Abstract
The mammalian circadian clock plays a central role in the temporal coordination of physiology across the 24-h light-dark cycle. A major function of the clock is to maintain energy constancy in anticipation of alternating periods of fasting and feeding that correspond with sleep and wakefulness. While it has long been recognized that humans exhibit robust variation in glucose tolerance and insulin sensitivity across the sleep-wake cycle, experimental genetic analysis has now revealed that the clock transcription cycle plays an essential role in insulin secretion and metabolic function within pancreatic beta cells. This review addresses how studies of the beta cell clock may elucidate the etiology of subtypes of diabetes associated with circadian and sleep cycle disruption, in addition to more general forms of the disease.
Keywords: pancreatic beta cell, diabetes, circadian transcription, genomics, mouse genetics
Circadian clocks (from circa diem, “about a day”) are found in all photosensitive organisms, where they maintain temporal organization of cell and system physiology in synchrony with the rotation of the earth (Edgar et al., 2012; Milev and Reddy, 2015; Bass, 2012). In mammals, circadian clocks are organized hierarchically, with light-responsive “master” pacemaker clocks located within the suprachiasmatic nucleus (SCN) of the hypothalamus, which in turn drive rhythmic cycles within extra-SCN neurons and peripheral tissues. The SCN aligns peripheral tissue clocks with the environmental light cycle through a combination of direct autonomic nervous system efferent and neuroendocrine signals (such as through control of 24-h cycles of hypothalamic-pituitary-adrenal axis and rhythmic growth hormone production) and feeding-derived cues (la Fleur et al., 2000; Bartness et al., 2001; Buijs et al., 2001; Buijs et al., 2003; Dibner et al., 2010; Gerber et al., 2013). The core circadian clock within both brain and peripheral cells is encoded by a negative transcription-translation feedback loop composed of transcription factors in the forward limb (CLOCK, NPAS2, and BMAL1) that activate repressors within the negative limb (PER1/2/3 and CRY1/2), which feedback to repress the activators in a cycle that repeats itself every 24 h (Partch et al., 2014).
A long-recognized aspect of circadian physiology in humans is the daily variation in blood glucose and insulin levels across the day (Polonsky et al., 1988; Gagliardino et al., 1984; Van Cauter et al., 1997). Clinical investigation in human subjects has shown that glucose clearance and insulin sensitivity in response to orally administered isocaloric glucose challenge peak in the early morning and decline in the afternoon and evening (Aparicio et al., 1974; Bowen and Reeves, 1967; Carroll and Nestel, 1973; Jarrett and Keen, 1969; Roberts, 1964); moreover, both ultradian and circadian variation in glucose tolerance has been observed in response to intravenous glucose delivered in either continuous or oscillatory infusion (Shapiro et al., 1988; Sturis et al., 1995). The daily variation in response to isocaloric feeding arises due to both changes in insulin sensitivity and beta cell insulin secretion and has been demonstrated using isotopic labeling and modeling approaches (Saad et al., 2012). These observations suggest that intrinsic meal-independent processes give rise to daily cycles in glucose metabolism; however, the underlying molecular basis for this observation has only recently emerged. Experimental genetic studies have now demonstrated that cell-autonomous circadian transcription cycles within pancreatic beta cells produce rhythmic cycles in nutrient-dependent insulin secretion through interactions with cell-type-specific regions of open chromatin (Perelis et al., 2015). Here, we focus on the genomic and physiologic basis for the circadian control of glucose metabolism across tissues and the application of this information toward understanding the pathophysiology of human diabetes mellitus.
CIRCADIAN CONTROL OF INSULIN SECRETION
Insulin-producing beta cells exist in specialized organoids called islets of Langerhans within the pancreas, and their dysregulation and/or destruction is central to the pathogenesis of type 1 and type 2 diabetes (Muoio and Newgard, 2008). Genetic analyses in mice first pointed to a key role for clock gene function in beta cells when it was discovered that multitissue ClockΔ19/Δ19 mice, originally isolated from an ENU mutagenesis screen for circadian behavior phenotypes (Vitaterna et al., 1994), developed obesity and hyperglycemia by 6 to 8 weeks of age (Turek et al., 2005) in parallel with reduced circadian variation in blood glucose levels (Rudic et al., 2004). Paradoxically, ClockΔ19/Δ19 mice did not display hyperinsulinemia, a hallmark of obesity-associated diabetes, providing the first clue that clock deficiency leads to beta cell failure (Turek et al., 2005). Subsequent studies showed that multitissue circadian disruption in both ClockΔ19/Δ19 and Bmal1−/− mutant mice displayed reduced glucose tolerance and glucose-stimulated insulin secretion both in vivo and in isolated islets in vitro (Lamia et al., 2008, Sadacca et al., 2011; Marcheva et al., 2010; Lee et al., 2011), further implicating clock genes in the regulation of pancreatic insulin production. However, a conundrum in these early studies was that loss-of-function mutations in the clock in liver also result in severe hypoglycemia during fasting (Lamia et al., 2008), which is likely due in part to mitochondrial dysfunction (Peek et al., 2013). Indeed, the idea that clock deficiency exerts distinct effects in different tissues emerged from the seemingly paradoxical observation that glucose levels vary according to nutrient state in multitissue mutants; thus, hyperglycemia is found in the postprandial condition whereas hypoglycemia occurs with prolonged fasting (Turek et al., 2005; Rudic et al., 2004).
