Abstract
Bladder cancer is one of the most frequent cancers among males, and a poor survival rate reflects problems with aggressiveness and chemo-resistance. Accumulating evidence indicates that SIRT1 is involved in bladder cancer tumorigenesis and is positively associated with chemo-resistance and poor prognosis. We recently synthesized water-soluble chemical derivatives of heliomycin, an antibiotic from Streptomyces resistomycificus, and demonstrated that they possess anticancer properties. In this present study, we used the cellular thermal shift assay (CETSA) in T24 bladder cancer cells to show that heliomycin (designated compound (H1)) and its 4-(tert-butylamino)methyl derivative (HD2) directly engaged with SIRT1 in the native cellular environment, whereas another derivative (HD3) did not. Upon binding, heliomycin downregulated SIRT1 protein expression without altering its transcript level, and subsequently induced autophagy. Interestingly, the derivative (HD2) triggered apoptosis. The interaction between SIRT1 protein and heliomycin or its derivatives was also speculated by a molecular docking simulation, suggesting heliomycin (H1) and derivative (HD2) acting with the different binding modes to SIRT1. Given the increased water-solubility, hydrogen bonds were found on Ala262 and Ile347 residues in the docked complex of derivative (HD2) to produce more steady interaction and initiate signaling pathways that were not observed in the case of heliomycin. Meanwhile, it is evident that derivative (HD3) did not engage with SIRT1 by CETSA or molecular docking studies, nor did it downregulate SIRT1 expression. Taken together, these findings clearly show that SIRT1 is targeted and downregulated by heliomycin and its water-soluble 4-aminomethylated derivative (HD2) possibly through autophagic and/or proteasomal degradation, leading to cell death and growth suppression of T24 bladder cancer cells.
Keywords: Apoptosis; autophagy; bladder cancer; cellular thermal shift assay; heliomycin; resistomycin; silent mating type information regulation 1 (Sirtuin 1, SIRT1)
Introduction
The updated figures on the global cancer load from GLOBOCAN 2020 approximate that 19.3 million new cancer cases and nearly 10 million cancer deaths occurred in 2020. Bladder cancer ranked as the 10th most frequently identified cancer internationally, representing roughly 573,000 new cases and 213,000 deaths in 2020 [1]. Bladder cancer is more common in men than in women, ranking as the 6th most prevalent cancer disease in men [1]. Although most cases are initially recognized as having non-muscle-invasive bladder cancer, many patients eventually develop muscle-invasive tumors that display aggressive and metastatic clinical behavior even after treatment [2]. At present, surgical procedures and cisplatin-based chemotherapy represent the standard of care for bladder cancer. Unfortunately, however, chemo-resistance often evolves, leading to poor prognosis in patients [3,4]. Thus, researchers continue to seek effective treatments for bladder cancer.
Silent mating type information regulation 1 (Sirtuin 1, SIRT1) is a mammalian homolog of yeast silent information regulator 2. It belongs to the sirtuin protein family of histone deacetylases (HDACs), which target histones and nonhistone proteins for post-translational modifications to critically modulate many cellular functions under metabolic and environmental stresses [5-11]. Emerging evidence suggests that SIRT1 overexpression is positively associated with poor prognosis in different types of solid tumors, including liver, lung, breast, prostate, and bladder cancers [12-15]. Interestingly, metabolic reprogramming through SIRT1 was shown to upregulate GLUT1 and promote glycolysis and cell proliferation in bladder cancer cells, while the application of a SIRT1-specific inhibitor produced opposite effects on glucose uptake and cell growth [16]. Studies identified a complex regulatory network involving a peroxisome proliferator-activated receptor-γ (PPARγ)-SIRT1 feedback loop and its downstream targets, further supporting the idea that SIRT1 plays key roles in the tumorigenesis of bladder cancer cells [17,18]. The importance of SIRT1 in bladder cancer tumorigenesis and progression was further supported by a loss-of-function study showing that SIRT1 deficiency attenuated bladder cell proliferation and migration, and also increased cell cycle arrest through FOXO3a-related pathways [19]. Moreover, a positive correlation was established between the chemo-sensitivity of bladder cancer and the level of miR-34a, which is a downstream effector of p53 that targets and represses SIRT1 and Cdk6 [20]. Together, these lines of evidence indicate that SIRT1 critically impacts cellular processes and could be a promising therapeutic target for bladder cancer management [21].
Heliomycin (also known as resistomycin) is an antibiotic that was initially isolated from marine sponges. It is generated by Streptomyces resistomycificus as a secondary metabolite, and reportedly possesses different activities, including anticancer activities [22-27]. Given that heliomycin was previously suggested to be an HDAC inhibitor [28] and SIRT1 belongs to the class III HDACs, we hypothesized that heliomycin could obstruct SIRT1 in the context of cancer. Heliomycin exhibits extremely low solubility in an aqueous solution, limiting its medical applications. To improve the aqueous solubility of heliomycin, we previously used two new synthetic approaches to generate a series of aminoalkyl derivatives of heliomycin [29,30]. Importantly, heliomycin and some of its novel derivatives exhibited comparable cytotoxicities against a wide range of cancer cell lines, including drug-resistant variants [29]. We further validated that the introduction of aminoalkyl side chains to the parental compound enhanced the affinity of the derivatives to DNA secondary structures, compared to that of heliomycin. This resulted in attenuation of topoisomerase 1 (Top1) activity and induction of apoptosis by the water-soluble derivatives [29,30]. However, the anticancer mechanism of heliomycin remained unknown.
In the present study, we show that heliomycin (designated compound H1 herein) and one of the derivatives tested herein (HD2, but not HD3) directly targeted SIRT1, as assessed by cellular thermal shift assay (CETSA) and a molecular docking simulation. Upon direct binding, heliomycin suppressed SIRT1 expression to enhance autophagic influx and autophagy in bladder cancer cells. In contrast, as suggested in our previous study [29], the water-soluble 4-(tert-butylamino)methyl derivative (HD2) targeted SIRT1 to provoke apoptosis in bladder cancer cells.
Materials and methods
Cell culture and reagents
The anti-SIRT1, anti-Atg5, anti-p53, anti-acetyl-p53, anti-phosphorylated mTOR, anti-mTOR, anti-phosphorylated Akt, anti-PARP, anti-Bak, anti-Puma, anti-Noxa, anti-Bcl2, anti-c-Flip, anti-c-Myc, anti-cleaved caspase-3, and anti-caspase-8 antibodies were purchased from Cell Signaling Technology, Inc. (Beverly, MA, USA). The anti-β-actin and anti-acetyl-c-Myc antibodies were from Millipore Corp. (Temecula, CA, USA). The anti-Akt antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The anti-atg6/Beclin 1, anti-Atg7, and anti-LC3 antibodies were obtained from Novus Biologicals (Centennial, CO, USA). Other chemicals were purchased from the Sigma Chemical Company (St. Louis, MO, USA), unless otherwise specified.
