Abstract
Aim of the study
AT-rich interactive domain 1A (ARID1A) is a subunit of the switch/sucrose non-fermentable chromatin remodeling complex, which is commonly mutated in human cancers. The clinical and pathological significance of ARID1A alteration in hepatocellular carcinoma (HCC) has not yet been clarified. The present study aimed to evaluate the clinical significance of the ARID1A gene signature in HCC and its relation to the likelihood of tumor recurrence after microwave ablation (MWA).
Material and methods
This study included 50 patients with cirrhotic HCC of Barcelona Clinic Liver Cancer stages 0/A eligible for MWA. Tumor and peri-tumor biopsies were obtained just prior to MWA and assessed for tumor pathological grade and ARID1A expression by immunohistochemistry. Patients were followed for one year after complete tumor ablation to detect any recurrence.
Results
Tumor size (MCp = 0.010) and α-fetoprotein level (p = 0.013) can effectively predict the response to MWA. Nuclear expression of ARID1A was significantly lower in HCC compared to the corresponding peri-tumor cirrhotic liver tissues (p = 0.002), but no significant difference in ARID1A cytoplasmic expression was found. Nuclear ARID1A expression level in HCC showed a significantly negative relation to tumor size (MCp = 0.006), pathological grade (MCp = 0.046) and post-MWA tumor recurrence (FEp = 0.041).
Conclusions
ARID1A loss may enhance HCC aggressiveness and post-MWA tumor recurrence. ARID1A could be a potential target to select HCC patients for future therapies.
Keywords: ARID1A, hepatocellular carcinoma, microwave ablation, tumor recurrence
Introduction
Liver cancer is the sixth most commonly diagnosed cancer and the fourth leading cause of cancer-related deaths worldwide [1]. Liver cancer is a highly fatal tumor with most cases detected at late stages and an incidence-to-mortality ratio that approaches 1 [2]. Hepatocellular carcinoma (HCC) represents about 75-85% of primary liver cancers and constitutes a major health problem worldwide [3]. Despite the recent progress in HCC diagnosis and intervention over the last decade, only one third of patients are candidates for curative or life-extending loco-regional therapies, and the overall prognosis still remains unsatisfactory due to the major obstacles of metastasis and tumor recurrence that may occur after any type of treatment [4-6].
Microwave ablation (MWA) is a new modality of thermal ablation which seems strikingly promising for treatment of hepatic malignancies [7]. It depends on production of electromagnetic waves which force dipolar water molecules to continuously rotate billions of times per second with the oscillating electric field typically at 900 to 2500 MHz, leading to production of heat and induction of coagulative necrosis [7, 8]. MWA displays several theoretical advantages over radiofrequency ablation including deeper and faster tissue penetration with efficient production of a larger ablation zone within a shorter ablation time, and the reduced “heat sink” effect [9-11]. Furthermore, the modern shaft cooling system prevents overheating of the shaft, which consecutively prevents skin burns, increases the amount of energy delivered to the target tissue, and facilitates the creation of large ablation zones [12]. Eventually, larger lesions can be treated by repositioning antennae or using multiple antennae simultaneously [13, 14]. In fact, using the new system leads to over 80% complete ablation in HCC nodules up to 8 cm in diameter [15].
One of the important roadblocks to treatment efficacy is the lack of understanding of the mechanisms involved in HCC pathogenesis and progression. Elucidating the molecular mechanisms of tumor growth would allow the development of effective molecule-targeted therapies, and ultimately help patients with HCC to achieve a favorable prognosis [16, 17]. Therefore, there is an urgent need to find novel biomarkers with high efficacy for accurate prediction of tumor recurrence and progression.
The role of epigenetics in cancer progression has been emphasized ever since the introduction of cancer genome-wide sequencing, which revealed significant alteration in genes responsible for modifying chromatin structure. Chromatin remodeling genes, such as AT-rich interacting domain containing protein 1A (ARID1A) and BRCA1-associated protein 1 (BAP1), along with other well-known genes such as TERT, TP53, and CTNNB1 are frequently mutated in a broad spectrum of cancer types [18, 19]. ARID1A belongs to a family of 15 proteins in humans all of which contain a characteristic 100-amino acid DNA-binding ARID domain. The ARID domain of ARID1A does not show sequence-specific DNA binding and the only other protein homology domain, located within the C-terminus, has unknown function [20]. ARID1A is located on chromosome 1p, migrates at approximately 250 kDa, is widely expressed and is present primarily (perhaps exclusively) in the nucleus. It is also found in the cytoplasm and nuclear ARID1A is less stable than cytoplasmic ARID1A because it is rapidly degraded by the ubiquitin-proteasome system in the nucleus. ARID1A has been implicated in numerous protein-protein interactions, and the most widely known and studied are those which make ARID1A a part of switching defective/sucrose non-fermenting (SWI/SNF) chromatin-remodeling complexes. As a member of SWI/SNF complexes, ARID1A is thought to contribute to specific recruitment of its chromatin remodeling activity by binding transcription factors and transcriptional co-activator/co-repressor complexes [21, 22].
Accumulating data have implicated ARID1A as a key member of the SWI/SNF chromatin-remodeling complex, which acts as a tumor suppressor in a broad spectrum of human cancers [23]. Genome sequencing and comparative genomic hybridization studies have also detected ARID1A insertion/deletion mutations at high frequency in a multitude of cancer types including hepato-pancreatic, utero-ovarian and breast cancers [24-28]. Many ARID1A insertion/deletion mutations with downregulation of the encoded protein were observed in 10-16.8% of liver tumors, and in 13% of hepatitis B virus-associated HCC [29-31]. Excitingly, ARID1A has a context-specific role in liver cancer, whereby ARID1A up-regulation promotes tumor initiation, while ARID1A down-regulation in established tumors increases metastasis in mouse models [32, 33]. Nevertheless, the clinical significance of ARID1A and its biological function in HCC are still controversial and have to be clarified.
