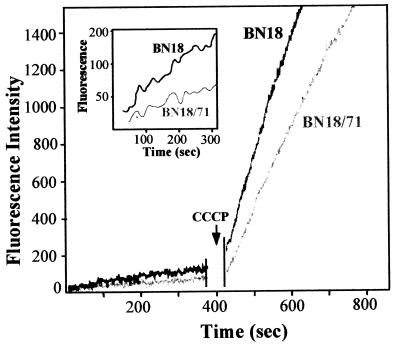
We used a fluorimetric method to investigate if the suspected active efflux could also decrease the level of incorporation in the mutants of ethidium bromide, a toxic hydrophobic cation known to be a substrate of efflux pumps (25). The levels of accumulation of ethidium bromide by parent strain BN18 and by the most resistant mutant, mutant BN18/71, are compared in Fig. 2.
FIG. 2.
Accumulation of ethidium bromide by parent strain BN/18 and mutant strain BN18/71. CCCP (100 μM) was added at the time indicated by the arrow. The inset shows the accumulation before addition of CCCP on an enlarged scale.
After 400 s the accumulation of ethidium bromide in BN18/71 was approximately 50% of that observed in BN18. In contrast to the results obtained with ciprofloxacin accumulation, addition of CCCP induced a drastic increase in ethidium bromide uptake in both strains. However, the entry of ethidium bromide appeared to be faster in BN18 than in BN18/71. These results confirmed that a proton motive force-dependent process was able to limit the incorporation of a toxic compound structurally unrelated to fluoroquinolones. This process appeared to be much more efficient in the fluoroquinolone-selected mutants than in the parent strain. Moreover, the apparently slower penetration of ethidium bromide in the mutant strain after addition of CCCP also suggested a lower permeability of the cell envelope.
(iii) Acr pump expression analysis.
Immunoblotting experiments revealed that AcrA, the periplasmic component of the AcrAB efflux system, faintly detected in strain BN18, was highly overproduced in all the derived mutants (Fig. 3). The AcrA expression levels appeared to correlate with the ciprofloxacin MICs for these mutants. This correlation was confirmed by an enzyme immunoassay (ELISA): AcrA production increased gradually with the number of selection steps on enrofloxacin, reaching a 10-fold increase in mutant BN18/71 (Table 1).
FIG. 3.
Immunoblot analysis of AcrA proteins of the parent strain and the three mutants. SH5014 (parent strain) and HN891 (the corresponding AcrA-overproducing mutant) are control strains described in Materials and Methods. K, kilodaltons.
Evidence for decreased outer membrane permeability.
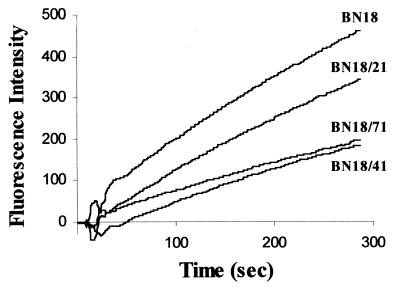
In order to assess the alteration of outer membrane permeability in the resistant mutants, the level of accumulation of ethidium bromide was measured after inhibition of active efflux by CCCP. The rate of accumulation of ethidium bromide was slower for the first mutant, BN18/21, and further decreased for mutants BN18/41 and BN18/71 (Fig. 4). This result indicated a decreased outer membrane permeability in the mutants.
FIG. 4.
Rate of accumulation of ethidium bromide by parent strain BN18 and three mutants after inhibition of active efflux by CCCP.
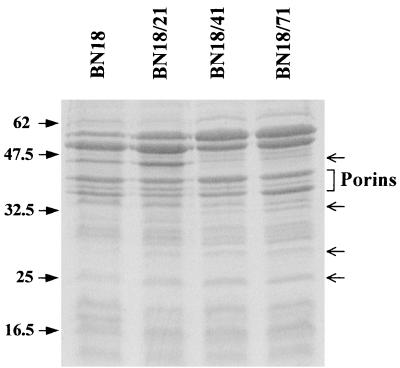
SDS-PAGE analysis of the OMPs revealed no major alteration in the expression of porins (Fig. 5). Expression of a protein band that migrated at 40 to 45 kDa was strongly repressed in mutant strains BN18/41 and BN18/71. The same observation was made with other fluoroquinolone-resistant mutants obtained independently in another selection line (data not shown). Slighter modifications were observed with bands with smaller molecular masses. In particular, bands that migrated at approximately at 25, 28, and 32 kDa appeared to be more strongly expressed in the mutants than in the parent strain.