The T24 cell line was established from a human urinary bladder cancer patient with high-grade and invasive transitional cell carcinoma and was grown in RPMI medium (The Bioresource Collection and Research Center, BCRC, Hsinchu, Taiwan). Media were supplemented with 10% FBS, 100 units/ml penicillin, and 50 µg/ml streptomycin. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 in air, and the media were replaced every 2-3 days. Cells were treated with different concentrations of compounds, dissolved in DMSO (for heliomycin) or water (for HD2 or HD3), or with the same volume of vehicle control.
Chemistry
Heliomycin (H1) was generated at Gause Institute of New Antibiotics (Moscow, Russian Federation) using Actynomyces variabilis var. heliomycini [22]. 4-((tert-butylamino)methyl)-heliomycin hydrochloride (HD2) and 4-(2-(S)-(ethoxycarbonyl)pyrrolidine-1-yl)methyl)-heliomycin hydrochloride (HD3) were prepared by the previously reported method [29]. The structure of heliomycin and its derivatives are illustrated in Figure 1. The purities of the tested compounds of H1, HD2, and HD3 were 97, 99, and 99%, respectively, as examined by HPLC analysis.
Figure 1.
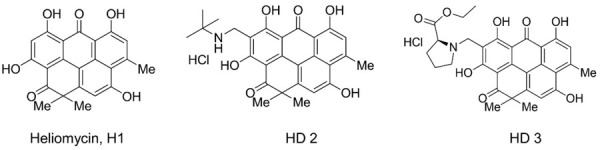
Structures of heliomycin (H1) and aminomethylated derivatives (HD2 and HD3).
Continuous observation of cell proliferation by cell impedance determinations
For continuous monitoring of changes in cell proliferation, cells (104 cells/well) were plated onto E-plates and incubated for 30 min at room temperature, after which E-plates were placed onto the xCELLigence System (Roche, Mannhein, Germany). Cells were grown overnight before being exposed to heliomycin (dissolved in DMSO) or derivatives (dissolved in water) and impedance was measured every hour, as previously described [31]. Cell impedance is defined by the cell index (CI) = (Zi - Z0) [Ohm]/15 [Ohm], where Z0 is background resistance and Zi is the resistance at an individual time point. A normalized cell index was determined as the cell index at a certain time point (CIti) divided by the cell index at the normalization time point (CInml_time).
Analysis of the sub-G1 phases
In brief, after treatments, 106 cells were collected and washed in PBS, slowly fixed in 75% ethanol, and kept at -20°C overnight. Next, the cell pellet was washed with PBS, centrifuged at 500× g for 5 min, then resuspended in 200 μl cold PBS, and then the nuclear DNA was stained with propidium iodide (PI) solution (20 mM Tris pH 8.0, 1 mM NaCl, 0.1% NP-40, 1.4 mg/mL RNase A, 0.05 mg/mL PI) for 30 min on ice in the dark. The total cellular DNA content was analyzed with an FC500 flow cytometer (Beckman Coulter Inc. Brea, CA, USA).
Autophagy determination
Autophagosomes-acidic intracellular compartments that mediate the degradation of cytoplasmic materials during autophagy-were visualized by staining with Acridine Orange (AO; Sigma Chemical Co.). After incubation, cells were washed with PBS and stained with 2 mg/ml AO for 10 min at 37°C. AO-stained cells were then washed, trypsinized, and analyzed using a Beckman Coulter FC500. The results are expressed as a percentage of total cells.
Reverse transcriptase-polymerase chain reaction (RT-PCR)
Total RNA from bladder cancer cells was isolated using the TRIzol reagent (Gibco, Carlsbad, CA, USA). First-strand cDNA was synthesized from 1 μg of total RNA using Superscript II (Life Technologies, Rockville, MD, USA). The following primers sets were used for PCR amplifications: SIRT1, 5’-GCCAGTGGATTCGCTCTTT-3” (sense) and 5’-GCTCTATCCTCCTCATCACTTTCAC-3’ (antisense), and β-actin, 5’-ACTCACCTTGGTGGTGCATA-3’ (sense) and 5’-ACACCTTGATGGGAAAGGTGG-3’ (antisense). The reaction conditions consisted of 30 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min, followed by a final extension of 5 min at 72°C. The obtained PCR products were resolved by 1.4% agarose gels electrophoresis and visualized by ethidium bromide staining.
Cellular target identification of heliomycin by cellular thermal shift assay (CETSA)
Intracellular SIRT1 as a cellular target of heliomycin and its derivatives were established by CETSA. Samples were prepared from control and compound-exposed cells. For each set, 2×107 cells were seeded in a 10-cm cultured dish. After 24 h of culture, the cells were pretreated with 10 μM MG132 for 1 h, washed with PBS, treated with trypsin, and collected. Samples were centrifuged at 12,000 rpm for 3 min at room temperature, the pellets were gently resuspended with 1 mL of PBS, and the samples were centrifuged at 7,500 rpm for 3 min at room temperature. The pellets were resuspended with 1 mL of PBS containing 20 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 2 mM phenylmethylsulfonyl fluoride (PMSF), 10 ng/ml leupeptin, and 10 μg/ml aprotinin. The samples were transferred to Eppendorf tubes and subjected to three freeze-thaw cycles; for each cycle, they were exposed to liquid nitrogen for 3 min, placed in a heating block at 37°C for 3 min, and vortexed briefly. For the experimental sample set, heliomycin or its derivative was added to a final concentration of 20 μM; the same volume of vesicle solvent was added. The samples were heated at 37°C for 1 h and dispensed to 100 μl aliquots. Pairs consisting of one control aliquot and one experimental aliquot were heated at 40°C, 43°C, 46°C, 49°C, 52°C, 55°C, 58°C, 61°C, or 67°C for 3 min. Insoluble proteins were separated by centrifugation at 12,000 rpm for 30 min at 4°C, and the supernatants with soluble proteins were used for SDS-PAGE and Western blot analysis using commercially available SIRT1 antibodies. β-actin was used as the control.
Immunoblotting
Cell extracts were prepared in lysis buffer (20 mM Tris-HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 2 mM phenylmethylsulfonyl fluoride (PMSF), 10 ng/ml leupeptin, 10 μg/ml aprotinin). Volumes of extract containing equal amounts of proteins (40 µg) were applied to SDS-PAGE gels, and resolved proteins were transferred to PVDF membranes (Schleicher & Schuell, Keene, NH, USA). The membranes were blocked, washed, and probed with primary antibodies. After washing to remove unbound primary antibody, membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 2 hours. The blots were washed again and developed using enhanced chemiluminescence (ECL) reagents, according to the manufacturer’s protocol (Amersham Biosciences, Piscataway, NJ, USA).
