Abstract
Transitions to terrestriality have been associated with major animal radiations including land snails and slugs in Stylommatophora (>20 000 described species), the most successful lineage of ‘pulmonates’ (a non-monophyletic assemblage of air-breathing gastropods). However, phylogenomic studies have failed to robustly resolve relationships among traditional pulmonates and affiliated marine lineages that comprise clade Panpulmonata (Mollusca, Gastropoda), especially two key taxa: Sacoglossa, a group including photosynthetic sea slugs, and Siphonarioidea, intertidal limpet-like snails with a non-contractile pneumostome (narrow opening to a vascularized pallial cavity). To clarify the evolutionary history of the panpulmonate radiation, we performed phylogenomic analyses on datasets of up to 1160 nuclear protein-coding genes for 110 gastropods, including 40 new transcriptomes for Sacoglossa and Siphonarioidea. All 18 analyses recovered Sacoglossa as the sister group to a clade we named Pneumopulmonata, within which Siphonarioidea was sister to the remaining lineages in most analyses. Comparative modelling indicated shifts to marginal habitat (estuarine, mangrove and intertidal zones) preceded and accelerated the evolution of a pneumostome, present in the pneumopulmonate ancestor along with a one-sided plicate gill. These findings highlight key intermediate stages in the evolution of air-breathing snails, supporting the hypothesis that adaptation to marginal zones played an important role in major sea-to-land transitions.
Keywords: adaptive radiation, Euthyneura, Gastropoda, heterobranch, Sacoglossa, Siphonaria
1. Introduction
Transitions from marine to terrestrial habitats occurred rarely over the history of animals due to the challenges of adapting to dry land and episodic oxygen limitation, yet ultimately produced spectacular radiations in groups including arachnids, hexapods and tetrapods [1–3]. Freshwater has not typically been an intermediate habitat for lineages shifting from sea to land, but transitional zones along coastal margins such as high intertidal, estuarine or mangrove (hereafter, ‘marginal’) habitats may be key intermediate environments during the evolution of terrestriality [1,4,5]. The ability to respire in both air and water may be favoured for amphibious lineages adapted to marginal habitats, which could facilitate shifts to land [4]. However, little work has formally tested for evolutionary correlations between habitat and traits involved in air-breathing that may drive such shifts and thus promote adaptive radiations.
Gastropods have been exceptionally successful at transitioning from marine to terrestrial habitats, with at least 10 independent origins of terrestriality distributed among subclasses Neritimorpha, Caenogastropoda and Heterobranchia [6,7]. Within Heterobranchia, clade Euthyneura encompasses diverse ‘sea slug’ lineages plus traditional ‘pulmonates', a non-monophyletic group of air-breathing snails and slugs including the explosive radiation of land snails and slugs in Stylommatophora (>20 000 spp.) [8]. Resolving phylogenetic relationships of euthyneurans is essential to resolve the interplay of key innovations and ecological transitions that fuelled this radiation of land gastropods. Traditional but less diverse ‘pulmonate’ groups that may be critical to understanding stylommatophoran success include Hygrophila (approx. 500 spp. [9,10]) from freshwater, and Ellobiida (approx. 250 spp. [11]) and Systellommatophora (approx. 110 spp. [12]) from mangroves and estuaries. A derived euthyneuran clade was named Panpulmonata based on molecular studies, encompassing traditional pulmonates as well as mostly (Acochlidiacea, approx. 50 spp.) or entirely (Pyramidelloidea, >6000 spp.) marine groups [13–15]. Panpulmonates account for over a third of described molluscan species, although this diversity is threatened by human activity [8,9,16,17]. However, relationships among major lineages are unresolved and the early branching order within Panpulmonata remains enigmatic [18–20]. This obscures our understanding of the evolutionary origins of air breathing and terrestriality in panpulmonates, and potentially of lineages that returned to the sea [21,22].
Previous analyses of rDNA-dominated datasets recovered Panpulmonata nested within heterobranch sea slugs as a polytomy comprising: (i) Siphonarioidea, mostly intertidal limpet-like snails; (ii) Sacoglossa, sea slugs that feed suctorially on algae using a uniseriate radula (one tooth per row); and (iii) the remaining panpulmonate groups (electronic supplementary material, figure S1A) [11–15,18–20]. Siphonariids respire using both a plicate gill and a lung with a non-contractile pneumostome [23,24]. Sacoglossa includes shelled superfamily Oxynooidea with a one-sided plicatidium (gill) and unshelled superfamily Plakobranchacea noted for photosynthesis by chloroplast-retaining family Plakobranchoidea and Costasiella [25]. Given the distinctive plicatidium shared by siphonariids and shelled sacoglossans, and the presence of a pneumostome in siphonariids, resolving the relationship of these lineages to other sea slugs and eupulmonates is crucial to reconstruct the evolution of air breathing and habitat transitions in Panpulmonata [20–24,26,27].
Some early analyses of mitochondrial genomes recovered a sister relationship for Sacoglossa and Siphonarioidea (‘Siphoglossa’ hypothesis) nested within a monophyletic ‘Opisthobranchia’ (sea slugs) and Pyramidelloidea grouping with pulmonates (electronic supplementary material, figure S1B) [28,29]. With increased lineage sampling, maximum-likelihood (ML) analyses of mitogenomes supported an ‘inverted’ topology: pulmonates were paraphyletic with respect to a monophyletic Opisthobranchia, while lower heterobranchs and Nudipleura occupied a derived position suggesting a rooting artefact (electronic supplementary material, figure S1C) [30,31]. The first phylogenomic analysis focused on euthyneuran relationships instead recovered Sacoglossa, then Siphonarioidea, as the earliest-branching panpulmonate lineages (electronic supplementary material, figure S1D) [32]. However, in more recent analyses of reduced-representation genomic datasets, Siphonarioidea either nested within traditional pulmonates [33] or was unstable among analyses [34], and Bayesian inference (BI) analyses of mitogenomes reversed the branching order of Siphonarioidea and Sacoglossa (electronic supplementary material, figures S1E–F) [31].
Resolving evolutionary relationships at the root of the panpulmonate radiation will permit comparative analyses of traits that facilitated the transition to non-marine habitats and promoted lineage diversification, including lungs, osmoregulatory systems and direct development [6,11,26,27,35,36]. Here, we greatly expand on prior phylogenomic analyses which included 0–1 siphonariids and 1–3 sacoglossans [32–34], improving taxonomic sampling for these key lineages. We sequenced new transcriptomes from representative lineages in Siphonarioidea (n = 4) and Sacoglossa (n = 34), including the enigmatic Cylindrobulla which has a multiseriate radula like most gastropods, and was variously recovered sister to [37] or within [38,39] Sacoglossa. We generated a new phylogenetic hypothesis for Euthyneura, then performed comparative tests to explore how habitat transitions influenced the evolution of the pneumostome, and how other key traits were gained or lost across this hyperdiverse radiation.