Therefore, the present study was designed to evaluate the clinical significance of the ARID1A gene signature in HCC and its relation to the likelihood of tumor recurrence after MWA treatment.
Material and methods
This study included 50 patients with definite newly diagnosed HCC on top of hepatitis C virus (HCV)- related liver cirrhosis who were referred to the Hepatobiliary Unit at the Alexandria University Hospitals. Patients with preserved liver function and early stages of HCC (Barcelona Clinic Liver Cancer [BCLC] stage 0: single lesion less than 2 cm and BCLC stage A: single lesion less than 5 cm or up to three lesions not greater than 3 cm each) were included in the study. Patients with active HCV infection received direct acting antivirals after HCC treatment. Patient collection lasted from May 2018 until June 2019 with longitudinal follow-up for a year. Exclusion criteria were cirrhotic patients of Child-Turcotte-Pugh (CTP) class C, patients with tumor-in-vein thrombus or metastasis, or those who received previous HCC treatment. Patients were assigned to MWA and tissue samples were obtained from the tumors and peri-tumor cirrhotic liver just prior to MWA. The research was conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines. The study protocol was approved by the Ethics Committee of the Alexandria Faculty of Medicine (IRB No. 00007555). Informed consent was obtained from all subjects included in the study.
All patients included in the study were evaluated clinically as regards manifestations of liver cirrhosis and malignancy, with complete clinical examination and routine laboratory investigations including complete blood picture and liver test assay. The severity of chronic liver disease was determined on the basis of CTP classification. Serum AFP levels were measured using enzyme-linked immunosorbent assay (ELISA) provided by Diagnostic Automation/Cortez Diagnostics (Catalog No. 5101Z) [34]. Radiological evaluation depended on a recent triphasic computed tomography (CT) abdomen and/or dynamic magnetic resonance imaging (MRI) performed within 4 weeks before intervention to confirm the diagnosis of HCC based on the characteristic enhancement pattern of contrast hyper-enhancement in the arterial phase “wash-in” and hypo-enhancement in the portal or delayed phase “wash-out”, and to determine the site, size and number of tumors and to exclude vascular invasion or metastasis [7]. The stage of HCC was determined according to the BCLC staging classification [35].
Ablation procedure
All ablations were performed by the same hepatologist (the author named A.A. who has about 20 years of experience in local ablation therapy of HCC). Thermal ablation of the tumor was done using a 14-gauge disposable MWA probe (AMICA probe MW) and a 2.45 GHz generator (AMICA GEN AGN-H-1.2, Italy) [36]. Wattage and duration used for ablation were chosen based on the manufacturer’s guidelines. Ultrasound-guided fine needle aspiration cytology (FNAC) smears from the tumors and peri-tumor cirrhotic liver tissues were obtained just prior to MWA for histological examination.
Histopathology and immunohistochemistry
All histologic evaluations were performed by the same pathologist who was blinded to the patients’ code (the author named N.B. with almost 30 years of experience in pathological examination. Two pathologists independently analyzed the immunohistochemical reactions, and the presented result was the mean if both readings were not the same). Tissue biopsies obtained from patients were fixed in 10% formalin solution, embedded in paraffin, sectioned (5 µm), dehydrated, cleared and subsequently cover slipped using DPX as a mounting medium. HCC was classified into trabecular, pseudo-glandular and compact morphology based on criteria laid down by the World Health Organization (WHO) [37]. HCC grade was assigned as regards the criteria of the Edmondson and Steiner (E-S) grading system [38]. Concerning the patients who had multiple focal lesions, the biopsy was taken from the largest tumor nodule.
Smeared slides were permeabilized by briefly dipping in a 0.1% solution of Triton X100 with gentle agitation (Thermo Scientific, Carpinteria, CA, USA). Endogenous peroxidases were blocked with 3% hydrogen peroxide. No antigen retrieval was performed. Smears were incubated with the peroxidase blocking agent for 10 minutes at room temperature. Subsequently, sections were incubated overnight with ARID1A monoclonal antibody (anti-ARID1A antibody, clone 3H2, Sigma-Aldrich, USA) at an optimal dilution of 1 : 25. Tissue sections were then incubated with the one step secondary antibody for 30 minutes at room temperature. Immune complexes were visualized using 3,3’-diaminobenzidine (DAB) (Dako, Carpinteria, CA, USA). Slides were counterstained with light hematoxylin, dehydrated, and cover-slipped. All incubations were performed at room temperature in a humidity chamber, unless otherwise stated. Positive tissue controls were included with each run.
A semi-quantitative scoring system considering the staining intensity and proportion was used to estimate the expression of ARID1A using an Olympus magnifying microscope (CH 20 BIMF 200, Olympus, China) [39]. Each slide was evaluated by the intensity of the cytoplasmic and nuclear staining (no staining = 0, weak staining = 1, moderate staining = 2, and strong staining = 3) and the proportion of stained cells (0% = 0, 1-30% = 1, 31-60% = 2, and 61-100% = 3). The sum ranging from 0 (the minimum score) to six (the maximum score) was the final immuno-reactive score. This was classified as negative expression (0), weak expression (1-2), moderate expression (3-4), and strong expression (5-6). We considered a final immunoreactive score of 1 or higher to be positive.