Molecular docking simulation
A crystal structure of the SIRT1 catalytic domain bound to an EX527 analog (PDB: 4I5I) [32] was employed for the molecular docking study. EX-527 is a nanomolar SIRT1 inhibitor with an IC50 value as low as 38 nM [33], and therefore its co-crystal structure of SIRT1 protein is suitable in the docking analysis. The water molecule and ligand molecule of the initial crystal structure were removed using the PyMOL program (https://pymol.org/2021) to prepare for the docking analysis. Molecular docking was performed using the AutoDock Vina package [34] in the PyRx software [35] to assess the probably binding modes of heliomycin and its derivatives in the SIRT1 catalytic domain. The docking site was determined according to the inhibitor binding site of EX527 analog in the crystal structure of SIRT1, as the setting in the grid selection. Each compound was optimized in molecular geometry, torsional barriers, and intermolecular-interaction geometry using the MMFF94 forcefield in CHARMM [36]. The best pose of each compound was chosen based on the lowest binding affinity calculated in kcal/mol by AutoDock Vina, for further interaction analyses. The post-docking analyses were conducted using the LigPlot+ software [37] to identify the ligand-protein hydrogen bonds and hydrophobic contacts. The docking results were visualized using the PyMOL program.
Statistics
All data are expressed as the means ± SEs of three independent experiments. The significance of differences between control and treatment groups was calculated using a one-way ANOVA.
Results
Heliomycin and its water-soluble derivatives attenuate cell proliferation in bladder cancer cells
We previously reported that heliomycin and its 4-aminomethylated derivatives exhibited anticancer activity against several cancer cell lines, including drug-resistance lines [29]. Cellular function studies and protein analyses indicated that the cytotoxicity of the water-soluble aminomethylated derivatives arose from their apoptotic activity [29]. However, the mechanisms underlying the cytotoxicity of heliomycin itself remained unknown. Among the first series of derivatives, heliomycin (H1) and the 4-(tert-butylamino)methyl derivative (HD2) demonstrated high antiproliferative potency on HCT116 p53-null and K562 cells, but not with the 4-(2-(S)-(ethoxycarbonyl)pyrrolidine-1-yl)methyl derivative (HD3) in our previous study (Figure 1) [29]. Here, we explored the molecular mechanisms underpinning the antitumor activity of these three compounds against bladder cancer cells. Analysis with an electrical impedance-based xCELLigence System showed that heliomycin (H1) and its water-soluble 4-(tert-butylamino)methyl derivative (HD2) exhibited considerable cytotoxicity at 1 μM in T24 bladder cancer cells, as evidenced by reduced cell index values (Figure 2A and 2B). However, the derivative (HD3) showed less cytotoxicity compared to the other two compounds against T24 bladder cancer cells (Figure 2C).
Figure 2.
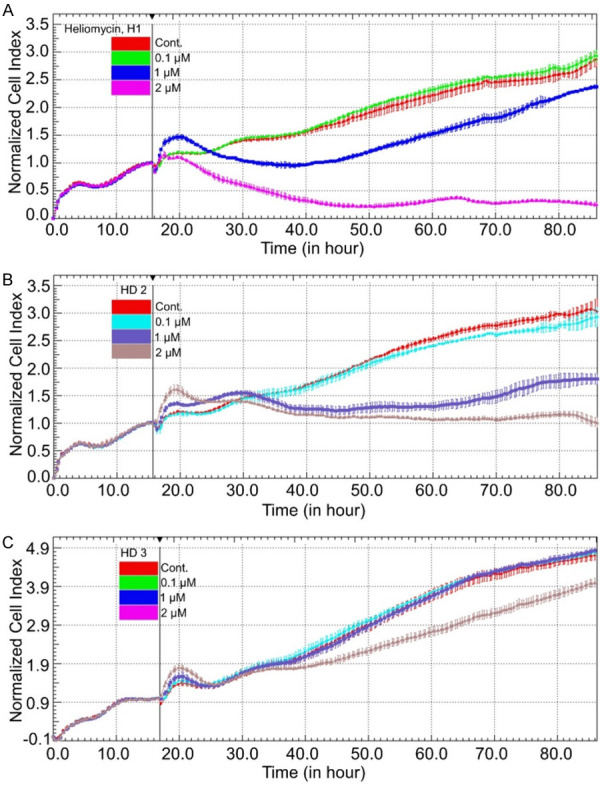
Heliomycin and its aminomethylated derivative suppress the growth of bladder cancer cells. Cell proliferation was dynamically monitored by impedance measurements in T24 cells, as described in the Materials and Methods. Shown are normalized cell index values measured. Cells were treated with various concentrations of heliomycin (H1) (A), water-soluble aminomethylated derivatives (HD2) (B), and (HD3) (C).
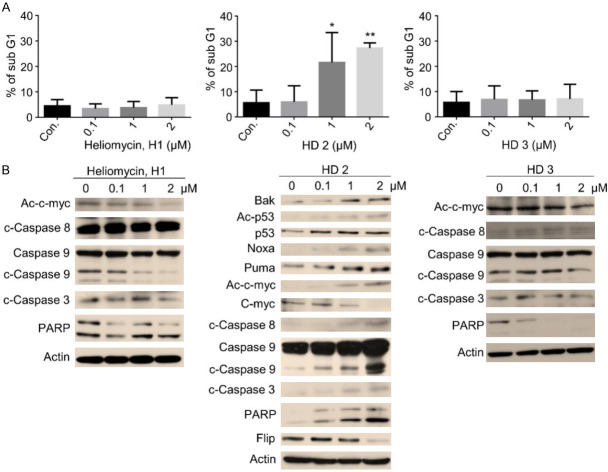
Since the aminomethylated derivative (HD2), but not heliomycin (H1), was previously demonstrated to induce apoptosis [29], we assessed whether cell death contributed to the cytotoxicity of these compounds in bladder cancer cells. Consistent with our previous report, the water-soluble derivative (HD2), but not heliomycin (H1) or (HD3), induced marked changes in the sub-G1 population percentage assessed by PI staining and flow cytometry, indicating that apoptosis was induced (Figure 3A). Furthermore, protein analysis showed that derivative (HD2) enhanced the expression levels of pro-apoptotic Bak, acetylated p53/p53, Noxa, Puma, cleaved caspase-8, 9, and 3, and cleaved PARP, all indicative of apoptosis induction (Figure 3B). The anticancer activity of the derivative (HD2) was also supported by the downregulation of anti-apoptotic Flip in these bladder cancer cells (Figure 3B). However, these indications were not observed in cells treated with heliomycin (H1) or derivative (HD3) (Figure 3B).
Figure 3.
The aminomethylated derivative (HD2) preferentially provokes apoptosis in T24 cancer cells. Cells were exposed to compounds at various concentrations for 24 hours. A. The percentage of sub-G1 cells was determined by flow cytometry, and the results are presented as the percentage of sub-G1 cells. Values (means ± SDs) are from at least three independent experiments (*P<0.05, **P<0.01). B. T24 cells were treated with compounds, DMSO (vesicle for heliomycin), or H2O (vesicle for the derivatives HD2 and HD3) for various concentrations, and aliquots of cell lysates were resolved by SDS-PAGE and analyzed for protein expression by Western blotting. β-actin was used as an internal loading control to monitor for equal loading. Representative images are provided from at least three independent experiments.