2. Material and methods
(a) . Sample collection
For full methodological details see electronic supplementary material, appendix S1 and tables S1 and S2. Briefly, specimens were collected under permits by the authors or colleagues and identified by morphology and placement in reference phylogenies. Undescribed species were provisionally numbered [39,40]. Specimens were maintained without food for several days to clear their digestive system, preserved in RNAlater and frozen at −80°C prior to extraction. New transcriptomes were generated for sacoglossans representing 24 of 34 widely accepted genera, including two Cylindrobulla spp.; ‘Ercolania boodlea’ is non-Ercolania Trinchese, 1872 [39], and ‘Stiliger sp.’ is of uncertain affinity given a polyphyletic Stiliger [40]. The only unsampled sacoglossan genera with ≥5 spp. were Ascobulla and Ercolania s.s. We included 12 representatives of chloroplast-retaining clade Plakobranchoidea and two Costasiella spp. to test for multiple origins of kleptoplasty. A public transcriptome attributed to Oxynoe viridis [33] was reassigned to O. jacksoni [41] by comparison of barcoding markers. Sampled siphonariids represent divergent lineages (e.g. Williamia) as well as multiple exemplars of species-rich Siphonaria. We also sequenced an unidentified Ganitus sp. (Acochlidiacea) and the nudipleuran Pleurobranchus areolatus, yielding 40 total new transcriptomes.
(b) . Library preparation and sequencing
Total RNA was extracted from tissue using a Trizol protocol (UC Davis) or the Omega Bio-Tek EZNA Mollusc RNA Kit (Univ. of Alabama). RNA concentration was measured with a Qubit 3 fluorometer and purity with a Nanodrop spectrophotometer; RNA quality was assessed from Bioanalyzer traces (Agilent). At UC Davis, cDNA libraries were prepared using the NEBNext Ultra II Directional RNA Kit (New England Biolabs); library concentrations were assessed by qPCR and an equi-molar solution prepared for 150 bp paired-end sequencing on an Illumina HiSeq 4000 (four runs). At the University of Alabama, cDNA libraries were prepared using the Takara SMART-Seq HT Plus kit and sequenced by Illumina NovaSeq.
(c) . Transcriptome assembly and decontamination
After removing adapters and quality trimming reads in Trimmomatic [42], transcriptomes were assembled de novo in Trinity [43]. Quality and completeness of assemblies were assessed against metazoan and eukaryote databases using BUSCO 3.0.2 [44,45]. HiSeq runs were individually decontaminated from ‘barcode hopping’ using CroCo [46].
(d) . Orthology assessment, alignment and matrix construction
Our approach followed the bioinformatic pipeline of [47]. Briefly, original data were combined with public data for 28 panpulmonate taxa (including four sacoglossans and one siphonariid), 22 other heterobranchs and 20 caenogastropods. We used OrthoFinder 2.4.0 [48] to identify putatively orthologous sequences among taxa, removing sequences of <100 amino acids (a.a.) from fasta files and keeping the longest non-redundant sequence. Fasta files sampled for ≥ 75% of taxa were aligned with MAFFT 7.310 [49], putatively mistranslated regions removed with HmmCleaner [50] and alignments trimmed to remove ambiguously aligned regions with BMGE 1.12.2 [51]. Approximately ML trees were constructed for each alignment with FastTree 2 [52], and PhyloPyPruner 0.9.5 (https://pypi.org/project/phylopypruner) was used to identify strictly orthologous sequences. We also selected the best 600 and 900 genes based on seven properties calculated in genesortR [53] and analysed those subsampled datasets separately from the complete 1 160-gene dataset.
(e) . Phylogenomic analyses
We initially performed ML analyses with IQ-Tree 2 [54] on the three datasets using the best-fitting model of a.a. substitution for each partition (-m MFP), assessing topological support with 1000 rapid bootstraps. A second ML approach used the PMSF model [55] in IQ-TREE 2, fitting profile mixture models (C20, C40, C60) to each supermatrix, with a guide tree generated for each analysis using the best-fitting site-homogeneous model for each partition. A final ML analysis implemented GHOST [56], an edge-unlinked mixture model accommodating heterotachy among lineages without data partitioning. The four sets of branch lengths were inspected individually. Analyses were performed on the University of Alabama High-Performance Computing cluster (MFP) or using the CIPRES web portal (PMSF, GHOST) [57].
Phylogenetic analyses using BI were performed via CIPRES with PhyloBayes-MPI 4 [58] on the 1160 gene dataset, using site-heterogeneous models to account for site-specific a.a. frequencies. The first analysis ran four chains for up to 2400 cycles using the CAT-f81 (Poisson) model, with four gamma-distributed rate classes [59]. We assessed convergence using the tracecomp command and by inspecting outputs in Tracer 1.7 [60]. One chain was discarded because outgroup taxon Semisulcospira grouped with pulmonates for one-third of the run, impeding convergence and mixing. We combined the last 50% of trees from three chains, generated a 50% majority-rule consensus tree and determined posterior probability (Pp) of nodes. A second BI analysis used the more computationally intensive CAT-GTR + G model, running four chains for 1200 cycles each. One chain became trapped in a local minimum and was excluded; parameters for the remaining three chains showed acceptable convergence (effective size > 100, relative difference < 0.3) although chains stabilized on alternative topologies with respect to the sister group of Stylommatophora. As placement of these taxa did not affect our primary conclusions, results of BI analyses were deemed interpretable [59]; the final 600 trees from each chain were pooled to generate a consensus tree and Pp scores.
Evolutionary relationships were also evaluated under the multi-species coalescent (MSC) using ASTRAL [61] and FASTRAL [62], which infer species trees from gene trees in contrast with concatenation approaches. Nodal support was assessed by bootstrapping (ASTRAL) or local Pp (both methods).
(f) . Comparative analyses
Trait evolution was modelled in BayesTraits 3.0.2 [63], correcting for phylogenetic uncertainty with the post-burnin sample from the CAT-GTR analysis. Characters were coded based on morphological analyses of sampled taxa or their closest congener in published datasets (electronic supplementary material, table S3). For ancestral state reconstruction (ASR) and model-fitting tests, priors were drawn from an exponential distribution generated using a hyperprior. MCMC chains were run for 5 × 107 iterations, sampling every 105 iterations and discarding the first 20% as burnin. Marginal likelihoods were estimated using the stepping-stone sampler, running 200 stones for 2 × 105 iterations and performing three runs per analysis. A reverse-jump (RJ) MCMC approach was used to reduce the sampled set of model parameters [64]. Using log-Bayes Factor (BF) tests, we compared support for alternative models by taking twice the difference between the median log (L) marginal likelihood (lk) score of three runs and treating values >2 as positive evidence and >5 as strong evidence favouring one model [65].
Habitat was coded as freshwater, marine, terrestrial (grouped as ‘stable’ for binary analyses) or ‘marginal’ (estuarine, mangrove or high intertidal zones with substantial aerial exposure, sensu [5]). Ancestral habitat was reconstructed across all key nodes, treating a node as confidently reconstructed if the median Pp for one state was twice the null probability (25% given four states); ‘transitional’ branches connect nodes with different inferred states [66]. Support for alternative states at select nodes was also compared by log-BF tests.