Follow-up and monitoring of response to treatment
All radiological evaluations were performed by the same radiologist who was blinded to the patients’ code and the post-treatment response of tumors was assessed on the basis of the modified Response Evaluation Criteria in Solid Tumors (mRECIST) criteria [40]. Tumor response to MWA was assessed by triphasic CT done 4 weeks after intervention. All patients were followed after complete ablation using triphasic CT every 3 months for 1 year to detect any tumor recurrence. Patients with residual activity were retreated by another session of MWA, and an extra follow-up triphasic CT was performed after another 4 weeks. Complete response was defined as disappearance of any intra-tumoral arterial phase enhancement in all target lesions. Partial response was defined as at least 30% decrease in the sum of the diameters of viable target lesions with arterial phase enhancement, taking as a reference the baseline sum of the diameters of target lesions. Local hepatic recurrence of tumor was defined as reactivation of the ablated target tumor lesion recorded since treatment started. Distant hepatic recurrence of tumor was defined as the appearance at other hepatic sites of a new target tumor lesion unrecorded since treatment started.
Statistical analysis
Data were fed to the computer and analyzed using the SPSS software version 20.0. (Armonk, NY: IBM Corporation). Quantitative data were described as range, mean ± standard deviation and median. Qualitative data were described as number and percentage. The Kolmogorov-Smirnov test was used to verify the normality of data distribution. Statistical significance of the obtained results was judged at the level p < 0.05. All calculated p values were two-tailed. The chi-square (χ2) test was used for comparison between different groups with respect to categorical variables, and Fisher’s exact test (FE) with Monte Carlo (MC) correction was appropriately applied when more than 20% of the cells had an expected count less than 5. Comparisons between two groups for normally distributed numerical variables were done using Student’s t-test. The Mann-Whitney U-test was used to compare between two groups for non-normally distributed numerical variables. The Wilcoxon signed rank test was applied to compare between two periods for non-normally distributed numerical variables. Univariate and multivariate logistic regression analysis was used to predict the categorical dependent variable using a given set of independent variables.
Results
Baseline clinical, biochemical, radiological and pathological data of patients included in the study are shown in Table 1.
Table 1.
Distribution of clinical, biochemical, radiological and pathological data of patients included in the study
| Baseline characteristics | Distribution (n = 50) Mean ±SD or n (%) |
|
|---|---|---|
| Age (years) | 56.13 ±6.77 | |
| Gender | ||
| Male | 38 (76) | |
| Female | 12 (24) | |
| Child-Turcotte-Pugh class | ||
| Class A | 24 (48) | |
| Class B | 26 (52) | |
| Hemoglobin concentration (g/dl) | 11.66 ±1.55 | |
| Platelet count (×103/mm3) | 119.73 ±35.68 | |
| Leucocyte count (×103/mm3) | 5.41 ±1.88 | |
| Serum aspartate aminotransferase (U/l) | 56.08 ±46.17 | |
| Serum alanine aminotransferase (U/l) | 36.19 ±25.98 | |
| Serum albumin (g/dl) | 3.09 ±0.60 | |
| Serum bilirubin (mg/dl) | 1.27 ±0.47 | |
| International normalized ratio | 1.26 ±0.15 | |
| Serum α-fetoprotein (ng/dl) | 235.41 ±384.16 | |
| Sum number of tumors | 1.72 ±1.02 | |
| Distribution | ||
| 1 nodule | 30 (60) | |
| 2 nodules | 8 (16) | |
| 3 nodules | 12 (24) | |
| Sum size of tumors | 4.46 ±1.39 | |
| Distribution | ||
| < 3 cm | 26 (29.6) | |
| 3-6 cm | 56 (63.6) | |
| > 6-< 9 cm | 6 (6.8) | |
| Pathological grade of tumors | ||
| Grade I | 4 (8.0) | |
| Grade II | 16 (32.0) | |
| Grade III | 30 (60.0) | |
Response to MWA therapy in relation to different clinico-chemical parameters and tumor characteristics
The mean number of MWA sessions needed per patient was 2.38 ±1.02 and patients with an incomplete response underwent another MWA session(s) to achieve complete ablation of the target lesions. Within 12 months of follow-up, 24 (48%) patients were recurrence-free, while 26 (52%) patients of the studied cohort witnessed tumor recurrence.
There was no statistically significant difference between patients who were recurrence-free after MWA and those who showed recurrence as regards gender and age of the patients (χ2 = 4.338, FEp = 0.065 and t = 0.405, p = 0.692 respectively), CTP class (χ2 = 1.330, p = 0.249), pathological grade of the tumor (χ2 = 0.181, p = 0.671) and the maximum number of tumors (χ2 = 4.154, MCp = 0.282). However, the maximum size of tumors (χ2 = 8.863, MCp = 0.010) and the level of serum AFP (U = 36.0, p = 0.013) were found to be significantly related to the response of HCC to MWA, where larger tumor nodules and higher AFP levels were found among patients who experienced tumor recurrence (Table 2).
Table 2.