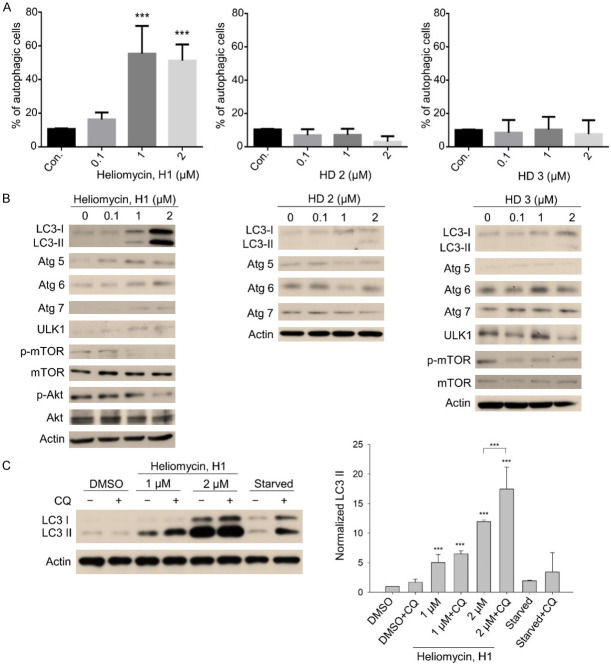
To explore other types of cells death, we utilized AO staining, which detects autophagolysosomes, to determine whether the tested compounds could induce autophagy. Interestingly, our results suggested that heliomycin triggered autophagy, whereas derivatives (HD2) and (HD3) did not (Figure 4A). Western blot analyses illustrated that heliomycin-mediated autophagy was associated with upregulation of the autophagy-related proteins, Atg 5, 6, 7, and ULK1, along with cleaved LC-3 II, and downregulation of the phosphorylation/activation of mTOR and Akt (Figure 4B). The changes in these protein markers were much less obvious in cells treated with derivative (HD2) and (HD3) compared to those of their controls (Figure 4B). We also examined the autophagic influx using the autophagosome-lysosome binding inhibitor, chloroquine (CQ). As expected, heliomycin exposure markedly enhanced LC-3 II expression compared to that seen in control cells (Figure 4B and 4C). Western blot analysis showed that co-treatment with CQ and heliomycin further augmented the level of LC-3 II beyond that seen in the heliomycin-alone group, indicating an increased accumulation of autophagosomes and enhancement of autophagic flux (Figure 4C). These lines of evidence support the idea that heliomycin provokes autophagy while the water-soluble derivative (HD2) induces apoptosis in a bladder cancer cell line.
Figure 4.
Heliomycin (H1) favorably induces autophagy in T24 cancer cells. Cells were exposed to compounds at various concentrations for 18 hours. A. Autophagy was determined by AO staining using flow cytometry, analysis, and the results are expressed as a percentage of autophagic cells. Values (means ± SDs) are from at least three independent experiments (***P<0.001). B. T24 cells were treated with the individual compound, DMSO (vesicle for heliomycin), or H2O (vesicle for the derivatives) for various concentrations, and aliquots of cell lysates were resolved by SDS-PAGE and analyzed for protein expression by Western blotting. β-actin was used as an internal loading control to monitor for equal loading. Representative images are provided from at least three independent experiments. C. T24 cells were exposed to DMSO or heliomycin for 18 hours, and the autophagosome-lysosome binding inhibitor CQ was added to the culture at 50 μM 2 hours before harvesting cells. Aliquots of cell lysates were then resolved by SDS-PAGE and analyzed for protein expression by Western blotting. β-actin was used as an internal loading control to monitor for equal loading. Representative images are provided from at least three independent experiments. The expression of LC-3 II was normalized to the expression of β-actin and values (means ± SDs) are from at least three independent experiments (***P<0.001).
Cellular target identification of heliomycin or its water-soluble 4-aminomethylated derivative (HD2) with SIRT1 by CETSA
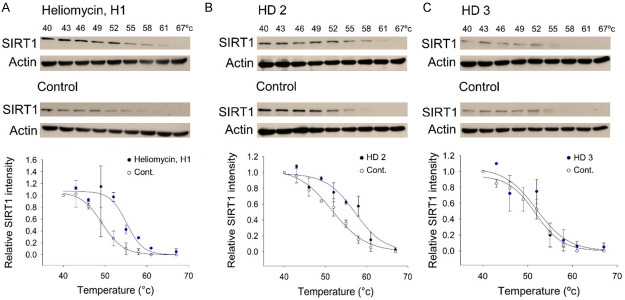
To validate SIRT1 as a potential protein target of heliomycin, we used the cellular thermal shift assay (CETSA), which has emerged as a powerful label-free method to assess target engagement for compounds in the native cellular environment [38-40]. Since ligand binding thermally stabilizes a target protein, the relative intensity of SIRT1 protein identified by Western blot was plotted against the temperature to generate CETSA melting curves. Based on these curves, we calculated the melting temperature, T m, as the temperature at which 50% of SIRT1 proteins were precipitated. Our results demonstrated that heliomycin treatment increased the T m from 49.3°C (control) to 55.1°C (heliomycin (H1)-treated), indicating that there was direct binding between SIRT1 and heliomycin in T24 bladder cells (Figure 5A). The aminomethylated derivative (HD2) was also found to target SIRT1, as evidenced by T m increasing from 52.1°C (control) to 57.7°C (HD2-treated) (Figure 5B). On the contrary, the CETSA melting curve of derivative (HD3)-exposed cells was similar to that of the control, suggesting that there was no direct binding between this compound and SIRT1 (Figure 5C).
Figure 5.
CETSA-based identification of direct binding between SIRT1 protein and heliomycin and its derivatives. Top panel: the immunoblot intensity of SIRT1 in T24 cells in the presence and absence of heliomycin (H1) (A), derivative (HD2) (B), and derivative (HD3) (C) in the CETSA experiments as described in the Material and Methods. Aliquots of cell lysates were resolved by SDS-PAGE and analyzed for protein expression by Western blotting. β-actin was used as an internal loading control to monitor for equal loading. Representative images are shown. Bottom panel: CETSA-melting curves of SIRT1 in the presence and absence of heliomycin (H1) (A), HD2 (B), and HD3 (C) in T24 cells. The immunoblot intensity was normalized to the intensity of the 40°C samples.