Fit of constrained versus unconstrained models was compared to test whether transitions to terrestrial or freshwater habitat occurred more rapidly from marginal habitat. A covarion model was supported over homogeneous evolution (log-BF = 3.9), so transition rates among habitats were first estimated by fitting an RJ Multistate + Covarion model. Unconstrained model fit was compared to models constraining different pairs of transition rates to be equal: (i) marginal to terrestrial (qET) = marine to terrestrial (qMT) habitats and (ii) marginal to freshwater (qEF) = marine to freshwater (qMF) habitats; reduced support for a constrained model indicates those transition rates are not equal.
To test for correlated evolution between habitat and the pneumostome (a key character in the evolution of air breathing), independent versus dependent models were fit to two binary traits: (i) habitat, coded as marginal versus stable and (ii) pneumostome present/absent. A homogeneous-rates RJ-dependent model was used, given weak support for a covarion model (log-BF = 1.0) [64]. Fit of an unconstrained dependent model was also compared to a dependent model with rates of pneumostome gain constrained to be equal in both habitats (q12 = q34), testing whether marginal habitat was a selective force in pneumostome evolution.
ASRs for additional morphological characters (electronic supplementary material, table S3) were also performed to identify cases of parallel trait evolution.
3. Results and discussion
(a) . Phylogenomic relationships in Euthyneura
The ≥75% taxon-occupancy matrix consisted of 1160 genes with 170 277 amino acid positions and 73.6% matrix occupancy. Based on genesortR results (electronic supplementary material, figure S2), we compared analyses of the complete matrix with the 600 and 900 best genes. Except where noted, all ML and BI analyses (n = 15) of concatenated datasets yielded highly congruent tree topologies. For complete results, see electronic supplementary material, appendix S2. Within Caenogastropoda, relationships among major lineages were well resolved and varied little among analyses, except ML and BI analyses recovered different relationships within Neogastropoda among Buccinoidea, Muricidae and Conidae. In Heterobranchia, all but 10 nodes received complete support in every analysis (figure 1; electronic supplementary material, figures S3–S6). All ML analyses supported the sampled lower heterobranch lineages as sister to a monophyletic Euthyneura, a clade not recovered in recent studies [33,34,67]; only two analyses returned less than complete support for Euthyneura: PMSF-C40 analysis of the best 600 genes (99%) and GHOST (88%) (electronic supplementary material, figure S4). However, both BI analyses recovered the lower heterobranchs as sister to Nudipleura with full support (electronic supplementary material, figure S5; see also [33]), whereas coalescent analyses did not resolve the placement of the lower heterobranchs (electronic supplementary material, figure S6A). All analyses fully supported the monophyly of Nudipleura (Nudibranchia + Pleurobranchaea), in contrast with [32], as well as Tectipleura (Euopisthobranchia + Panpulmonata) and Euopisthobranchia (figure 1; electronic supplementary material, figures S3–S6) (see also [33,34,67]).
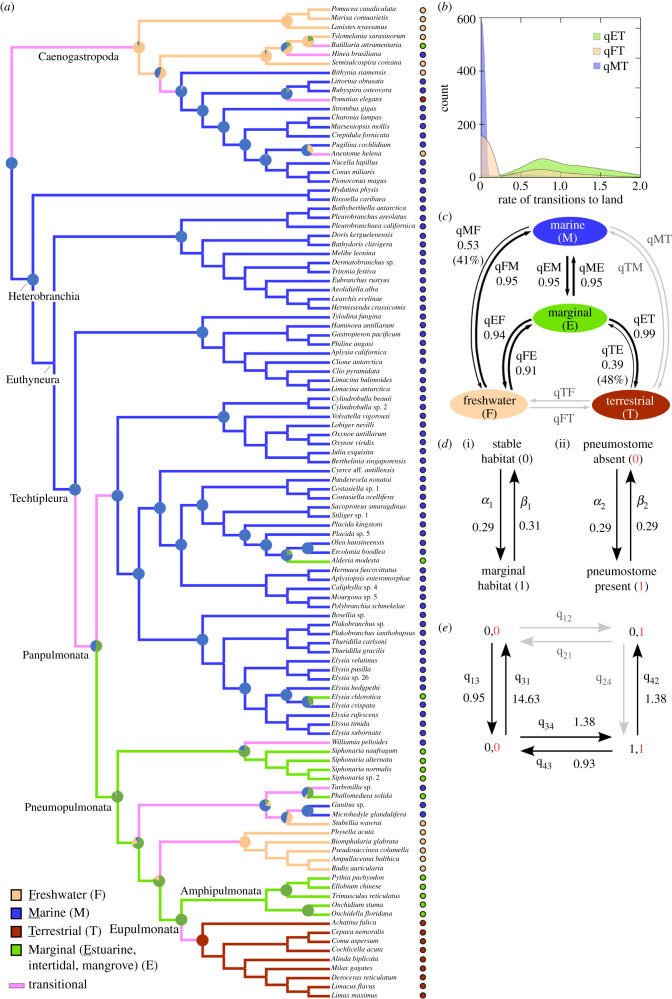
Figure 1.
Phylogeny of Euthyneura based on amino acid sequences from 1160 nuclear protein-coding genes, showing ML topology and branch lengths from the PMSF-C60 analysis. All nodes received complete support in four analyses of the concatenated dataset unless values are given (order per inset box); asterisk = 100% BS or 1.0 Pp support; dash = non-significant (BS < 70%, Pp < 0.9). Some families or genera collapsed where indicated. Bolded terminal names indicate original transcriptomes sequenced for this study (including members of collapsed lineages).
Sacoglossa was recovered as sister to the remaining panpulmonate taxa in all partitions of the dataset using ML analysis (figure 1; electronic supplementary material, figures S3–S4), in BI analyses (electronic supplementary material figure S5), and in coalescent analyses (electronic supplementary material figure S6). No analysis supported Siphoglossa (Siphonarioidea + Sacoglossa), or any alternative sister group for Sacoglossa. Traditional suborders Oxynooidea (shelled) and Plakobranchacea (shell-less) were separated by branch lengths exceeding those distinguishing major panpulmonate lineages; superfamilies Limapontioidea (bearing dorsal cerata) and Plakobranchoidea were sister groups in all analyses (figure 1; electronic supplementary material, figure S4B, S5–S7). Most shell-less taxa are currently placed in superfamily Plakobranchoidea [68] because prior molecular studies recovered a paraphyletic Limapontioidea [25,39–40]. Our results support the monophyly of Limapontioidea, the traditional clade name for predominantly ceratiform families, and Plakobranchacea, the traditional clade name for Limapontioidea + Plakobranchoidea (electronic supplementary material, figure S7A) [37,69]. Contrary to the hypothesis that Cylindrobullidae was sister to Sacoglossa [33], all analyses recovered Cylindrobulla sister to the remaining shelled sacoglossans (figure 1; electronic supplementary material, figure S7). Among shell-less taxa, family Caliphyllidae was polyphyletic: most analyses recovered Cyerce (cerata lacking digestive glands) as sister to the remaining taxa in Limapontioidea. Placida was also non-monophyletic, highlighting the need for systematic revision of Sacoglossa (see electronic supplementary material, appendix S2 for full results).