FIG. 5.
OMP profiles prepared from parent strain BN18 and three mutants. A total of 20 μg of OMPs was loaded per lane. Arrows on the right side indicate the protein bands discussed in the text. Molecular masses in kilodaltons are indicated on the left.
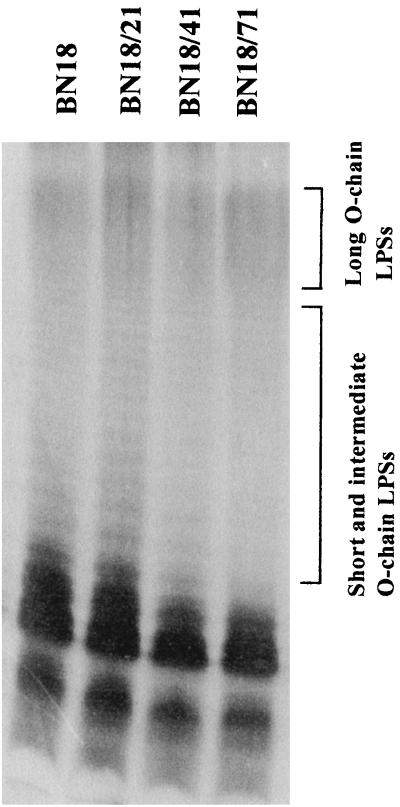
LPS profiles (Fig. 6) revealed significant alterations of the LPS: mutants BN18/41 and BN18/71 exhibited LPSs with a smaller amounts of short and intermediate-length O-polysaccharidic chains. As a consequence, more long O-chain LPSs were detected in mutants BN18/41 and BN18/71 than in the parent strain.
FIG. 6.
LPS profiles of the parent strain and the fluoroquinolone-selected mutants.
The cell wall alterations (OMPs and LPS) observed in the mutants appeared to correlate with their decreased outer membrane permeability.
DISCUSSION
Our previous study (9) had shown that genetic changes in the genes that code for the target proteins of fluoroquinolones, i.e., gyrase and topoisomerase IV, could not totally account for the resistance phenotypes of S. enterica serovar Typhimurium mutants selected in vivo or in vitro. Therefore, we investigated the possible involvement of an active efflux mechanism, which acts synergistically or not, with alterations of the bacterial cell wall. The observation that the fluoroquinolone-selected mutants had also gained resistance to unrelated antibiotics such as tetracycline, chloramphenicol, and β-lactams made us suspect that we had selected Mar (multiple antibiotic resistance) mutants. This broad-spectrum resistance phenotype has been shown in E. coli to result from increased active efflux (16). It was also demonstrated in E. coli that the AcrAB multidrug efflux system plays a major role in the drug resistance phenotype of Mar mutants (24). In serovar Typhimurium, overproduction of the Acr pump was also directly linked to increased rates of resistance to a wide range of compounds including quinolones such as nalidixic acid or norfloxacin (23). However, in the selection process that can lead to clinical resistance, it was not clear whether (i) decreased susceptibility to fluoroquinolones due to enhanced active efflux could be consistently selected in a predetermined way before the occurrence of gyrA mutations or (ii) the level of expression of the Acr pump was correlated to the fluoroquinolone MICs for strains in which gyrase mutations alone could not totally explain the resistance phenotype.
Time course ciprofloxacin uptake experiments were performed with parent and mutant strains that exhibited increasing rates of resistance, one of which had no mutation in the QRDRs of gyrA, gyrB, parC, and parE and the two others of which had only one mutation in gyrA that led to the Gly81Cys substitution (Table 1). A very large difference in the amount of cell-associated ciprofloxacin, which was nearly reversed by addition of the protonophore CCCP, was observed between the parent strain and the three derived mutants (Fig. 1). This result provided evidence of an active efflux process that uses the proton motive force as an energy source. However, no differences in the levels of accumulation could be seen for the different mutants. In another fluorimetric assay, the monitoring of the accumulation of ethidium bromide, a known substrate for the efflux pump (1), by the parent strain and the most resistant mutant also gave evidence for the existence of an active efflux process (Fig. 2). As is expected for Mar mutants, we could confirm using an anti-AcrA antibody the overproduction of the Acr pump in the mutants. Interestingly, this overproduction already reached 3.5-fold that for the parent strain in mutant BN18/21 (ciprofloxacin MIC, 0.125 μg/ml), which was not yet mutated in the gyrA gene. In mutants BN18/41 and BN18/71, for which the ciprofloxacin MICs reached 2 and 8 μg/ml, respectively, AcrA expression levels were about 7- and 10-fold higher than those in BN18. Similarly, an overexpression of AcrA was observed in clinical mutants that were resistant to enrofloxacin and that had decreased susceptibility to ciprofloxacin (data not shown). The AcrAB pump is known to be partly responsible for the intrinsic level of resistance of gram-negative bacteria when it is expressed at its basic expression level (21). These results suggested that its overproduction could participate, independently from gyrA mutations, in a first decrease in susceptibility to fluoroquinolones and in the accession to high fluoroquinolone resistance levels.