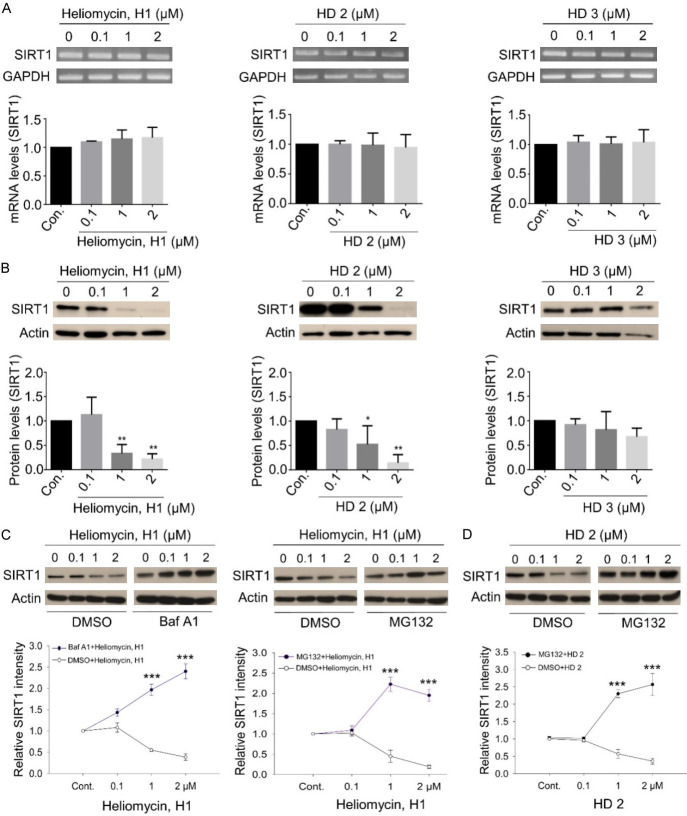
We next evaluated how the binding of heliomycin or its derivatives affected SIRT1 expression at the protein and transcript levels. Our data suggested that the transcript level of SIRT1 was not significantly affected by heliomycin or its derivatives in T24 cells (Figure 6A). In contrast, protein analysis indicated that treatment with 1 or 2 μM heliomycin (H1) or derivative (HD2) markedly attenuated SIRT1 protein expression, whereas no such effect was seen for the other derivative (HD3) (Figure 6B). To further validate these results, we showed that the pretreatment with the autophagy inhibitor bafilomycin A1 (Baf A1) markedly reversed SIRT1 downregulation, indicating that the autophagic degradation pathway might be involved in this heliomycin-mediated SIRT1 suppression. Similarly, SIRT1 downregulation was prevented by treating T24 cells with the proteasome inhibitor MG132, insinuating a possible proteasomal degradation of SIRT1 (Figure 6C). Given that significant apoptosis rather than autophagy predominated in T24 cells treated by the water-soluble derivative (HD2), we found that MG132 effectively inhibited SIRT1 downregulation, conceivably through a proteasomal mechanism (Figure 6D). These lines of evidence point towards that heliomycin and its water-soluble derivative (HD2) mediated multiple pathways to degrade SIRT1 protein.
Figure 6.
SIRT1 downregulation is mediated by heliomycin (H1) and its water-soluble derivative HD2, but not HD3. A. The transcript level of SIRT1 was not affected by heliomycin or its derivatives analyzed by RT-PCR. B. Heliomycin (H1) and its derivative (HD2) significantly attenuated SIRT1 protein expression analyzed by Western blotting, whereas no such effect was seen for derivative (HD3). C. Heliomycin-mediated SIRT1 downregulation was reinstated by the autophagy inhibitor Baf A1 or the proteasome inhibitor, MG132. D. HD2-inhibited SIRT1 expression was restored by the proteasome inhibitor, MG132. Aliquots of cell lysates were resolved by SDS-PAGE and analyzed by Western blotting. β-actin was detected as an internal control. Representative images are shown. Values (means ± SDs) are from at least three independent experiments (**P<0.01, ***P<0.001).
The docking of heliomycin or its water-soluble 4-aminomethylated derivative (HD2) into SIRT1 by a molecular docking simulation
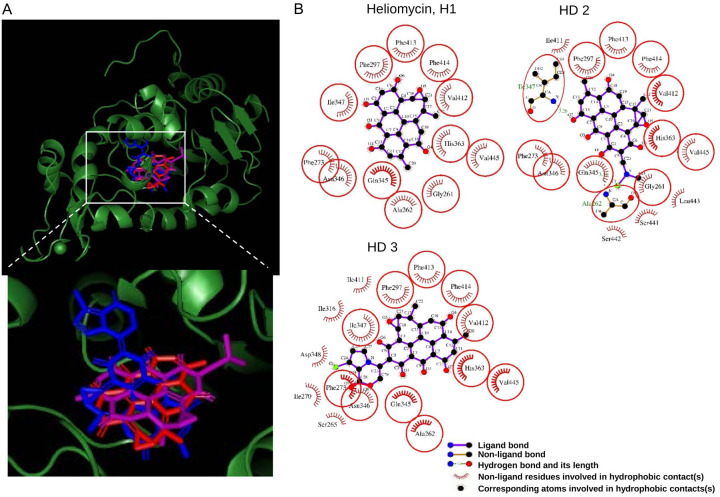
Additionally, by a molecular docking study, we took it a step further to explore the binding mode of heliomycin and its derivatives in the inhibitor binding pocket of the crystal structure of the SIRT1 catalytic domain [32]. The binding poses of heliomycin (H1) and its derivatives were generated by molecular docking, which could be observed to superimpose with obviously different orientations (Figure 7A). The docking energy score for the heliomycin (H1) complex (-9.5 kcal/mol) and the derivative (HD2) complex (-9.1 kcal/mol) indicated that showed significantly more affinity than the derivative (HD3) complex (-0.6 kcal/mol). Moreover, further analysis showed that the hydrophobic interactions were found in several consistent residues surrounding the binding pocket of SIRT1 protein among the docking orientations of heliomycin and its derivatives (Figure 7B). The consistent interaction residues in the three docked complexes are shown in red circles and ellipses, including Gly261, Ala262, Phe273, Phe297, Gln345, Asn346, Ile347, His363, Val412, Phe413, Phe414, and Val445. It can be seen from the post docking analysis that the higher affinities of heliomycin (H1) and derivatives (HD2) to SIRT1 were supported by the hydrophobic moieties surrounding the whole molecules. Furthermore, the hydrogen bonds were also found on Ala262 and Ile347 residues in the docked complex of water-soluble derivative (HD2) to produce steady interaction. On the other hand, derivative (HD3) was formed the hydrophobic moieties on almost all consistent residues, except for Gly 261 of SIRT1. By this result, we speculate that the lack of interaction residues surrounded the derivative (HD3) leading to the lower affinity to SIRT1.
Figure 7.
The binding modes of heliomycin and its derivatives after docking into the catalytic domain of SIRT1. A. Superimposition of the docked heliomycin (H1) (red), derivative (HD2) (purple), and derivative (HD3) (blue). B. Schematic presentations of the putative SIRT1 binding modes with heliomycin and its derivatives. The key residues surrounding the binding pocket of SIRT1 were identified through the best docking pose of each compound. The red circles and ellipses represent the consistent residues that interacted with heliomycin and its derivatives.