Most ML analyses recovered Siphonarioidea as sister to the remaining panpulmonate lineages with ≥95% support (figure 1), except for the PMSF-C20 (<70%) and PMSF-C60 analyses (85%) of the best 900 genes (electronic supplementary material, figure S3), and the GHOST analysis (79%). Under the GHOST model, three sets of branch lengths returned topologies congruent with the ‘main’ topology from partitioned ML analyses (figure 1; electronic supplementary material, figure S4); in the 4th set, evolutionary rates were tenfold slower and major panpulmonate lineages formed a polytomy, suggesting this subset of sites evolves too slowly to resolve sampled gastropod diversity. All BI analyses recovered Siphonarioidea sister to the remaining ‘pulmonates’ (sensu [70]) (electronic supplementary material, figure S5), but coalescent analyses failed to resolve the phylogenetic placement of Siphonarioidea (electronic supplementary material, figure S6). No analysis supported alternative relationships proposed for Siphonarioidea, such as Basommatophora (Siphonarioidea + Hygrophila + Amphiboloidea) or Thalassophila (Siphonarioidea + Amphiboloidea). Within Siphonarioidea, the subtidal Williamia was sister to a clade comprising the four high intertidal Siphonaria spp.
Our concatenated analyses are the first since [32] to support Siphonarioidea as sister to the remaining panpulmonate lineages aside from Sacoglossa, echoing some morphological analyses [26,27]. Our findings contrast with studies in which a single siphonariid exemplar nested among ‘pulmonates’ [33] or was unstable across data partitions [34]. Presently, there is no agreed-upon clade name for traditional ‘pulmonates’ plus affiliated groups (Pyramidelloidea, Acochlidiacea) [70]. We propose Pneumopulmonata for the clade comprising all descendants of the last common ancestor (LCA) shared by Siphonarioidea, Amphiboloidea and Stylommatophora, to reflect our findings that the LCA of Pneumopulmonata possessed a pneumostome (see below) as do extant representatives of those lineages.
The remaining panpulmonate lineages formed a clade with a core topology consistently recovered across ML and BI analyses (figure 1; ‘main topology’, electronic supplementary material, figure S3B). Most ML analyses supported (Pyramidelloidea + Amphiboloidea) as sister to Acochlidiacea (electronic supplementary material, figure S3A), although these relationships were unresolved in BI and coalescent analyses (electronic supplementary material, figures S5 and S6). We could not fully test the monophyly of Pylopulmonata as transcriptomic data for Glacidorboidea, a potential sister group to Pyramidelloidea, were unavailable [13,34]. Hygrophila (freshwater snails) was fully supported in all analyses and was sister to Eupulmonata with full support in MFP and PMSF analyses of the complete dataset (figure 1; electronic supplementary material, figure S3A), and with strong support in 4 of 8 ML analyses using 600 or 900 loci (electronic supplementary material, figure S3A); under the GHOST model (electronic supplementary material, figure S4); in BI analyses (figure 1; electronic supplementary material, figure S5) and under the MSC (electronic supplementary material, figure S6). Within Hygrophila, the three species of Lymnaeidae formed a clade in all analyses, with Physidae sister to (Lymnaeidae + Planorbidae), consistent with other recent studies [10]. Finally, the major subclade ((Pyramidelloidea + Amphiboloidea) + Acochlidiacea) was recovered as sister to (Hygrophila + Eupulmonata) in all ML analyses of the complete dataset with ≥99% support and with strong support in some analyses of the best 600 genes (figure 1; electronic supplementary material, figure S3). These relationships were also recovered in analyses under the GHOST (electronic supplementary material, figure S4) and CAT + GTR models (electronic supplementary material, figure S5A), albeit without significant support.
Eupulmonata was fully supported as monophyletic in all analyses. ML analyses of the complete dataset strongly supported Stylommatophora sister to Amphipulmonata [70], in which Ellobiida and Systellommatophora (primarily from marginal habitats) were sister groups (≥98%) (figure 1). Amphipulmonata was also supported in five of eight ML analyses using the best 600 or 900 genes (electronic supplementary material, figure S3E), under GHOST (electronic supplementary material, figure S4B), and by ASTRAL (electronic supplementary material, figure S6B). BI analyses either recovered Ellobiida sister to Stylommatophora (CAT + f81) or a polytomy (CAT + GTR) (electronic supplementary material, figure S5). Geophila (Systellommatophora + Stylommatophora) was weakly supported in only one analysis (electronic supplementary material, figure S3C). The sister relationship of Achatina to the rest of Stylommatophora and other internal relationships were fully supported in every analysis.
(b) . Habitat shifts and correlated evolution of the pneumostome
For the LCA of Panpulmonata, Pp support was almost evenly divided between marine and marginal habitat, but comparative tests favoured a marine ancestor (log-BF = 4.1). Marginal habitat was strongly supported for the LCA of Pneumopulmonata and Eupulmonata (figure 2a), consistent with [5]. Transitions to terrestriality occurred more often from marginal (qET) than from marine (qMT) or freshwater (qFT) habitats, with the latter two rates frequently set to 0 by RJ MCMC chains (figure 2b). The only likely path to terrestriality occurred through marginal habitat, with median values of qMT = qFT = 0 based on the pooled posterior sample of three runs (figure 2c). An unconstrained model was favoured over a model constraining qET = qMT (log-BF = 3.3), indicating terrestriality originated more often from marginal habitat than from a marine ancestor. Marginal habitat was thus a key evolutionary intermediate, facilitating transitions to all other habitat types at roughly equal rates; in contrast, most transition rates between stable habitats were much lower (qMF) or approximately 0 (qMT/TM, qFT/TF) (figure 2c).
Figure 2.
Comparative analyses of habitat and pneumostome evolution in Euthyneura. (a) Cladogram plotting Pp for four habitat states on key nodes; ML topology and branch lengths from figure 1. Branches joining nodes with the same ancestral state (ASR Pp ≥ 0.5) were coloured as retaining that state; branches joining nodes differing in ASR were coded ‘transitional’. (b) Frequency kernels for transition rates (95% CI) sampled for shifts to terrestriality from marginal (qET), freshwater (qFT) or marine (qMT) habitat. (c) Transition rates among habitats, with line width proportional to median rate; % of time rate = 0 given in parentheses, unless < 15% (not shown) or ≥ 50% (greyed out; median = 0). (d) Independent model of evolution for two binary traits: (i) stable versus marginal habitat; (ii) pneumostome present versus absent. Median rate estimates were pooled from three runs. (e) Model of dependent evolution supporting an evolutionary correlation between marginal habitat and gaining a pneumostome. Median transition rates given (black) unless assigned to the zero bin >80% of the time (grey; median = 0). (Online version in colour.)