Response of the tumor to microwave ablation in relation to the clinical, biochemical, radiological and pathological parameters
| Parameter | Response | Test of sig. | P value | ||||
|---|---|---|---|---|---|---|---|
| Responder (n = 24) |
Non-responder (n = 26) |
||||||
| n | % | n | % | ||||
| Gender | |||||||
| Male | 22 | 91.6 | 16 | 61.5 | χ2 = 4.338 | FEp = 0.065 | |
| Female | 2 | 8.4 | 10 | 38.5 | |||
| Age (years) | |||||||
| Min.-Max. | 51.0-60.0 | 43.0-70.0 | t = 0.405 | 0.692 | |||
| Mean ±SD | 56.57 ±2.68 | 55.67 ±7.33 | |||||
| Median | 56.0 | 55.0 | |||||
| Child-Turcotte-Pugh class | |||||||
| A | 6 | 25.0 | 18 | 69.2 | χ2 = 1.330 | 0.249 | |
| B | 18 | 75.0 | 8 | 30.8 | |||
| Pathological grade of tumors | |||||||
| Grade I | 2 | 8.33 | 2 | 7.7 | χ2 = 0.181 | 0.671 | |
| Grade II | 10 | 41.66 | 10 | 38.4 | |||
| Grade III | 12 | 50.0 | 14 | 53.9 | |||
| Sum number of tumors | |||||||
| 1 nodule | 18 | 75.0 | 12 | 46.2 | χ2 = 4.154 | MCp = 0.282 | |
| 2 nodules | 2 | 8.3 | 6 | 23.1 | |||
| 3 nodules | 4 | 16.6 | 8 | 30.7 | |||
| Sum size of tumors | (n = 62) | (n = 26) | |||||
| < 3 cm | 24 | 37.5 | 2 | 8.3 | χ2 = 8.863* | MCp = 0.010* | |
| 3-6 cm | 38 | 62.5 | 18 | 66.7 | |||
| > 6-< 9 cm | 0 | 0.0 | 6 | 25.0 | |||
| Serum α-fetoprotein | |||||||
| Min.-Max. | 4.20-215.8 | 2.40-1250.0 | U = 36.0* | 0.013* | |||
| Mean ±SD | 45.62 ±62.67 | 456.8 ±481.1 | |||||
| Median | 12.90 | 196.4 | |||||
χ2 – Chi square test, FE – Fisher exact, MC – Monte Carlo correction, U – Mann-Whitney test, t – Student t-test, p – p value for association between different categories,
*statistically significant at p ≤ 0.05
ARID1A expression in HCC and peri-tumor tissues
ARID1A expression was observed in the form of nuclear staining in the cellular population of HCC and peri-tumor tissues stained with the antibody (Fig. 1). Nuclear ARID1A immunostaining in the tumor tissue was negative in up to 20 (40%) cases, while 20 (40%) cases showed weak expression, 6 (12%) cases showed moderate expression and only 4 (8.0%) cases showed strong expression. However, nuclear ARID1A immunostaining in the peri-tumor tissue was evident in all patients where 28 (56%) cases showed weak expression, 10 (20%) cases showed moderate (20%) and 12 (24%) cases showed strong expression.
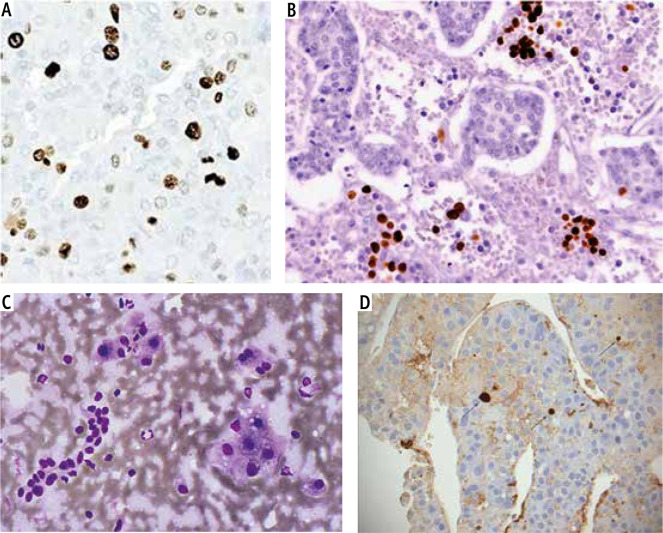
Fig. 1.
ARID1A immunohistochemistry. A) Grade I HCC showing positive ARID1A staining (ARID1A Monoclonal Antibody, Streptavidin-Peroxidase technique, 200×). B) Grade II HCC showing positive nuclear ARID1A staining (ARID1A Monoclonal Antibody, Streptavidin-Peroxidase technique, 400×). C) Grade III HCC showing negative ARID1A staining (ARID1A Monoclonal Antibody, Streptavidin-Peroxidase technique, 100×). C) Peri-tumor cirrhotic liver tissue showing positive ARID1A staining (ARID1A Monoclonal Antibody, Streptavidin-Peroxidase technique, 100×)
The nuclear expression of ARID1A was significantly lower in HCC compared to the corresponding peri-tumor cirrhotic liver tissues (Z = 4.066, p < 0.001), but there was no significant difference as regards ARID1A cytoplasmic expression between tumor and corresponding peri-tumor tissues (p = 1.00) (Table 3).
Table 3.
ARID1A expression in hepatocellular carcinoma and peri-tumor cirrhotic liver tissues
| Expression level | ARID1A | |||||||
|---|---|---|---|---|---|---|---|---|
| Nuclear | Cytoplasmic | |||||||
| Tumor (n = 50) |
Peri-tumor (n = 50) |
Tumor (n = 50) |
Peri-tumor (n = 50) |
|||||
| n | % | n | % | n | % | n | % | |
| Negative | 20 | 40.0 | 0 | 0.0 | 22 | 44.0 | 22 | 44.0 |
| Weak | 20 | 40.0 | 28 | 56.0 | 10 | 20.0 | 10 | 20.0 |
| Moderate | 6 | 12.0 | 10 | 20.0 | 12 | 24.0 | 12 | 24.0 |
| Strong | 4 | 8.0 | 12 | 24.0 | 6 | 12.0 | 6 | 12.0 |
| Test of sig. (p value) | Z = 4.066* (< 0.001*) | Z = 0.0 (1.000) | ||||||
Z – Wilcoxon signed ranks test, p – p value for comparing between tumor and peri-tumor expression, *statistically significant at p ≤ 0.05
ARID1A expression level in HCC in relation to tumor characteristics and post-treatment tumor recurrence
No significant relation was found between the nuclear or the cytoplasmic ARID1A expression levels in HCC and the maximum number of tumors on subgrouping the patients as those with a single nodule vs. others with multiple nodules (Table 4).