Discussion
The growing interest in sirtuins (SIRTs) largely arises from studies showing that these highly conserved NAD+-dependent histone deacetylases, commonly known as the type III HDACs, are important for an array of cellular events and characteristics, including transcriptional regulation, metabolism, and longevity [7,8,41,42]. SIRT1, which is the most highly studied member of the SIRT family, has been shown to utilize NAD+ as a cofactor to deacetylate histone and non-histone proteins at different stages of cancer and is thus poised to affect tumorigenesis [9,10,43]. Many substrate proteins involved in a wide range of pathways are post-translational modified by SIRT1, such that its tight regulation may offer a potential therapeutic strategy against cancer. Here, we reveal that the heliomycin-induced downregulation of SIRT1 reduces cell growth and enhances autophagy in bladder cancer cells, providing additional evidence for the oncogenic functions of SIRT1. SIRT1 was previously suggested to regulate autophagy through deacetylation of the important autophagic components, ATG5, 7 and 8 [44], or via deacetylation of the FOXO transcription factors, leading to enhanced expression of the autophagic machinery [45].
Heliomycin (also known as resistomycin) was first identified more than 60 years ago as a metabolite generated by marine sponges; it possesses many biological activities, including antibacterial and antifungal activities [23-26], anticancer properties [24,27,46,47], and HDAC inhibitor activity [28]. Here, we explored whether heliomycin could directly target SIRT1 by cellular thermal shift assay (CETSA). In the presence of heliomycin, the SIRT1 protein maintained its native antibody-recognized configuration at a higher temperature compared with that observed in the absence of heliomycin (H1) (Figure 5). Upon binding, heliomycin suppressed SIRT1 protein expression to enhance autophagy and attenuate the growth of a bladder cancer cell line. We further validated that SIRT1 downregulation by heliomycin might be via autophagic degradation and/or proteasomal degradation, based on our inhibitor experiments (Figure 6). With limited information on the homeostasis of SIRT1 at the protein level, our findings replicated in a recent study suggest autophagy plays an imperative role in SIRT1 degradation [48,49]. Equally important, SIRT1 is reported to be ubiquitinated and degraded through proteasome pathways in response to stresses [50-52].
Although heliomycin is demonstrated to possess multiple biological activities, its medical applications have been limited due to the extremely low solubility. We previously synthesized a series of novel water-soluble heliomycin derivatives that exhibit chemotherapeutic potential [29,30]. Among them, the water-soluble 4-aminomethylated derivative (HD2) of heliomycin was demonstrated to be apoptotic, but its cellular target remained unknown [29]. Here, we verified the apoptotic activity of the derivative (HD2) and used CETSA to further identify SIRT1 as a cellular protein target of this derivative (Figure 5). The binding between the aminomethylated derivative (HD2) and SIRT1 inhibited SIRT1 to increase the acetylations of C-myc and p53, which contributed to apoptotic pathways (Figure 3). A potent and selective SIRT1 inhibitor, EX527, was shown to induce apoptosis through enhancing p53 acetylation in glioma cancer cell lines [53]. This same inhibitor was highly effective in sensitizing multidrug-resistant human cancer cells to therapeutic treatments, suggesting that targeting SIRT1 could serve as a novel strategy to overcome multidrug resistance [54,55]. A second example is BZD9L1, the novel sirtuin inhibitor, which was recently shown to exhibit anticancer properties in colorectal cancer through the induction of apoptosis [56]. Furthermore, key molecular targets of BZD9L1 were identified by both experimental and computational modeling approaches, and were found to be associated with the p53-dependent apoptotic pathways in colorectal cancer [57]. Combination treatment with BZD9L1 plus 5-FU exerted synergistic cytotoxic effects and diminished cancer growth both in vitro and in vivo, further highlighting the therapeutic potential of SIRT1 inhibitors [58]. These small molecule inhibitors, as well as heliomycin and its water-soluble derivative, have been used to modulate SIRT1 activity that would assist in the elucidation of the complex biological function of SIRT1. At this point, we are not certain of the exact inhibition mechanism of heliomycin and its derivative on SIRT1. However, we speculate that extra hydrogen bonds formed between the water-soluble derivative (HD2) and Ala262 and Ile347 residues of SIRT1 protein might interfere with its substrate binding. Subsequently, depending on the bound substrate, the interaction would initiate a distinct cell death pathway that differs from the one triggered by heliomycin. To confirm this hypothesis, it is necessary to conduct further structure determination of the SIRT1/inhibitor complexes or the complexes containing reaction intermediates.
In sum, we herein employed CETSA to show that heliomycin targets SIRT1 to provoke autophagy of bladder cancer cells through SIRT1 downregulation, whereas a water-soluble aminomethylated derivative of heliomycin (HD2) also targets SIRT1 and induces apoptosis. Our experimental and computational evidence clearly suggest that SIRT1 is a promising drug target, and that heliomycin and its water-soluble derivative may warrant further development as potential cancer remedies.
Acknowledgements
This work was supported by grants from the Ministry of Sciences and Technology, Taiwan (MOST 106-2320-B-005-008-MY3 to PJC and MOST 108-2923-B-005-001-MY3 to PJC), the Russian Foundation for Basic Research (Project 19-53-52008 to AES), and An Nan Hospital, China Medical University, Tainan, Taiwan (ANHRF108-05 and ANHRF109-11).
Disclosure of conflict of interest
None.