It has previously been demonstrated in E. coli that the AcrAB efflux system is essentially regulated by the global regulator loci marRAB or soxRS (17, 24), which also regulates the synthesis of the major porin OmpF (5). Therefore, we examined whether OMP expression had been altered in our S. enterica serovar Typhimurium mutants. We also investigated the possible modification of the LPS, since it has been shown in E. coli that LPS alterations could affect the correct folding (6) and trimerization (29, 30) of porins and also since quinolone resistance has been associated with changes in the LPSs of gram-negative bacteria such as Burkholderia cepacia (26). Cell envelope modifications in both the OMPs and the LPS, possibly in relation to decreased outer membrane permeability, were found in the fluoroquinolone-selected mutants. These modifications appeared to be much more important in the two most resistant mutants, BN18/41 and BN18/71, which had mutations in the gyrA gene, than in the BN18/21 mutant, which exhibited only enhanced active efflux. In contrast to what can be expected from Mar mutants (22), no porin expression decrease was observed. However, expression of several proteins was altered: one protein band at 40 to 45 kDa was strongly repressed, while other bands appeared to be highly expressed in the resistant mutants. In the absence of identification of these protein bands, it is difficult to determine whether their altered expression is involved in the resistance phenotype. As a matter of fact, mutations in a global regulator locus such as the mar locus that affect the resistance phenotype can also have pleiotropic effects on the expression of genes not necessarily involved in the resistance phenotype. Work is in progress to identify proteins whose expression is altered in the mutants by using two-dimensional gel electrophoresis and N-terminal amino acid microsequencing.
Examination of the LPS profile revealed an increase in the proportion of the long O-polysaccharidic chains. The role of the composition of the LPS on the accumulation of quinolones has been extensively studied previously, but it remains unclear, as contradictory results have been obtained (3, 8, 10, 18, 19). It is thought that hydrophilic quinolones like ciprofloxacin preferentially use the porin pathway to penetrate the cells (3). Therefore, accessibility to the porin through the LPS is expected to be determinant. It has been hypothesized in quinolone-selected P. aeruginosa strains that increased amounts of LPS form a permeability barrier which acts preferentially against hydrophilic quinolones (18). The lengthening of the O chains could also result in a lower level of accessibility to the outer membrane.
In summary, we demonstrated enhanced active efflux and gradually increased levels of production of the AcrAB pump in mutants increasingly resistant to ciprofloxacin and other antibiotics. This overproduction appeared to be early compared to the occurrence of a gyrA mutation and thus could be responsible for the first decrease in susceptibility to fluoroquinolones. Our study suggests that, in the absence of multiple mutations that affect the genes that code for gyrase and topoisomerase IV, as observed in E. coli, active efflux that possibly acts synergistically with a decrease in outer membrane permeability could play a major role in ciprofloxacin resistance in S. enterica serovar Typhimurium. Although no Salmonella strains resistant to ciprofloxacin could be isolated in the field (9), overexpression of AcrA observed in the clinical strains with decreased susceptibilities to ciprofloxacin suggests the generality of the efflux phenomenon.
ACKNOWLEDGMENTS
This work was supported by grants from the Conseil Régional de la Région Centre.
We thank C. Mouline and G. Flaujac for technical assistance, H. Nikaido (University of California, Berkeley) for providing the anti-AcrA antibody and strains HN891 and SH5014, and A. P. Teixeira-Gomes and I. Moriyon for helpful discussions.