The complex effects of multitissue circadian gene ablation on metabolism can mask an underlying functional defect; however, tissue-specific and inducible gene ablation has been used effectively to delineate organ-specific and developmental stage–specific clock gene functions. Tissue-specific mutagenesis has confirmed the concept that clock function within endocrine beta cells plays a role in insulin secretion after feeding, whereas within liver it is critical in mitochondrial oxidative metabolism during fasting. Specifically, the use of tissue-specific gene targeting within pancreas (PdxCre;Bmal1flx/flx) and beta cells (RipCre;Bmal1flx/flx) showed that Bmal1 deletion resulted in severe glucose intolerance and hypoinsulinemia as well as impaired glucose-stimulated insulin secretion in islets isolated from these animals, confirming the idea that cell-autonomous expression of clock genes within the pancreas is essential for insulin secretion and glucose homeostasis (Marcheva et al., 2010; Sadacca et al., 2011; Lee et al., 2013). Notably, disruption of genes encoding repressors in the negative limb of the circadian clock in Cry1/2−/− mice results in hyperglycemia and impaired glucose tolerance due in part to altered regulation of glucagon and glucocorticoid signaling in the liver; however, the effects of the mutations on insulin secretion have not been examined (Lamia et al., 2011; Zhang et al., 2010). Per2 mutant mice display reduced blood glucose levels and increased glucose-stimulated insulin secretion (Zhao et al., 2012), suggesting complex and distinct roles of individual clock components in regulating glucose homeostasis.
Recent work using beta cell–specific, tamoxifen-inducible Cre drivers (PdxCreER;Bmal1flx/flx or RipCreER;Bmal1flx/flx mice) to delete pancreatic Bmal1 specifically during adulthood has revealed an essential role for the clock transcription cycle in adult animals since abrogation of circadian transcription factor expression in 8- to 12-week-old mice administered tamoxifen and also in islets isolated ex vivo caused acute beta cell failure, independent of effects during development (Perelis et al., 2015; Rakshit et al., 2016). Surprisingly, altered insulin levels and glucose disposal were not observed in mice following deletion of Bmal1 in all tissues during adulthood in mice (Yang et al., 2016). However, it is key to note that just as in the constitutive Bmal1 knockout condition (throughout development), loss of Bmal1 in liver in mice with global adult-life Bmal1 ablation masks the adverse effects of its loss in pancreata, because the co-occurrence in these mice of reduced hepatic glucose production and beta cell failure confounds the analysis of integrated physiologic control of glucose homeostasis. As such, isolated analyses in islets from Bmal1 global mutants reveal hypoinsulinemia in response to glucose and incretin secretagogues. The key point from such observations is that the clock functions “in opposition” in different tissues and one must separate the effects of time of day, nutrient state, and tissue type in order to construct an accurate view of the physiologic role of the circadian system in the intact animal. Thus, genetic analyses of circadian mutant mice set the stage for understanding the molecular mechanisms by which the cell-autonomous pancreatic clock regulates insulin secretion in mammals.
Insight into how disruption of the pancreatic clock might lead to impaired insulin secretion on a cellular level emerged from the initial finding that islets from both the multitissue and pancreas-specific Bmal1 knockouts displayed normal insulin content in parallel with intact glucose-stimulated calcium influx, suggesting that the molecular pancreatic clock must control a late stage of insulin exocytosis. Exposure of PdxCre;Bmal1flx/flx islets to KCl, an insulin secretagogue that bypasses glucose metabolism by directly depolarizing the plasma membrane, reduced insulin secretion in the Bmal1 null islets, confirming an exocytotic defect in islets lacking an intact clock (Marcheva et al., 2010). However, studies using islets isolated from RipCre;Bmal1flx/flx have also found alterations in antioxidant gene expression and mitochondrial ATP production, indicating that multiple levels of clock regulation exist in the beta cell.