Habitat shifts occurred frequently near the base of the pneumopulmonate radiation. For instance, each habitat type was represented in the lineage comprising Acochlidiacea (mostly marine, some freshwater) + Pyramidelloidea (marine, parasitic) + Amphiboloidea (estuarine and mangrove). We did not sample Glacidorboidea (freshwater), which in prior analyses grouped with Pyramidelloidea and Amphiboloidea as Pylopulmonata [34]. Notably, our ASRs support a reversion to marine habitat for Pyramidelloidea (approx. 6000 spp.), which is more speciose than non-marine pylopulmonate lineages and could be an exception to the rule that secondarily marine lineages generally diversify less than non-marine sister groups [21]. Improved taxonomic sampling of Pylopulmonata should confirm this result and explore the combination of ecological and life-history traits (parasitism, specialization, dispersal) that expanded pyramidelloidean diversity.
A vascularized pallial cavity protected by a pneumostome may have been a key pre-adaptation favouring the colonization of terrestrial biomes by Stylommatophora [1,6,26]. A pneumostome was strongly favoured (log-BF = 6.8) in the LCA of Pneumopulmonata. Positive evidence favoured a correlation between habitat and pneumostome evolution (figure 2d–e); dependent models were favoured whether all transition rates were estimated independently (log-BF = 4.9) or reduced models were sampled by RJ MCMC (log-BF = 2.0). Based on transition rate estimates, transitions to marginal habitat preceded, and favoured, gaining a pneumostome. RJ chains assigned three rates to the zero bin >80% of the time: gaining or losing a pneumostome in stable habitats (q12 and q21); and shifts into marginal habitat after evolving a pneumostome (q24) (figure 2e; electronic supplementary material, figure S8). An unconstrained RJ-dependent model was strongly favoured over a model constraining q34 = q12 (log-BF = 5.0), supporting more frequent gains of a pneumostome in marginal habitat. These findings indicate marginal habitats were occupied before the pneumostome evolved (q13»q12), a key morphological change that in turn preceded the evolution of contractile pneumostomes and air-breathing in several lineages. A pneumostome also arose in marginal environments (q34 > 0) but not in other habitats (q12 ≈ 0), suggesting marginal habitat was a selective force behind the narrowing of the pallial opening.
(c) . Morphological trait evolution in Euthyneura
Our phylogenetic hypothesis provides a new framework for analysing character evolution across Euthyneura. For instance, the plicatidium (a derived gill) is two-sided in euopisthobranchs, whereas shelled sacoglossans have a one-sided plicatidium considered homologous with that of siphonariids, which also possess a pneumostome (non-contractile in most species) [23,24,26,27]. Log-BF tests favoured a one-sided over a two-sided plicatidium in the LCAs of both Panpulmonata and Pneumopulmonata (log-BF = 2.5; electronic supplementary material, figure S9), although ASRs differed in whether a plicatidium was more likely than no gill at all. The evolution of gill structure may thus be important to the tectipleuran radiation, culminating in a pneumopulmonate ancestor with both a one-sided gill and a pneumostome.
Other traits showed a high degree of evolutionary lability. Most sacoglossans have a uniseriate radula (one tooth per row), a presumed synapomorphy, whereas Cylindrobulla has a multiseriate radula [37,38]. ASR supported a uniseriate radula as the likely ancestral state for the LCAs of Sacoglossa and of Oxynooidea, but with Pp < 1.0, and ‘uniseriate’ was not favoured over ‘multiseriate’ for either node (log-BF < 1) (electronic supplementary material, figure S7A). It thus remains ambiguous if multiple origins of ‘uniseriate’ are favoured over a reversal to the multiseriate state in Cylindrobulla. A non-photosynthetic LCA was strongly favoured for all unshelled sacoglossans (Plakobranchacea; log-BF = 6.9) and for Limapontioidea (log-BF = 7.6), supporting independent origins of kleptoplasty in Costasiella versus Plakobranchiidae (electronic supplementary material, figure S7B); however, transitions could not be placed on branches due to ambiguous ASRs.
An operculum is present in larvae of most sampled gastropods, and most adult caenogastropods, but few adult heterobranchs. ASR supported at least three re-acquisitions of an adult operculum within Heterobranchia: Rissoellidae in the lower Heterobranchia; a subset of thecostome pteropods in Euopisthobranchia; and (Pyramidelloidea + Amphiboloidea) in Panpulmonata (electronic supplementary material, figure S10). The operculate Architectonicidae may represent an additional origin but exemplars were excluded due to phylogenetic instability, hence character states at the base of Heterobranchia could not be robustly inferred. The operculate Glacidorbidea was unsampled but likely shares a common origin with other pylopulmonates [34].
Within Eupulmonata, stalked eyes were traditionally considered a synapomorphy of Geophila (Stylommatophora + Systellommatophora), but our analyses did not recover Geophila and strongly supported ‘eyes at base of tentacles' in the LCA of Eupulmonata (log-BF = 8.5); thus, stalked eyes likely evolved independently in Stylommatophora and Systellommatophora. Globineurons in the procerebrum may be a synapomorphy for Eupulmonata [26], but many other characters are shared by eupulmonates and early branching groups including a dorsal jaw (Siphonarioidea) and gizzard pouch (Hygrophila, Amphiboloidea). Most character states shared by Pyramidelloidea and other panpulmonates are inferred to be plesiomorphic in Heterobranchia, but a re-examination of the CNS of Acochlidiacea and Pyramidelloidea is warranted to assess whether the procerebrum and dorsal bodies are possible synapomorphies for Pneumopulmonata [26,71]. Overall, our ASRs support frequent homoplasy consistent with the historical challenges identifying synapomorphies for major panpulmonate clades, highlighting the dynamic evolution of morphology in this radiation [19,26,27,70].
4. Conclusion and prospectus for future study
A pallial cavity with a pneumostome and a one-sided plicate gill were supported as plesiomorphic for the diverse radiation we term Pneumopulmonata, with Siphonarioidea as the sister taxon of the remaining lineages. Together with evidence that marginal habitat preceded and favoured the evolution of the pneumostome, our findings support the hypotheses that marginal environments facilitated transitions from sea to land, and that amphibious ancestors adapted to respire in both air and water were key intermediates. As shifts onto land triggered major adaptive radiations in several animal lineages, these results are important to understand general processes that contributed significantly to animal diversification. Adaptation to terrestriality depends partly on lineage-specific pre-adaptations (e.g. the specific air-breathing organ: book gills evolving into book lungs, and repeated evolution of tracheae in arthropods; a respiratory pharynx evolving into the tetrapod lung; a vascularized pallial cavity with a pneumostome evolving into the stylommatophoran lung [3]). However, our work highlights the initial drivers that promoted such change and allowed several groups to exploit niches available on dry land with remarkable success.