REFERENCES
- 1.Baranova N N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belland R J, Morrison S G, Ison C, Huang W M. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–380. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 3.Chapman J S, Georgopapadakou N H. Routes of quinolone permeation in Escherichia coli. Antimicrob Agents Chemother. 1988;32:438–442. doi: 10.1128/aac.32.4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherubin C E, Eng R H. Quinolones for the treatment of infections due to Salmonella. Rev Infect Dis. 1991;13:343–344. doi: 10.1093/clinids/13.2.343. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Cock H, Brandenburg K, Wiese A, Holst O, Seydel U. Nonlamellar structure and negative charges of lipopolysaccharides required for efficient folding of outer membrane protein PhoE of Escherichia coli. J Biol Chem. 1999;274:5114–5119. doi: 10.1074/jbc.274.8.5114. [DOI] [PubMed] [Google Scholar]
- 7.Deguchi T, Fukuoka A, Yasuda M, Nakano M, Ozeki S, Kanematsu E, Nishino Y, Ishihara S, Ban Y, Kawada Y. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1997;41:699–701. doi: 10.1128/aac.41.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denis A, Moreau N J. Mechanisms of quinolone resistance in clinical isolates: accumulation of sparfloxacin and of fluoroquinolones of various hydrophobicity, and analysis of membrane composition. J Antimicrob Chemother. 1993;32:379–392. doi: 10.1093/jac/32.3.379. [DOI] [PubMed] [Google Scholar]
- 9.Giraud E, Brisabois A, Martel J L, Chaslus-Dancla E. Comparative studies of mutations in animal isolates and experimental in-vitro and in-vivo mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob Agents Chemother. 1999;43:2131–2137. doi: 10.1128/aac.43.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 11.Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40:879–885. doi: 10.1128/aac.40.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heurtin-Le Corre C, Donnio P Y, Perrin M, Travert M F, Avril J L. Increasing incidence and comparison of nalidixic acid-resistant Salmonella enterica subsp. enterica serotype Typhimurium isolates from humans and animals. J Clin Microbiol. 1999;37:266–269. doi: 10.1128/jcm.37.1.266-269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jerse A E, Kaper J B. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect Immun. 1991;59:4302–4309. doi: 10.1128/iai.59.12.4302-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Levy S B. Active efflux mechanisms for antimicrobial resistance. Antimicrob Agents Chemother. 1992;36:695–703. doi: 10.1128/aac.36.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma D, Cook D N, Alberti M, Pon N G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 18.Michea-Hamzehpour M, Furet Y X, Pechere J C. Role of protein D2 and lipopolysaccharide in diffusion of quinolones through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:2091–2097. doi: 10.1128/aac.35.10.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitsuyama J, Itoh Y, Takahata M, Okamoto S, Yasuda T. In vitro antibacterial activities of tosufloxacin against and uptake of tosufloxacin by outer membrane mutants of Escherichia coli, Proteus mirabilis, and Salmonella typhimurium. Antimicrob Agents Chemother. 1992;36:2030–2036. doi: 10.1128/aac.36.9.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortimer P G, Piddock L J. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J Antimicrob Chemother. 1991;28:639–653. doi: 10.1093/jac/28.5.639. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikaido H. Multiple antibiotic resistance and efflux. Curr Opin Microbiol. 1998;1:516–523. doi: 10.1016/s1369-5274(98)80083-0. [DOI] [PubMed] [Google Scholar]
- 23.Nikaido H, Basina M, Nguyen V, Rosenberg E Y. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those beta-lactam antibiotics containing lipophilic side chains. J Bacteriol. 1998;180:4686–4692. doi: 10.1128/jb.180.17.4686-4692.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajyaguru J M, Muszynski M J. Association of resistance to trimethoprim/sulphamethoxazole, chloramphenicol and quinolones with changes in major outer membrane proteins and lipopolysaccharide in Burkholderia cepacia. J Antimicrob Chemother. 1997;40:803–809. doi: 10.1093/jac/40.6.803. [DOI] [PubMed] [Google Scholar]
- 27.Reid T M. The treatment of non-typhi salmonellosis. J Antimicrob Chemother. 1992;29:4–8. doi: 10.1093/jac/29.1.4. [DOI] [PubMed] [Google Scholar]
- 28.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 29.Sen K, Nikaido H. Lipopolysaccharide structure required for in vitro trimerization of Escherichia coli OmpF porin. J Bacteriol. 1991;173:926–928. doi: 10.1128/jb.173.2.926-928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sen K, Nikaido H. Trimerization of an in vitro synthesized OmpF porin of Escherichia coli outer membrane. J Biol Chem. 1991;266:11295–11300. [PubMed] [Google Scholar]