Further insight into the molecular defects underpinning the impaired exocytosis in circadian mutant islets arose from studies using signaling molecules generated from intermediates in glucose metabolism and second messenger signaling cascades that act on well-defined steps in insulin secretion, including 3′,5′-cyclic adenosine monophosphate (cAMP), which is synthesized by adenyl cyclase downstream of Gs-coupled receptor activation following stimulation with circulating nutrient-sensitive incretin hormones such as glucagon-like peptide-1 (GLP-1) and gastric inhibitory peptide (GIP). cAMP enhances insulin secretion in beta cells by activating protein kinase A (PKA) and rap guanine nucleotide exchange factor 3 (RAPGEF3, also known as EPAC1/2) signaling (Seino et al., 2011). Interestingly, islets from clock-deficient PdxCre;Bmal1flx/flx and adult-life-induced PdxCreER;Bmal1flx/flx mice displayed significantly impaired insulin secretion in response to the cAMP mimetic 8-Br-cAMP, the GLP-1 receptor agonist exenatide, and a downstream agonist of cAMP synthesis (forskolin) (Marcheva et al., 2010; Perelis et al., 2015), revealing that second messenger signaling cascades might be regulated by the circadian clock. Of note, despite these cAMP-related exocytotic defects, PdxCreER;Bmal1flx/flx islets secrete normal levels of insulin in the presence of both the Gq-coupled muscarinic receptor agonist carbachol and the diacyl glycerol (DAG) mimetic phorbol 12-myristate 13-acetate (PMA). Gq proteins, following activation of their receptors by acetylcholine, induce the hydrolysis of phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2) by phospholipase C to generate DAG and inositol 1,4,5-trisphosphate (IP3). DAG and IP3 enhance insulin secretion by activating protein kinase C (PKC) and insulin vesicle priming factors and stimulating Ca2+ release from the endoplasmic reticulum (ER) to induce activity of calcium-calmodulin kinases and synaptotagmins, respectively (Ruiz de Azua et al., 2011; Rodriguez-Diaz et al., 2011). Thus, while the pancreatic clock is a critical regulator of glucose-, incretin-, and Ca2+-stimulated insulin secretion, muscarinic receptor–induced DAG signaling either bypasses or compensates for deficiencies in clock-controlled pathways.
GENOME-WIDE CIRCADIAN TRANSCRIPTION IN ISLETS
In the last 15 years, genome-wide transcription profiling studies using cDNA microarray and RNA sequencing (RNA-seq) technologies have been applied to define the repertoire of circadian-controlled genes in a variety of mouse tissues in vivo, enabling elucidation of the molecular underpinnings of tissue-specific circadian control of physiology (Panda et al., 2002; Storch et al., 2002; Koike et al., 2012, Menet et al., 2012; Vollmers et al., 2012; Zhang et al., 2014). These studies have revealed rhythmic oscillations of up to 43% of mRNAs in mice (Zhang et al., 2014), many of which encode key rate-limiting proteins important in organ-specific functions (Panda et al., 2002; Storch et al., 2002; Zhang et al., 2014). One approach to delineate the role of tissue-autonomous clock gene expression on cell physiology, versus systemically driven oscillations of circulating serum factors or autonomic input (Gerber et al., 2013), has entailed application of tissue-specific genetic models of circadian disruption. Transgenic models for studies of intertissue circadian regulation of behavior and metabolism have included mice overexpressing the circadian repressor REV-ERBα within liver, which abolishes liver-specific core circadian gene activity (LAP-tTA/TRE-Rev-erbα mice) (Kornmann et al., 2007), or multitissue ClockΔ19/Δ19 mice with genetic rescue of the wild-type CLOCK protein within the brain (Scg2∷tTA;tetO∷ClockWT; ClockΔ19/Δ19 mice), which restores circadian behavior (Hughes et al., 2012). Microarray analysis of mRNA expression in livers of these animals revealed that while oscillation of most genes depended on a functional cell-autonomous hepatic clock, a small subset of genes, including the core circadian repressor Per2, continued to oscillate robustly in clock-deficient hepatocytes in the presence of a functioning brain clock, suggesting that Per2 cycles in liver are likely generated by systemic signals rather than cell-autonomous clock gene expression within the intact animal. Together, these studies demonstrate that a combination of cell-autonomous and nonautonomous processes drive rhythmic transcription within liver; however, how distinct cell physiologic functions might be rhythmically controlled is less well understood.