Abbreviations
- AO
Acridine orange
- CETSA
Cellular thermal shift assay
- ROS
Reactive oxygen species
- tNOX
tumor-associated NADH oxidase
- SIRT1
Sirtuin 1
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Shang PF, Kwong J, Wang ZP, Tian J, Jiang L, Yang K, Yue ZJ, Tian JQ. Intravesical Bacillus Calmette-Guerin versus epirubicin for Ta and T1 bladder cancer. Cochrane Database Syst Rev. 2011:CD006885. doi: 10.1002/14651858.CD006885.pub2. [DOI] [PubMed] [Google Scholar]
- 3.Mak RH, Hunt D, Shipley WU, Efstathiou JA, Tester WJ, Hagan MP, Kaufman DS, Heney NM, Zietman AL. Long-term outcomes in patients with muscle-invasive bladder cancer after selective bladder-preserving combined-modality therapy: a pooled analysis of Radiation Therapy Oncology Group Protocols 8802, 8903, 9506, 9706, 9906, and 0233. J. Clin. Oncol. 2014;32:3801–9. doi: 10.1200/JCO.2014.57.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gofrit ON. Re: long-term outcomes in patients with muscle-invasive bladder cancer after selective bladder-preserving combined-modality therapy: a pooled analysis of Radiation Therapy Oncology Group Protocols 8802, 8903, 9506, 9706, 9906, and 0233. Eur Urol. 2015;68:165–166. doi: 10.1016/j.eururo.2015.03.057. [DOI] [PubMed] [Google Scholar]
- 5.Toiber D, Sebastian C, Mostoslavsky R. Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. Handb Exp Pharmacol. 2011;206:189–224. doi: 10.1007/978-3-642-21631-2_9. [DOI] [PubMed] [Google Scholar]
- 6.Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD(+) and sirtuin-activating compounds. Nat Rev Mol Cell Biol. 2016;17:679–690. doi: 10.1038/nrm.2016.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 8.Vaziri H, Dessain SK, Eagon EN, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 9.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 10.Song NY, Surh YJ. Janus-faced role of SIRT1 in tumorigenesis. Ann N Y Acad Sci. 2012;1271:10–19. doi: 10.1111/j.1749-6632.2012.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choupani J, Mansoori Derakhshan S, Bayat S, Alivand MR, Shekari Khaniani M. Narrower insight to SIRT1 role in cancer: a potential therapeutic target to control epithelial-mesenchymal transition in cancer cells. J Cell Physiol. 2018;233:4443–4457. doi: 10.1002/jcp.26302. [DOI] [PubMed] [Google Scholar]
- 12.Wang CW, Yang W, Dong F, Guo YW, Tan J, Ruan SN, Huang T. The prognostic role of Sirt1 expression in solid malignancies: a meta-analysis. Oncotarget. 2017;8:66343–66351. doi: 10.18632/oncotarget.18494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan J, Liu YY, Maimaiti Y, Wang CW, Yan Y, Zhou J, Ruan SN, Huang T. Combination of SIRTI and Src overexpression suggests poor prognosis in luminal breast cancer. Onco Targets Ther. 2018;11:2051–2061. doi: 10.2147/OTT.S162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 15.Al-Maghrabi JA. Overexpression of SIRT1 in urothelial carcinoma of the urinary bladder is associated with local recurrence and poor survival. Saudi Med J. 2019;40:541–547. doi: 10.15537/smj.2019.6.24248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Cao L, Li ZQ, Li YW. SIRT1 promotes GLUT1 expression and bladder cancer progression via regulation of glucose uptake. Hum Cell. 2019;32:193–201. doi: 10.1007/s13577-019-00237-5. [DOI] [PubMed] [Google Scholar]
- 17.Cao R, Wang G, Qian KY, Chen L, Qian GF, Xie CH, Dan HC, Jiang W, Wu M, Wu CL, Xiao Y, Wang XH. Silencing of HJURP induces dysregulation of cell cycle and ROS metabolism in bladder cancer cells via PPAR gamma-SIRT1 feedback loop. J Cancer. 2017;8:2282–2295. doi: 10.7150/jca.19967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao R, Wang G, Qian KY, Chen L, Ju LA, Qian GF, Wu CL, Dan HC, Jiang W, Wu M, Xiao Y, Wang XH. TM4SF1 regulates apoptosis, cell cycle and ROS metabolism via the PPAR gamma-SIRT1 feedback loop in human bladder cancer cells. Cancer Lett. 2018;414:278–293. doi: 10.1016/j.canlet.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Hu QX, Wang G, Peng JP, Qian GF, Jiang W, Xie CH, Xiao Y, Wang XH. Knockdown of SIRT1 suppresses bladder cancer cell proliferation and migration and induces cell cycle arrest and antioxidant response through FOXO3a-mediated pathways. Biomed Res Int. 2017;2017:3781904. doi: 10.1155/2017/3781904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinall RL, Ripoll AZ, Wang SS, Pan CX, deVere White RW. MiR-34a chemosensitizes bladder cancer cells to cisplatin treatment regardless of p53-Rb pathway status. Int J Cancer. 2012;130:2526–2538. doi: 10.1002/ijc.26256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang JH, Yan HD, Zhuang SG. Histone deacetylases as targets for treatment of multiple diseases. Clin Sci (Lond) 2013;124:651–662. doi: 10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brazhnikova MG, Uspenskaia TA, Sokolova LB, Preobrazhenskaia TP, Gauze GF, Ukholina RS, Shorin VA, Rossolimo OK, Vertogradova TP. New anti-viral antibiotic heliomycin. Antibiotiki (Mosc) 1958;3:29–34. [PubMed] [Google Scholar]
- 23.Zhang YL, Li S, Jiang DH, Kong LC, Zhang PH, Xu JD. Antifungal activities of metabolites produced by a termite-associated Streptomyces canus BYB02. J Agric Food Chem. 2013;61:1521–1524. doi: 10.1021/jf305210u. [DOI] [PubMed] [Google Scholar]
- 24.Adinarayana G, Venkateshan MR, Bapiraju VV, Sujatha P, Premkumar J, Ellaiah P, Zeeck A. Cytotoxic compounds from the marine actinobacterium. Bioorg Khim. 2006;32:328–334. doi: 10.1134/s1068162006030125. [DOI] [PubMed] [Google Scholar]
- 25.Arora SK. Molecular-structure of heliomycin, an Inhibitor of RNA-synthesis. J Antibiot (Tokyo) 1985;38:113–115. doi: 10.7164/antibiotics.38.113. [DOI] [PubMed] [Google Scholar]
- 26.Roggo BE, Petersen F, Delmendo R, Jenny HB, Peter HH, Roesel J. 3-Alkanoyl-5-hydroxymethyl tetronic acid homologues and resistomycin: new inhibitors of HIV-1 protease. I. Fermentation, isolation and biological activity. J Antibiot (Tokyo) 1994;47:136–142. doi: 10.7164/antibiotics.47.136. [DOI] [PubMed] [Google Scholar]
- 27.Vijayabharathi R, Bruheim P, Andreassen T, Raja DS, Devi PB, Sathyabama S, Priyadarisini VB. Assessment of resistomycin, as an anticancer compound isolated and characterized from Streptomyces aurantiacus AAA5. J Microbiol. 2011;49:920–926. doi: 10.1007/s12275-011-1260-5. [DOI] [PubMed] [Google Scholar]