Table 4.
Tumor ARID1A expression level in relation to the maximum number of tumors
| Tumor ARID1A expression | Sum number of tumors | χ2 | MC p | ||||
|---|---|---|---|---|---|---|---|
| Single (n = 30) | Multiple (n = 20) | ||||||
| n | % | n | % | ||||
| Nuclear | |||||||
| Negative | 14 | 46.7 | 6 | 30.0 | 1.737 | 0.714 | |
| Weak | 12 | 40.0 | 8 | 40.0 | |||
| Moderate | 2 | 6.7 | 4 | 20.0 | |||
| Strong | 2 | 6.7 | 2 | 10.0 | |||
| Cytoplasmic | |||||||
| Negative | 14 | 46.7 | 8 | 40.0 | 2.083 | 0.684 | |
| Weak | 8 | 26.7 | 2 | 10.0 | |||
| Moderate | 6 | 20.0 | 6 | 30.0 | |||
| Strong | 2 | 6.7 | 4 | 20.0 | |||
χ2 – Chi square test, MC – Monte Carlo correction, p – p value for comparing between the two categories, *statistically significant at p ≤ 0.05
On classifying the patients as regards the maximum size of the tumor (max. size ≤ 3 cm vs. > 3 cm), the analysis revealed a significant negative relation between the nuclear, not the cytoplasmic, ARID1A expression level in HCC and the maximum size of the tumor (χ2 = 9.418, MCp = 0.006) and ARID1A negatively expressed lesions tended to have larger sizes than the positively expressed ones (Table 5).
Table 5.
Tumor ARID1A expression level in relation to the maximum size of tumors
| Tumor ARID1A expression | Sum size of tumors | χ2 | MC p | |||
|---|---|---|---|---|---|---|
| ≤ 3 cm (n = 8) | > 3 cm (n = 42) | |||||
| n | % | n | % | |||
| Nuclear | ||||||
| Negative | 0 | 0.0 | 20 | 47.6 | 9.418* | 0.006* |
| Weak | 2 | 25.0 | 18 | 42.9 | ||
| Moderate | 2 | 25.0 | 4 | 9.5 | ||
| Strong | 4 | 50.0 | 0 | 0.0 | ||
| Cytoplasmic | ||||||
| Negative | 0 | 0.0 | 22 | 52.4 | 4.887 | 0.147 |
| Weak | 2 | 25.0 | 8 | 19.0 | ||
| Moderate | 4 | 50.0 | 8 | 19.0 | ||
| Strong | 2 | 25.0 | 4 | 9.5 | ||
χ2 – Chi square test, MC – Monte Carlo correction, p – p value for comparing between the two categories, *statistically significant at p ≤ 0.05
A significant negative relation was found between the nuclear, not the cytoplasmic, ARID1A expression level in HCC and the pathological grade of the tumor (χ2 = 5.571, MCp = 0.046) and ARID1A negatively expressed lesions tended to have higher grades than the positively expressed ones (Table 6).
Table 6.
Tumor ARID1A expression level in relation to the pathological grade of tumors
| Pathological grade of tumors | Tumor ARID1A expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Nuclear | Cytoplasmic | |||||||
| Negative (n = 20) |
Positive (n = 30) |
Negative (n = 22) |
Positive (n = 28) |
|||||
| n | % | n | % | n | % | n | % | |
| Grade I | 0 | 0.0 | 4 | 13.3 | 0 | 0.0 | 4 | 14.3 |
| Grade II | 2 | 10.0 | 14 | 46.7 | 6 | 27.3 | 10 | 35.7 |
| Grade III | 18 | 90.0 | 12 | 40.0 | 16 | 72.7 | 14 | 50.0 |
| Test of sig. (MCp value) | χ2 = 5.751* (0.046*) | χ2 = 1.895 (0.577) | ||||||
| Pathological grade of tumors | Nuclear tumor ARID1A expression | |||||||
|
Negative
(n = 20) |
Weak
(n = 20) |
Moderate
(n = 6) |
Strong
(n = 4) |
|||||
| n | % | n | % | n | % | n | % | |
| Grade I | 0 | 0.0 | 0 | 0.0 | 2 | 33.3 | 2 | 50.0 |
| Grade II | 2 | 10.0 | 12 | 60.0 | 0 | 0.0 | 2 | 50.0 |
| Grade III | 18 | 90.0 | 8 | 40.0 | 4 | 66.7 | 0 | 0.0 |
| Test of sig. (MCp value) | χ2 = 7.188 (0.068) | |||||||
χ2 – Chi square test, MC – Monte Carlo correction, p – p value for comparing between the two categories, *statistically significant at p ≤ 0.05
No significant relation was found between the nuclear or the cytoplasmic ARID1A expression levels in HCC and the frequency of tumor recurrence after MWA. Among 26 patients who experienced post-treatment tumor recurrence, 16 (61.5%) cases showed negative nuclear ARID1A expression in HCC, and 14 (53.8%) cases showed negative cytoplasmic ARID1A expression in HCC (Table 7). On classifying the patients as regards the type of hepatic recurrence of the tumor (local vs. distant), the analysis revealed a significant negative relation between both the nuclear and the cytoplasmic ARID1A expression levels in HCC and the type of hepatic recurrence of the tumor (χ2 = 8.174, MCp = 0.035 and χ2 = 6.861, MCp = 0.030 respectively) and ARID1A negatively expressed lesions tended to develop post-MWA tumor reactivation more often than the positively expressed ones (Table 8). However, these results need to be interpreted with caution because of the small number of patients in the group of distant hepatic recurrence.
Table 7.