Circadian control of physiological processes at a cell- or tissue-autonomous level can also be defined by examining circadian gene transcription and cellular processes in organotypic tissue explants ex vivo (Peek et al., 2015; Peek et al., 2013). For example, one of the earliest studies examining circadian rhythms within isolated islet cells demonstrated that rat islets displayed a spontaneous circadian pattern of insulin release when continuously perfused with media containing a low concentration of glucose (Peschke and Peschke, 1998), suggesting that islet cell-autonomous circadian transcription might regulate beta cell function across the 24-h day-night cycle. Consistently, islets isolated from transgenic Per2Luc reporter mice, in which firefly luciferase is fused in frame with the endogenous Per2 locus, displayed self-sustained bioluminescent rhythms ex vivo (Marcheva et al., 2010; Sadacca et al., 2011), as did islets isolated from Per1∷LUC transgenic rats (Qian et al., 2013). More recently, isolated mouse islets synchronized with the cAMP agonist forskolin were found to display a pronounced time-of-day-dependent variation in the amount of insulin secreted in response to stimulatory concentrations of both glucose and KCl that corresponded with circadian Per2Luc expression in isolated islets (Perelis et al., 2015). These findings are consistent with the observation that both glucose- and KCl-stimulated insulin secretion are impaired in clock-deficient beta cells and indicate that circadian regulation affects the late stages of the insulin secretory pathway. One challenge of ex vivo studies of islet insulin secretion is extrapolation of the analyses to the intact animal. In this respect, in vivo, the acrophase of endogenous Per2 mRNA peaks within the pancreata and liver during the transition from the dark to light period, the time of day when mice are most glucose-tolerant and display the greatest magnitude of glucose-stimulated insulin secretion (Marcheva et al., 2010). Interestingly, the timing of maximal glucose and KCl responsiveness at the light-dark transition was similar to the time of maximal Per2Luc bioluminescence in isolated islets, suggesting that the islet cell–autonomous clock anticipates the time of day when insulin demand is highest, thereby priming the beta cell for maximal responsiveness at the start of the active period.
Next-generation sequencing of islet mRNA from wild-type and circadian mutant islets has provided detailed insight into the link between the core circadian transcription cycle and downstream gene regulatory networks that give rise to rhythmic insulin secretion. Wild-type islets displayed self-sustained oscillations in the expression of 27% (3909) of expressed mRNAs that were enriched for gene pathways involved in vesicle biogenesis, transport, and signaling-induced exocytosis (Perelis et al., 2015). Among these were genes encoding the coat protein complex II (COPII; Sec24a and Sec31a), which is important for vesicle biogenesis; soluble NSF attachment protein receptor (SNARE) components (Vamp1, Vamp5, Vamp8, Stx1a, Stx4a, and Stx8), which mediate the final stages of exocytosis; and cAMP targets (Pclo, Rims2, and Rab3b) and Ca2+-sensing synaptotagmins (Syt11, Syt14, and Syt16), which enhance insulin secretion (Seino et al., 2011). Whereas previous microarray analyses were performed in islets immediately following isolation, these more recent sequencing studies were performed in islets maintained ex vivo over 2 full circadian cycles, thus revealing a greater repertoire of mRNAs that underlie the diurnal variation in insulin secretion.