Gastropods repeatedly evolved terrestriality in several clades. Given this evolutionary replication, future work should sample all terrestrial neritomorph and caenogastropod lineages as well as related taxa from representative habitat types. This will facilitate tests of whether pre-adaptation to marginal habitat was important in repeated shifts from sea to land. Within Panpulmonata, present-day diversity is low for groups adapted to the stressful and fluctuating conditions of marginal zones (e.g. Siphonarioidea, Ellobiida and Onchidiidae), yet such environments facilitated transitions to stable biomes in which Hygrophila (approx. 500 spp., freshwater), Pyramidelloidea (>6000 spp., marine) and Stylommatophora (>20 000 spp., terrestrial) radiated. Pyramidelloidea may be secondarily marine, a rare case of diversification rate increasing after a return to the sea. Overall, the extraordinary diversity of Panpulmonata results from a complex interplay of pre-adaptation, ecological opportunity and the outcome of species-level selection triggered by lineage-specific traits; however, this radiation also provides broad insights into the evolutionary mechanisms that may have generally promoted animal diversity as lineages transitioned onto land.
Supplementary Material
Acknowledgements
We thank P. Bouchet, A. Labuda and N. Nakata for field assistance; A. Falconer, M. Middlebrooks and H. Moeller for samples; J. Goodheart for assistance collecting P. areolatus; and two reviewers whose feedback greatly improved this paper.
Ethics
All samples were collected with the permission of the state or country, including Special Activity Licenses from the State of Florida (P.J.K. and K.M.K.), Scientific Collecting Permits from the State of California (P.J.K. and S.C.) and a collecting permit from the state of Hawaii (P.J.K.). The specimens from New Caledonia were collected during the ‘Our Planet Reviewed’ New Caledonia expedition (2016–2019), a joint project of the Muséum national d'Histoire Naturelle (Paris) and Conservatoire d'espaces Naturels (CEN) de Nouvelle-Calédonie, operating under a permit issued by Direction du Développement Economique et de l'Environnement (DDEE) of Province Nord.
Data accessibility
All raw data files, output files from all analyses, and relevant code are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.q83bk3jkk [72]. All raw DNA sequences are available through the NCBI SRA database (electronic supplementary material, tables S1 and S2 [73]).
Authors' contributions
P.J.K.: conceptualization, formal analysis, funding acquisition, investigation, project administration and writing—original draft; S.A.C.: data curation, funding acquisition, investigation and writing—review and editing; K.A.: data curation, funding acquisition and investigation; K.T.: data curation, funding acquisition, investigation and writing—review and editing; A.A.V.: funding acquisition, investigation and writing—review and editing; R.W.: investigation and writing—review and editing; N.L.W.S.W.: investigation and writing—review and editing; D.J.E.: funding acquisition, investigation and writing—review and editing; K.M.K.: conceptualization, data curation, formal analysis, funding acquisition, investigation, project administration and writing—original draft.
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by Lounsbery Foundation, Gouvernement de la Nouvelle-Calédonie, Province Nord, Agence Française de la Biodiversité (AFB), US National Science Foundation (grant nos. ACI 1548562, DEB 1355230, DEB 1846174, HRD 1807387 and PRFB 1907177), Office des Postes et Télécommunications (OPT), CSU Council on Ocean Affairs, Science & Technology (grant no. GDP-2017-005) and U.S. National Institutes of Health (grant no. GM08228).
References
- 1.Vermeij GJ, Dudley R. 2000. Why are there so few evolutionary transitions between aquatic and terrestrial ecosystems? Biol. J. Linn. Soc. 70, 541-554. ( 10.1111/j.1095-8312.2000.tb00216.x) [DOI] [Google Scholar]
- 2.Ward P, Labandeira C, Laurin M, Berner RA. 2006. Confirmation of Romer's Gap as a low oxygen interval constraining the timing of initial arthropod and vertebrate terrestrialization. Proc. Natl Acad Sci. USA 103, 16 818-16 822. ( 10.1073/pnas.0607824103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsia CC, Schmitz A, Lambertz M, Perry S, Maina JN. 2013. Evolution of air breathing: oxygen homeostasis and the transitions from water to land and sky. Comp. Physiol. 3, 849-915. ( 10.1002/cphy.c120003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapman EG, Przhiboro AA, Harwood JD, Foote BA, Hoeh WR. 2012. Widespread and persistent invasions of terrestrial habitats coincident with larval feeding behavior transitions during snail-killing fly evolution (Diptera: Sciomyzidae). BMC Evol. Biol. 12, 1-22. ( 10.1186/1471-2148-12-175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klussmann-Kolb A, Dinapoli A, Kuhn K, Streit B, Albrecht C. 2008. From sea to land and beyond–new insights into the evolution of euthyneuran Gastropoda (Mollusca). BMC Evol. Biol. 8, 1-16. ( 10.1186/1471-2148-8-57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponder WF, Lindberg DR, Ponder JM. 2020. Biology and evolution of the Mollusca. Boca Raton, FL: CRC Press. [Google Scholar]
- 7.Mordan P, Wade C. 2008. Heterobranchia II: the Pulmonata. In Phylogeny and evolution of the Mollusca (eds Ponder WF, Lindberg DR), pp. 409-426. Berkeley, CA: University of California Press. [Google Scholar]
- 8.Rosenberg G. 2014. A new critical estimate of named species-level diversity of the recent Mollusca. Amer. Malacol. Bull. 32, 308-322. ( 10.4003/006.032.0204) [DOI] [Google Scholar]
- 9.Strong E, Gargominy O, Ponder W, Bouchet P. 2008. Global diversity of gastropods (Gastropoda; Mollusca) in freshwater. Hydrobiologia 595, 149-166. ( 10.1007/s10750-007-9012-6) [DOI] [Google Scholar]
- 10.Saadi AJ, Davison A, Wade CM. 2020. Molecular phylogeny of freshwater snails and limpets (Panpulmonata: Hygrophila). Zool. J. Linn. Soc. 190, 518-531. ( 10.1093/zoolinnean/zlz177) [DOI] [Google Scholar]
- 11.Romero PE, Pfenninger M, Kano Y, Klussmann-Kolb A. 2016. Molecular phylogeny of the Ellobiidae (Gastropoda: Panpulmonata) supports independent terrestrial invasions. Mol. Phylogen. Evol. 97, 43-54. ( 10.1016/j.ympev.2015.12.014) [DOI] [PubMed] [Google Scholar]
- 12.Dayrat B. 2009. Review of the current knowledge of the systematics of Onchidiidae (Mollusca: Gastropoda: Pulmonata) with a checklist of nominal species. Zootaxa 2068, 1-26. ( 10.11646/zootaxa.2068.1.1) [DOI] [Google Scholar]