- 28.Abdelfattah MS, Elmallah MIY, Faraag AHI, Hebishy AMS, Ali NH. Heliomycin and tetracinomycin D: anthraquinone derivatives with histone deacetylase inhibitory activity from marine sponge-associated Streptomyces sp SP9. 3 Biotech. 2018;8:282. doi: 10.1007/s13205-018-1304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nadysev GY, Tikhomirov AS, Lin MH, Yang YT, Dezhenkova LG, Chen HY, Kaluzhny DN, Schols D, Shtil AA, Shchekotikhin AE, Chueh PJ. Aminomethylation of heliomycin: preparation and anticancer characterization of the first series of semi-synthetic derivatives. Eur J Med Chem. 2018;143:1553–1562. doi: 10.1016/j.ejmech.2017.10.055. [DOI] [PubMed] [Google Scholar]
- 30.Tikhomirov AS, Abdelhamid MAS, Nadysev GY, Zatonsky GV, Bykov EE, Chueh PJ, Waller ZAE, Shchekotikhin AE. Water-soluble heliomycin derivatives to target i-Motif DNA. J Nat Prod. 2021;84:1617–1625. doi: 10.1021/acs.jnatprod.1c00162. [DOI] [PubMed] [Google Scholar]
- 31.Lee YH, Chen HY, Su LJ, Chueh PJ. Sirtuin 1 (SIRT1) deacetylase activity and NAD(+)/NADH ratio are imperative for capsaicin-mediated programmed cell death. J Agric Food Chem. 2015;63:7361–7370. doi: 10.1021/acs.jafc.5b02876. [DOI] [PubMed] [Google Scholar]
- 32.Zhao X, Allison D, Condon B, Zhang F, Gheyi T, Zhang A, Ashok S, Russell M, MacEwan I, Qian Y, Jamison JA, Luz JG. The 2.5 A crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J Med Chem. 2013;56:963–969. doi: 10.1021/jm301431y. [DOI] [PubMed] [Google Scholar]
- 33.Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 36.Brooks BR, Brooks CL 3rd, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 38.Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- 39.Martinez Molina D, Nordlund P. The cellular thermal shift assay: a novel biophysical assay for in situ drug target engagement and mechanistic biomarker studies. Annu Rev Pharmacol Toxicol. 2016;56:141–161. doi: 10.1146/annurev-pharmtox-010715-103715. [DOI] [PubMed] [Google Scholar]
- 40.Almqvist H, Axelsson H, Jafari R, Dan C, Mateus A, Haraldsson M, Larsson A, Martinez Molina D, Artursson P, Lundback T, Nordlund P. CETSA screening identifies known and novel thymidylate synthase inhibitors and slow intracellular activation of 5-fluorouracil. Nat Commun. 2016;7:11040. doi: 10.1038/ncomms11040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nat Genet. 1999;23:281–285. doi: 10.1038/15458. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Peterson LM, Li X. Trending topics of SIRT1 in tumorigenicity. Biochim Biophys Acta Gen Subj. 2021;1865:129952. doi: 10.1016/j.bbagen.2021.129952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng F, Tang BL. Sirtuins’ modulation of autophagy. J Cell Physiol. 2013;228:2262–2270. doi: 10.1002/jcp.24399. [DOI] [PubMed] [Google Scholar]
- 46.Riaz A, Rasul A, Hussain G, Saadullah M, Rasool B, Sarfraz I, Masood M, Asrar M, Jabeen F, Sultana T. Resistomycin, a pentacyclic polyketide, inhibits the growth of triple negative breast cancer cells through induction of apoptosis and mitochondrial dysfunction. Pak J Pharm Sci. 2020;33:1233–1238. [Google Scholar]
- 47.Liu X, Arai MA, Toume K, Ishibashi M. Isolation of resistomycin from a terrestrial actinomycete with TRAIL resistance-overcoming activity. Nat Prod Commun. 2018;13:65–66. [Google Scholar]
- 48.Xu CY, Wang L, Fozouni P, Evjen G, Chandra V, Jiang J, Lu CC, Nicastri M, Bretz C, Winkler JD, Amaravadi R, Garcia BA, Adams PD, Ott M, Tong W, Johansen T, Dou ZX, Berger SL. SIRT1 is downregulated by autophagy in senescence and ageing. Nat Cell Biol. 2020;22:1170–1179. doi: 10.1038/s41556-020-00579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang L, Xu C, Johansen T, Berger SL, Dou Z. SIRT1 - a new mammalian substrate of nuclear autophagy. Autophagy. 2021;17:593–595. doi: 10.1080/15548627.2020.1860541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren X, Chen N, Chen Y, Liu W, Hu Y. TRB3 stimulates SIRT1 degradation and induces insulin resistance by lipotoxicity via COP1. Exp Cell Res. 2019;382:111428. doi: 10.1016/j.yexcr.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Q, Zhang P, Qi GJ, Zhang Z, He F, Lv ZX, Peng X, Cai HW, Li TX, Wang XM, Tian B. Cdk5 suppression blocks SIRT1 degradation via the ubiquitin-proteasome pathway in Parkinson’s disease models. Biochim Biophys Acta Gen Subj. 2018;1862:1443–1451. doi: 10.1016/j.bbagen.2018.03.021. [DOI] [PubMed] [Google Scholar]
- 52.Huang CY, Kuo WW, Yeh YL, Ho TJ, Lin JY, Lin DY, Chu CH, Tsai FJ, Tsai CH, Huang CY. ANG II promotes IGF-IIR expression and cardiomyocyte apoptosis by inhibiting HSF1 via JNK activation and SIRT1 degradation. Cell Death Differ. 2014;21:1262–1274. doi: 10.1038/cdd.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang T, Li X, Sun SL. EX527, a Sirt-1 inhibitor, induces apoptosis in glioma via activating the p53 signaling pathway. Anticancer Drugs. 2020;31:19–26. doi: 10.1097/CAD.0000000000000824. [DOI] [PubMed] [Google Scholar]
- 54.Kim HB, Lee SH, Um JH, Oh WK, Kim DW, Kang CD, Kim SH. Sensitization of multidrug-resistant human cancer cells to Hsp90 inhibitors by down-regulation of SIRT1. Oncotarget. 2015;6:36202–36218. doi: 10.18632/oncotarget.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yun UJ, Lee IH, Lee JS, Shim J, Kim YN. Ginsenoside Rp1, a ginsenoside derivative, augments anti-cancer effects of actinomycin D via downregulation of an AKT-SIRT1 pathway. Cancers (Basel) 2020;12:605. doi: 10.3390/cancers12030605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan YJ, Lee YT, Yeong KY, Petersen SH, Kono K, Tan SC, Oon CE. Anticancer activities of a benzimidazole compound through sirtuin inhibition in colorectal cancer. Future Med Chem. 2018;10:2039–2057. doi: 10.4155/fmc-2018-0052. [DOI] [PubMed] [Google Scholar]
- 57.Tan YJ, Lee YT, Mancera RL, Oon CE. BZD9L1 sirtuin inhibitor: identification of key molecular targets and their biological functions in HCT 116 colorectal cancer cells. Life Sci. 2021;284:119747. doi: 10.1016/j.lfs.2021.119747. [DOI] [PubMed] [Google Scholar]
- 58.Tan YJ, Lee YT, Petersen SH, Kaur G, Kono K, Tan SC, Majid AMSA, Oon CE. BZD9L1 sirtuin inhibitor as a potential adjuvant for sensitization of colorectal cancer cells to 5-fluorouracil. Ther Adv Med Oncol. 2019;11:1758835919878977. doi: 10.1177/1758835919878977. [DOI] [PMC free article] [PubMed] [Google Scholar]