Tumor ARID1A expression level in relation to the frequency of hepatic recurrence of tumors
| Tumor ARID1A expression | Frequency of tumor recurrence | χ2 | MC p | ||||
|---|---|---|---|---|---|---|---|
| No recurrence (n = 24) |
Recurrence (n = 26) |
||||||
| n | % | n | % | ||||
| Nuclear | |||||||
| Negative | 4 | 16.7 | 16 | 61.5 | 5.629 | 0.105 | |
| Weak | 14 | 58.3 | 6 | 23.1 | |||
| Moderate | 4 | 16.7 | 2 | 7.7 | |||
| Strong | 2 | 8.3 | 2 | 7.7 | |||
| Cytoplasmic | |||||||
| Negative | 8 | 33.3 | 14 | 53.8 | 6.688 | 0.072 | |
| Weak | 10 | 41.7 | 0 | 0.0 | |||
| Moderate | 4 | 16.7 | 8 | 30.8 | |||
| Strong | 2 | 8.3 | 4 | 15.4 | |||
χ2 – Chi square test, MC – Monte Carlo correction, p – p value for comparing between the two categories, *statistically significant at p ≤ 0.05
Table 8.
Tumor ARID1A expression level in relation to the type of hepatic recurrence of tumors
| Tumor ARID1A expression | Type of tumor recurrence | χ2 | MCp | ||||
|---|---|---|---|---|---|---|---|
| Local (n = 20) | Distant (n = 6) | ||||||
| n | % | n | % | ||||
| Nuclear | |||||||
| Negative | 16 | 80.0 | 0 | 0.0 | 8.174* | 0.035* | |
| Weak | 2 | 10.0 | 4 | 66.7 | |||
| Moderate | 2 | 10.0 | 0 | 0.0 | |||
| Strong | 0 | 0.0 | 2 | 33.3 | |||
| Cytoplasmic | |||||||
| Negative | 14 | 70.0 | 0 | 0.0 | 6.861* | 0.030* | |
| Weak | 0 | 0.0 | 0 | 0.0 | |||
| Moderate | 6 | 30.0 | 2 | 33.3 | |||
| Strong | 0 | 0.0 | 4 | 66.7 | |||
χ2 – Chi square test, MC – Monte Carlo correction, p – p value for comparing between the two categories, *statistically significant at p ≤ 0.05
Predictors of tumor recurrence after MWA therapy
The set of potential variables was assessed for possible relations to post-MWA tumor recurrence by logistic regression analysis. In the univariate analysis, both CTP class B and a reduced nuclear ARID1A expression level in HCC showed a significant impact on tumor recurrence. In the multivariate analysis, however, reduced ARID1A expression was not found to be an independent predictor of post-MWA tumor recurrence (Table 9).
Table 9.
Univariate and multivariate logistic regression analysis of variables predicting tumor recurrence after microwave ablation
| Variables | Univariate | Multivariate# | |||
|---|---|---|---|---|---|
| p | OR (95% CI) | p | OR (95% CI) | ||
| Gender | 0.910 | 1.111 (0.177-6.970) | |||
| Child-Turcotte-Pugh class | 0.033* | 6.750 (1.162-39.200) | 0.999 | – | |
| Pathological grade of tumors | 0.881 | 1.098 (0.322-3.743) | |||
| Sum size of tumors | 0.251 | 1.453 (0.768-2.749) | |||
| Sum number of tumors | 0.515 | 1.308 (0.583-2.933) | |||
| Serum alpha fetoprotein | 0.173 | 1.006 (0.997-1.015) | |||
| ARID1A expression | |||||
| Tumor/Nuclear | 0.031* | 0.125 (0.019-0.823) | 0.999 | – | |
| Tumor/Cytoplasmic | 0.306 | 0.429 (0.085-2.169) | |||
| Peri-tumor/Nuclear$ | – | – | |||
| Peri-tumor/Cytoplasmic | 0.306 | 0.429 (0.085-2.169) | |||
OR – odds ratio, CI – confidence interval, #all variables with p < 0.05 were included in the multivariate analysis, $all cases were positive, *statistically significant at p ≤ 0.05
Discussion/Conclusions
The SWI/SNF complex represents a novel link between chromatin remodeling and tumor suppression. Recurrent mutations in subunits of the complex have been identified in various cancers [23]. ARID1A, a subunit of SWI/SNF complexes that controls how much “read access” the cellular transcription machinery has to DNA sequences, can have profound consequences for gene expression, and genes encoding chromatin-remodeling proteins are some of the most frequently mutated genes in human cancer [26, 27]. However, few studies of the association between ARID1Aand hepatocarcinogenesis have been published, and a recent study reported controversial results [32]. Therefore, our purpose was to identify the role of ARID1A as a predictor of outcome in HCC patients after MWA therapy with the intent of identifying the mechanism underlying the suppression of ARID1A.
Our study showed that CTP classification has no significant impact on prognosis of HCC after MWA. A similar observation was made by Wang et al. [41].
The present study showed the significant importance of serum AFP level and size of tumor nodules in predicting HCC recurrence after MWA. Similar observations were also reported by Ma et al. [36], who concluded that tumor > 5 cm in diameter, and higher serum AFP level (> 20 ng/ml) were significant unfavorable prognosticators of progression-free survival after MWA. In contrast, Liang et al. [42] reported that the pre-ablation serum AFP level was not related to the post-MWA prognosis or patient survival.
The current study concluded that the nuclear expression of ARID1A was significantly low in HCC tumor tissues compared to their corresponding non-tumor cirrhotic liver tissues, a finding implicating ARID1A loss in the pathogenesis of HCC. Similar observations were reported by He et al. [33], who performed a study on 64 patient-derived paired HCC and adjacent non-tumorous tissues and found strong ARID1A-positive nuclear staining in non-tumor tissues, whereas ARID1A-negative staining was seen in tumor tissues. These results go hand in hand with the findings of different studies performed on a substantial fraction of additional cancer subtypes where ARID1A mutations or deletions were detected [24-28].