- 13.Jörger KM, Stöger I, Kano Y, Fukuda H, Knebelsberger T, Schrödl M. 2010. On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evol. Biol. 10, 323. ( 10.1186/1471-2148-10-323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrödl M, Jörger K, Klussmann-Kolb A, Wilson NG. 2011. Bye bye ‘Opisthobranchia’! A review on the contribution of mesopsammic sea slugs to euthyneural systematics. Thalassas 27, 101-112. [Google Scholar]
- 15.Kano Y, Neusser T, Fukumori H, Jörger KM, Schrödl M. 2015. Sea-slug invasion of the land. Biol. J. Linn. Soc. 116, 253-259. ( 10.1111/bij.12578) [DOI] [Google Scholar]
- 16.Bouchet P, Bary S, Héros V, Marani G. 2016. How many species of molluscs are there in the world's oceans, and who is going to describe them? In Tropical deep-sea benthos, vol. 29 (eds Héros V, Strong E, Bouchet P), pp. 9-24. Paris, France: Muséum national d'Histoire naturelle. [Google Scholar]
- 17.Lydeard C, et al. 2004. The global decline of nonmarine mollusks. BioScience 54, 321-330. ( 10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2) [DOI] [Google Scholar]
- 18.Schrödl M, Jörger KM, Wilson NG. 2011. A reply to Medina et al. (2011): crawling through time: transition of snails to slugs dating back to the Paleozoic based on mitochondrial phylogenomics. Mar. Genomics 4, 301-303. ( 10.1016/j.margen.2011.07.003) [DOI] [PubMed] [Google Scholar]
- 19.Dinapoli A, Klussmann-Kolb A. 2010. The long way to diversity: phylogeny and evolution of the Heterobranchia (Mollusca: Gastropoda). Mol. Phylogenet. Evol. 55, 60-76. ( 10.1016/j.ympev.2009.09.019) [DOI] [PubMed] [Google Scholar]
- 20.Dayrat B, Conrad M, Balayan S, White T, Albrecht C, Golding R, Gomes S, Harasewych M, de Frias Martins AM.. 2011. Phylogenetic relationships and evolution of pulmonate gastropods (Mollusca): new insights from increased taxon sampling. Mol. Phylogenet. Evol. 59, 425-437. ( 10.1016/j.ympev.2011.02.014) [DOI] [PubMed] [Google Scholar]
- 21.Grosberg RK, Vermeij GJ, Wainwright PC. 2012. Biodiversity in water and on land. Curr. Biol. 22, R900-R903. ( 10.1016/j.cub.2012.09.050) [DOI] [PubMed] [Google Scholar]
- 22.Davison A. 2002. Land snails as a model to understand the role of history and selection in the origins of biodiversity. Popul. Ecol. 44, 129-136. ( 10.1007/s101440200016) [DOI] [Google Scholar]
- 23.Hodgson AN. 1999. The biology of siphonariid limpets (Gastropoda: Pulmonata). Oceanogr. Mar. Biol. Ann. Rev. 37, 245-314. [Google Scholar]
- 24.De Villiers CJ, Hodgson AN.. 1987. The structure of the secondary gills of Siphonaria capensis (Gastropoda: Pulmonata). J. Mollusc. Stud. 53, 129-138. ( 10.1093/mollus/53.2.129) [DOI] [Google Scholar]
- 25.Christa G, Händeler K, Kück P, Vleugels M, Franken J, Karmeinski D, Wägele H. 2015. Phylogenetic evidence for multiple independent origins of functional kleptoplasty in Sacoglossa (Heterobranchia, Gastropoda). Org. Divers. Evol. 15, 23-36. ( 10.1007/s13127-014-0189-z) [DOI] [Google Scholar]
- 26.Barker GM. 2001. Gastropods on land: phylogeny, diversity and adaptive morphology. In The biology of terrestrial Molluscs (ed. Parker GM), pp. 1-146. Wallingford, UK: CABI Publishing. [Google Scholar]
- 27.Dayrat B, Tillier S. 2002. Evolutionary relationships of euthyneuran gastropods (Mollusca): a cladistic re-evaluation of morphological characters. Zool. J. Linn. Soc. 135, 403-470. ( 10.1046/j.1096-3642.2002.00018.x) [DOI] [Google Scholar]
- 28.Grande C, Templado J, Zardoya R. 2008. Evolution of gastropod mitochondrial genome arrangements. BMC Evol. Biol. 8, 1-5. ( 10.1186/1471-2148-8-61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medina M, Lal S, Vallès Y, Takaoka TL, Dayrat BA, Boore JL, Gosliner T. 2011. Crawling through time: transition of snails to slugs dating back to the Paleozoic, based on mitochondrial phylogenomics. Mar. Genomics 4, 51-59. ( 10.1016/j.margen.2010.12.006) [DOI] [PubMed] [Google Scholar]
- 30.White TR, Conrad MM, Tseng R, Balayan S, Golding R, de Martins AMF, Dayrat BA. 2011. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evol. Biol. 11, 295. ( 10.1186/1471-2148-11-295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varney RM, Brenzinger B, Malaquias M, Meyer C, Schrödl M, Kocot KM. 2021. Assessment of mitochondrial genomes for heterobranch gastropod phylogenetics. BMC Ecol. Evol. 21, 1-14. ( 10.1186/s12862-020-01734-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kocot KM, Halanych KM, Krug PJ. 2013. Phylogenomics supports Panpulmonata: opisthobranch paraphyly and key evolutionary steps in a major radiation of gastropod molluscs. Mol. Phylogen. Evol. 69, 764-771. ( 10.1016/j.ympev.2013.07.001) [DOI] [PubMed] [Google Scholar]
- 33.Zapata F, Wilson NG, Howison M, Andrade SC, Jörger KM, Schrödl M, Goetz FE, Giribet G, Dunn CW. 2014. Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc. R. Soc. B 281, 20141739. ( 10.1098/rspb.2014.1739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teasdale LC. 2017. Phylogenomics of the pulmonated land snails. PhD dissertation, University of Melbourne. [Google Scholar]
- 35.Ruthensteiner B. 1997. Homology of the pallial and pulmonary cavity of gastropods. J. Moll. Stud. 63, 353-367. ( 10.1093/mollus/63.3.353) [DOI] [Google Scholar]
- 36.Collin R, Wise JB. 1997. Morphology and development of Odostomia columbiana Dall and Bartsch (Pyramidellidae): implications for the evolution of gastropod development. Biol. Bull. 192, 243-252. ( 10.2307/1542718) [DOI] [PubMed] [Google Scholar]
- 37.Jensen K. 1996. Phylogenetic systematics and classification of the Sacoglossa (Mollusca, Gastropoda, Opithobranchia). Phil. Trans. R. Soc. Lond. B 351, 91-122. ( 10.1098/rstb.1996.0006) [DOI] [Google Scholar]
- 38.Mikkelsen PM. 1998. Cylindrobulla and Ascobulla in the western Atlantic (Gastropoda, Opisthobranchia, Sacoglossa): systematic review, description of a new species, and phylogenetic reanalysis. Zool. Scripta 27, 49-71. ( 10.1111/j.1463-6409.1998.tb00428.x) [DOI] [Google Scholar]
- 39.Krug PJ, et al. 2015. Species selection favors dispersive life histories in sea slugs, but higher per-offspring investment drives shifts to short-lived larvae. Syst. Biol. 64, 983-999. ( 10.1093/sysbio/syv046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krug PJ, Wong NL, Medina M, Gosliner T, Valdés AA. 2018. Cryptic speciation yields remarkable mimics: a new genus of sea slugs that masquerade as toxic algae (Caulerpa spp.). Zool. Scripta 47, 699-713. ( 10.1111/zsc.12310) [DOI] [Google Scholar]
- 41.Krug PJ, Berriman J, Valdés AA. 2018. Phylogenetic systematics of the shelled sea slug genus Oxynoe Rafinesque, 1814 (Heterobranchia: Sacoglossa), with integrative descriptions of seven new species. Invert. Syst. 32, 950-1003. ( 10.1071/IS17080) [DOI] [Google Scholar]
- 42.Faircloth BC. 2013. Illumiprocessor: a trimmomatic wrapper for parallel adapter and quality trimming. See 10.6079/J9ILL (accessed 4 November 2016). [DOI]