In the present study, a significant inverse correlation was found between tumor ARID1A expression and size of tumors and the larger tumor nodules showed more negative expression than the smaller ones. Also a statistically significant negative relation between tumor nuclear ARID1A expression and pathological grade of the tumor was noted wherein ARID1A negatively expressed lesions tended to have higher grades than the positively expressed ones. These findings are consistent with the results reported by Yan et al. [43], who found that reduced ARID1A expression was associated with tumor infiltration, lymph node metastasis and poor prognosis in patients with gastric carcinoma. In contrast, Abe et al. [44], who demonstrated loss of ARID1A expression in 3.8% of cases, found that altered ARID1A expression was associated with well or moderately differentiated histology, but was associated with larger tumor size, which is accordant with the results of this study.
It was found in the present study that ARIDA1A expression can effectively predict the response to MWA among HCC patients as tumor ARID1A expression was significantly lower in patients who experienced recurrence than those who did not experience recurrence after MWA, which proves the prognostic significance of ARID1A expression. Similar results were obtained by Peng et al. [45]. Consistently, He et al. [33] also found that patients with tumors with low ARID1A expression showed a significantly worse prognosis compared with those with high ARID1A expression. In addition, low ARID1A expression in tumors was significantly correlated with a higher metastatic rate including local lymph node and distant metastases. ARID1A knockdown also promoted HCC cell proliferation, while overexpression of ARID1A inhibited proliferation and impaired clonal formation in HCC cells, but recurrence-free survival and overall survival failed to show a significant correlation with ARID1A expression status [44].
Agreeing with the bad prognosis of ARID1A mutation in HCC, Hu et al. [46] also concluded that ARID1A deficiency occurs in advanced human HCC and was associated with increased vessel density. Mechanistically they found that loss of ARID1A causes aberrant histone H3K27ac deposition at the angiopoietin-2 (Ang2) enhancer and promoter, which eventually leads to ectopic expression of Ang2 and promotes HCC progression.
Recently, Iseda et al. [47] found that 31.7% of the studied HCC samples were negative for ARID1A, and negative ARID1A expression was significantly associated with male sex, high AFP level, large tumor size, high rate of poor differentiation and microscopic intrahepatic metastasis. In addition they revealed that negative ARID1A expression was an independent predictor for recurrence-free survival, overall survival and poor clinical outcome.
In conclusion, the present study postulated that ARID1A expression in cancer cells may decelerate HCC progression and post-treatment tumor recurrence. It is evident that ARID1A gene expression not only serves as an indicator that could distinguish correctly and accurately between patients who are at low and high risk of HCC recurrence after MWA, but also can provide more information about the tumor behavior making the identification of aggressive tumors possible, even with considerable tumor size and number. Collectively, the serum AFP level together with tumor size and ARID1A expression could be used as pre-treatment indicators for the likelihood of tumor recurrence after HCC therapy (especially MWA). Excitingly, ARID1A may be a potential biomarker for the selection of HCC patients for future therapies.
Since ARID1A loss is implicated in the pathogenesis of HCC, it may be recommended that the validity of ARID1A expression to predict HCC aggressiveness and tumor recurrence after MWA should be extensively studied in prospective longitudinal studies with a large-scale population and longer follow-up period including patients with etiologies of chronic liver disease other than HCV infection and the use of other modalities of locoregional treatment of HCC. Furthermore, it is imperative to explore how ARID1A suppresses HCC development and whether ARID1A deficiency could be exploited for therapy to prevent or slow the progression of the disease and improve the patients’ survival.
Acknowledgments
The authors would like to deeply thank Prof. Sherif Shama (who has almost 20 years of experience in diagnostic and interventional radiology) for his great help and support in radiological evaluation of HCC and assessment of response to therapy.
Disclosure
The authors declare no conflict of interest.
References
- 1.Wang H, Naghavi M, Allen C, et al. Global Burden of Disease 2015 Mortality and Causes of Death Collaborators . Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016; 388: 1459-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akinyemiju T, Abera S, Ahmed M, et al. Global Burden of Disease Liver Cancer Collaboration. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol 2017; 3: 1683-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394-424. [DOI] [PubMed] [Google Scholar]
- 4.Bosch FX, Ribes J, Diaz M, et al. Primary liver cancer: worldwide incidence and trends. Gastroenterology 2004; 127 (5 Suppl 1): S5-S16. [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016; 2: 16018. [DOI] [PubMed] [Google Scholar]
- 6.Nakashima O, Kojiro M. Recurrence of hepatocellular carcinoma: multicentric occurrence or intrahepatic metastasis? A view point in terms of pathology. J Hepatobiliary Pancreat Surg 2001; 8: 404-409. [DOI] [PubMed] [Google Scholar]
- 7.Gale PR, Forner A, Llovet JM, et al. European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol 2018; 69: 182-236. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka M, Sato M. Microwave heating of water, ice, and saline solution: molecular dynamics study. J Chem Phys 2007; 126: 034509. [DOI] [PubMed] [Google Scholar]