- 43.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494-1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva E, Zdobnov E. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210-3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 45.Waterhouse RM, Seppey M, Simão F, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva E, Zdobnov E. 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543-548. ( 10.1093/molbev/msx319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simion P, Belkhir K, François C, Veyssier J, Rink JC, Manuel M, Philippe H, Telford MJ. 2018. A software tool ‘CroCo' detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 16, 1-9. ( 10.1186/s12915-018-0486-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kocot KM, et al. 2017. Phylogenomics of Lophotrochozoa with consideration of systematic error. Syst. Biol. 66, 256-282. [DOI] [PubMed] [Google Scholar]
- 48.Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. ( 10.1186/s13059-019-1832-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nuc. Acids Res. 33, 511-518. ( 10.1093/nar/gki198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Franco A, Poujol R, Baurain D, Philippe H.. 2019. Evaluating the usefulness of alignment filtering methods to reduce the impact of errors on evolutionary inferences. BMC Evol. Biol. 19, 1-17. ( 10.1186/s12862-019-1350-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Criscuolo A, Gribaldo S. 2010. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 1-21. ( 10.1186/1471-2148-10-210) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490. ( 10.1371/journal.pone.0009490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koch NM. 2021. Phylogenomic subsampling and the search for phylogenetically reliable loci. Mol. Biol. Evol. 38, 4025-4038. ( 10.1093/molbev/msab151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R.. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530-1534. ( 10.1093/molbev/msaa015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang HC, Minh BQ, Susko E, Roger AJ. 2018. Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 67, 216-235. ( 10.1093/sysbio/syx068) [DOI] [PubMed] [Google Scholar]
- 56.Crotty SM, Minh BQ, Bean NG, Holland BR, Tuke J, Jermiin LS, von Haeseler A.. 2020. GHOST: recovering historical signal from heterotachously-evolved sequence alignments. Syst. Biol. 69, 249-264. [DOI] [PubMed] [Google Scholar]
- 57.Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O'Leary MA. 2015. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol. Bioinform. 11, 43-48. ( 10.4137/EBO.S21501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lartillot N, Rodrigue N, Stubbs D, Richer J. 2013. PhyloBayes MPI: phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611-615. ( 10.1093/sysbio/syt022) [DOI] [PubMed] [Google Scholar]
- 59.Lartillot N. 2020. PhyloBayes: Bayesian phylogenetics using site-heterogeneous models. In Phylogenetics in the genomic era (eds Scornavacca C, Delsuc F, Galtier N), pp. 1.5:1-1.5:16. See https://hal.archives-ouvertes.fr/hal-02535342. [Google Scholar]
- 60.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901-904. ( 10.1093/sysbio/syy032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sayyari E, Mirarab S. 2016. Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 33, 1654-1668. ( 10.1093/molbev/msw079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dibaeinia P, Bordbar ST, Warnow T. 2021. FASTRAL: improving scalability of phylogenomic analysis. Bioinformatics 37, 2317-2324. ( 10.1093/bioinformatics/btab093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53, 673-684. ( 10.1080/10635150490522232) [DOI] [PubMed] [Google Scholar]
- 64.Pagel M, Meade A. 2006. Bayesian analysis of correlated evolution of discrete characters by reversible-jump Markov chain Monte Carlo. Am. Nat. 167, 808-825. ( 10.1086/503444) [DOI] [PubMed] [Google Scholar]
- 65.Raftery AE. 1996. Hypothesis testing and model selection. In Markov chain Monte Carlo in practice (eds Gilks WR, Richardson S, Spiegelhalter D), pp. 163-185. Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- 66.Kubo T, Sakamoto M, Meade A, Venditti C. 2019. Transitions between foot postures are associated with elevated rates of body size evolution in mammals. Proc. Natl Acad Sci. USA 116, 2618-2623. ( 10.1073/pnas.1814329116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cunha TJ, Giribet G. 2019. A congruent topology for deep gastropod relationships. Proc. R. Soc. B 286, 20182776. ( 10.1098/rspb.2018.2776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bouchet P, Rocroi JP, Hausdorf B, Kaim A, Kano Y, Nützel A, Parkhaev P, Schrödl M, Strong EE. 2017. Revised classification, nomenclator and typification of gastropod and monoplacophoran families. Malacologia 61, 1-526. ( 10.4002/040.061.0201) [DOI] [Google Scholar]
- 69.Marcus EDB. 1982. Systematics of the genera of the order Ascoglossa (Gastropoda). J. Moll. Stud. 48 (supl. 10), 1-31. [Google Scholar]
- 70.Schrödl M. 2014. Time to say ‘Bye-bye Pulmonata’. Spixiana 37, 161-164. [Google Scholar]
- 71.Haszprunar G, Huber G. 1990. On the central nervous system of Smeagolidae and Rhodopidae, two families questionably allied with the Gymnomorpha (Gastropoda: Euthyneura). J. Zool. 220, 185-199. ( 10.1111/j.1469-7998.1990.tb04302.x) [DOI] [Google Scholar]
- 72.Krug PJ, Caplins SA, Algoso K, Thomas K, Valdés ÁA, Wade R, Wong NLWS, Eernisse DJ, Kocot KM. 2022. Data from: Phylogenomic resolution of the root of Panpulmonata, a hyperdiverse radiation of gastropods: new insight into the evolution of air breathing. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- 73.Krug PJ, Caplins SA, Algoso K, Thomas K, Valdés ÁA, Wade R, Wong NLWS, Eernisse DJ, Kocot KM. 2022. Phylogenomic resolution of the root of Panpulmonata, a hyperdiverse radiation of gastropods: new insight into the evolution of air breathing. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Krug PJ, Caplins SA, Algoso K, Thomas K, Valdés ÁA, Wade R, Wong NLWS, Eernisse DJ, Kocot KM. 2022. Data from: Phylogenomic resolution of the root of Panpulmonata, a hyperdiverse radiation of gastropods: new insight into the evolution of air breathing. Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- Krug PJ, Caplins SA, Algoso K, Thomas K, Valdés ÁA, Wade R, Wong NLWS, Eernisse DJ, Kocot KM. 2022. Phylogenomic resolution of the root of Panpulmonata, a hyperdiverse radiation of gastropods: new insight into the evolution of air breathing. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All raw data files, output files from all analyses, and relevant code are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.q83bk3jkk [72]. All raw DNA sequences are available through the NCBI SRA database (electronic supplementary material, tables S1 and S2 [73]).