- 9.Lucchina N, Tsetis D, Ierardi AM, et al. Current role of microwave ablation in the treatment of small hepatocellular carcinomas. Ann Gastroenterol 2016; 29: 460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minami Y, Kudo M. Radiofrequency ablation of hepatocellular carcinoma: Current status. World J Radiol 2010; 2: 417-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, Liang P, Yu X, et al. A comparison of microwave ablation and bipolar radiofrequency ablation both with an internally cooled probe: results in ex vivo and in vivo porcine livers. Eur J Radiol 2011; 79: 124-130. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Zheng Y, Li S, et al. Percutaneous microwave ablation of larger hepatocellular carcinoma. Clin Radiol 2013; 68: 21-26. [DOI] [PubMed] [Google Scholar]
- 13.Tang XY, Wang Z, Wang T, et al. Efficacy, safety and feasibility of ultrasound-guided percutaneous microwave ablation for large hepatic hemangioma. J Dig Dis 2015; 16: 525-530. [DOI] [PubMed] [Google Scholar]
- 14.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet 2012; 379: 1245-1255. [DOI] [PubMed] [Google Scholar]
- 15.Zhang NN, Lu W, Cheng XJ, et al. High-powered microwave ablation of larger hepatocellular carcinoma: evaluation of recurrence rate and factors related to recurrence. Clin Radiol 2015; 70: 1237-1243. [DOI] [PubMed] [Google Scholar]
- 16.Hao K, Luk JM, Lee NP, et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer 2009; 9: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llovet JM, Villanueva A, Lachenmayer A, et al. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol 2015; 12: 408-424. [DOI] [PubMed] [Google Scholar]
- 18.Ally A, Balasundaram M, Carlsen R, et al. Cancer Genome Atlas Research Network. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017; 169: 1327-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson BG, Roberts CWM. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11: 481-492. [DOI] [PubMed] [Google Scholar]
- 20.Dallas PB, Pacchione S, Wilsker D, et al. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol Cell Biol 2000; 20: 3137-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nie Z, Xue Y, Yang D, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol 2000; 20: 8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu RC, Wang TL, Shih IM. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther 2014; 15: 655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012; 44: 694-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shain AH, Giacomini CP, Matsukuma K, et al. Convergent structural alterations define SWItch/Sucrose Non Fermentable (SWI/SNF) chromatin remodeler as a central tumor suppressive complex in pancreatic cancer. Proc Natl Acad Sci 2012; 109: E252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones S, Wang TL, Shih IM, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 2010; 330: 228-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guan B, Mao TL, Panuganti PK, et al. Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am J Surg Pathol 2011; 35: 625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamo A, Cavallone L, Tuzmen S, et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene 2012; 31: 2090-2100. [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 2012; 44: 760-764. [DOI] [PubMed] [Google Scholar]
- 30.Tornesello ML, Buonaguro L, Tatangelo F, et al. Mutations in TP53, CTNNB1 and PIK3CA genes in hepatocellular carcinoma associated with hepatitis B and hepatitis C virus infections. Genomics 2013; 102: 74-83. [DOI] [PubMed] [Google Scholar]
- 31.Huang J, Deng Q, Wang Q, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet 2012; 44: 1117-1121. [DOI] [PubMed] [Google Scholar]
- 32.Sun X, Wang SC, Wei Y, et al. Arid1a has context-dependent oncogenic and tumor suppressor functions in liver cancer. Cancer Cell 2017; 32: 574-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F, Li J, Xu JF, et al. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J Exp Clin Cancer Res 2015; 34: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol 2006; 12: 1175-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 1999; 19: 329-338. [DOI] [PubMed] [Google Scholar]
- 36.Ma S, Ding M, Li J, et al. Ultrasound-guided percutaneous microwave ablation for hepatocellular carcinoma: clinical outcomes and prognostic factors. J Cancer Res Clin Oncol 2017; 143: 131-142. [DOI] [PubMed] [Google Scholar]
- 37.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumours of the digestive system. In: Bosman FT, Carneiro F, Hruban RH, Theise ND (eds.). WHO Classification of Tumours. IARC Press, Lyon: 2010; 217-225. [Google Scholar]
- 38.Edmondson HA, Steiner PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer 1954; 7: 462-503. [DOI] [PubMed] [Google Scholar]
- 39.Li K, Wang RW, Jiang YG, et al. Overexpression of Sox3 is associated with diminished prognosis in esophageal squamous cell carcinoma. Ann Surg Oncol 2013; 20 (Suppl 3): S459-466. [DOI] [PubMed] [Google Scholar]
- 40.Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis 2010; 30: 52-60. [DOI] [PubMed] [Google Scholar]
- 41.Wang ZL, Liang P, Dong BW, et al. Prognostic factors and recurrence of small hepatocellular carcinoma after hepatic resection or microwave ablation: a retrospective study. J Gastrointest Surg 2008; 12: 327-337. [DOI] [PubMed] [Google Scholar]
- 42.Liang P, Wang Y. Microwave ablation of hepatocellular carcinoma. Oncology 2007; 72 (Suppl 1): 124-131. [DOI] [PubMed] [Google Scholar]
- 43.Yan HB, Wang XF, Zhang Q, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis 2014; 35: 867-876. [DOI] [PubMed] [Google Scholar]
- 44.Abe H, Hayashi A, Kunita A, et al. Altered expression of AT-rich interactive domain 1A in hepatocellular carcinoma. Int J Clin Exp Pathol 2015; 8: 2763-2770. [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Y, Gao B, Zhou Z, et al. Hepatocellular carcinoma with ARID1A mutation is associated with higher TMB and poor survival. J Clin Oncol 2020; 38: e16667. [Google Scholar]
- 46.Hu C, Li W, Tian F, et al. Arid1a regulates response to anti-angiogenic therapy in advanced hepatocellular carcinoma. J Hepatol 2018; 68: 465-475. [DOI] [PubMed] [Google Scholar]
- 47.Iseda N, Itoh S, Yoshizumi T, et al. ARID1A deficiency is associated with high programmed death ligand 1 expression in hepatocellular carcinoma. Hepatol Commun 2021; 5: 675-688. [DOI] [PMC free article] [PubMed] [Google Scholar]