The observation that cell-autonomous circadian regulation gives rise to widespread rhythmic oscillation of the transcriptome within various tissues, including the beta cell, raises the question as to how clock transcription factors mediate cell-type-specific physiology (Perelis et al., 2015; Kornmann et al., 2007; Panda et al., 2002; Ueda et al., 2002; Zhang et al., 2014). Genetic studies originally demonstrated that the core clock transcription mechanism governs both behavior and physiology through the binding of circadian transcriptional heterodimer CLOCK/BMAL1 to E-box elements in their own repressors, whereas more recent genomic studies in liver suggest that core clock factor binding within enhancer regions determines the phase of downstream oscillatory RNAs (Koike et al., 2012; Rey et al., 2011; Menet et al., 2012). Yet what has remained somewhat mysterious is the mechanism through which the core clock cycle affects differential physiologic functions within distinct tissues. The best-characterized tissue for studies of clock transcription in mammals has been the liver, where rhythmic binding of both CLOCK/BMAL1 and the downstream circadian repressors REV-ERBα and E4BP4 to enhancers has been shown to regulate the transcription of enhancer-derived RNAs (eRNAs) and predicts phase-specific transcription of metabolic genes (Fang et al., 2014). Cistromic profiling by chromatin immunoprecipitation sequencing (ChIP-seq) in mouse beta cells revealed that CLOCK and BMAL1 predominantly bind to distal enhancers to regulate rhythmic islet gene expression (Perelis et al., 2015). Surprisingly, the BMAL1 binding sites within enhancers in beta cells were divergent from BMAL1 sites identified in liver, even among those genes found to be rhythmic in both tissues (Perelis et al., 2015), suggesting that clock proteins cooperate with distinct transcription factors and coactivators to drive rhythmic gene expression in different organs. Indeed, a recent comparative analysis of cycling genes across 12 mouse tissues found that while 43% of all protein-coding genes were rhythmic in at least 1 organ, only 10 genes are commonly rhythmic in all tissues (Zhang et al., 2014). One possible explanation for the tissue-specificity of circadian transcriptomes is that the availability of binding sites for CLOCK/BMAL1 and other rhythmic transcription is limited by the developmentally established enhancer repertoire of a given cell type. Consistent with this idea, epigenetically defined active enhancers marked by co-occupancy of H3K4Me2 and H3K27Ac in cycling islet genes exhibited reduced H3K27Ac at corresponding sites in liver (Perelis et al., 2015), indicating reduced transcriptional activity (Creyghton et al., 2010), and were enriched for the transcription factor PDX1, a critical regulator of pancreas organogenesis (Stoffers et al., 1997). Therefore, in vitro studies in isolated islets and beta cells suggest that clock transcription factors bind to tissue-specific enhancers to promote the transcription of genes that sensitize beta cells to maximally secrete insulin at specific time windows throughout the circadian cycle. However, the small subset of sites in which CLOCK/BMAL1 co-localize in liver and the beta cell are all components of the core clock itself (Perelis et al., 2015), suggesting that the generation of rhythm occurs through common elements, whereas the output of the central oscillator localizes to unique tissue-type specific loci. Importantly, not all cycling islet genes are direct targets of CLOCK/BMAL1. For example, the components of the circadian negative feedback loop CRY1, CRY2, and PER2 bind to a large number of unique chromatin sites independently of CLOCK/BMAL1 and may regulate downstream oscillating transcription networks (Koike et al., 2012). Further studies are needed to understand how other cycling transcription factors contribute to the diurnal control of insulin secretion.
ROLE OF PANCREATIC CLOCK IN ORGANISMAL GLUCOSE HOMEOSTASIS IN VIVO
While in vitro studies have enabled the identification of molecular pathways regulating insulin secretion controlled by islet cell–autonomous clocks, as noted above, it has been challenging to tease apart the opposing effects of circadian disruption across distinct peripheral metabolic tissues in multitissue circadian mutants in vivo. For example, the diabetic phenotype in multitissue ClockΔ19/Δ19 and Bmal1 mutant mice is not as pronounced as in islet-specific circadian mutant mice. Despite significantly impaired glucose-stimulated insulin secretion observed in isolated pancreatic islets, early metabolic analyses of young multitissue ClockΔ19/Δ19 mice did not detect hyperglycemia (Turek et al., 2005). This was later attributed to masking effects of clock disruption in the liver, which compensated for the reduced beta cell insulin secretion by impairing hepatic glucose production and increasing insulin sensitivity (Lamia et al., 2008; Zhang et al., 2010). Of note, when clock mutations were isolated to the liver, mice displayed hypoglycemia at times when the animals were inactive and fasting (Lamia et al., 2008; Peek et al., 2013), in contrast to the hyperglycemia and impaired insulin exocytosis observed predominantly in the feeding period in pancreas-specific clock mutants (Marcheva et al., 2010). Thus, the composite effects of clock function in different tissues in vivo give rise to the overall “net” glucose phenotype. Importantly, electrolytic ablation of the SCN pacemaker cells in mice also impaired hepatic insulin sensitivity and glucose production (Coomans et al., 2013), indicating that the brain clock also regulates hepatic glucose metabolism. Ultimately, pancreatic, hepatic, and pacemaker clocks therefore appear to cooperate to maintain whole body glucose homeostasis throughout the day by promoting insulin-stimulated glucose clearance during the feeding periods while sustaining glucose levels by enhancing mitochondrial oxidative metabolism and glucose production in liver during periods of fasting (Fig. 1).
Figure 1.
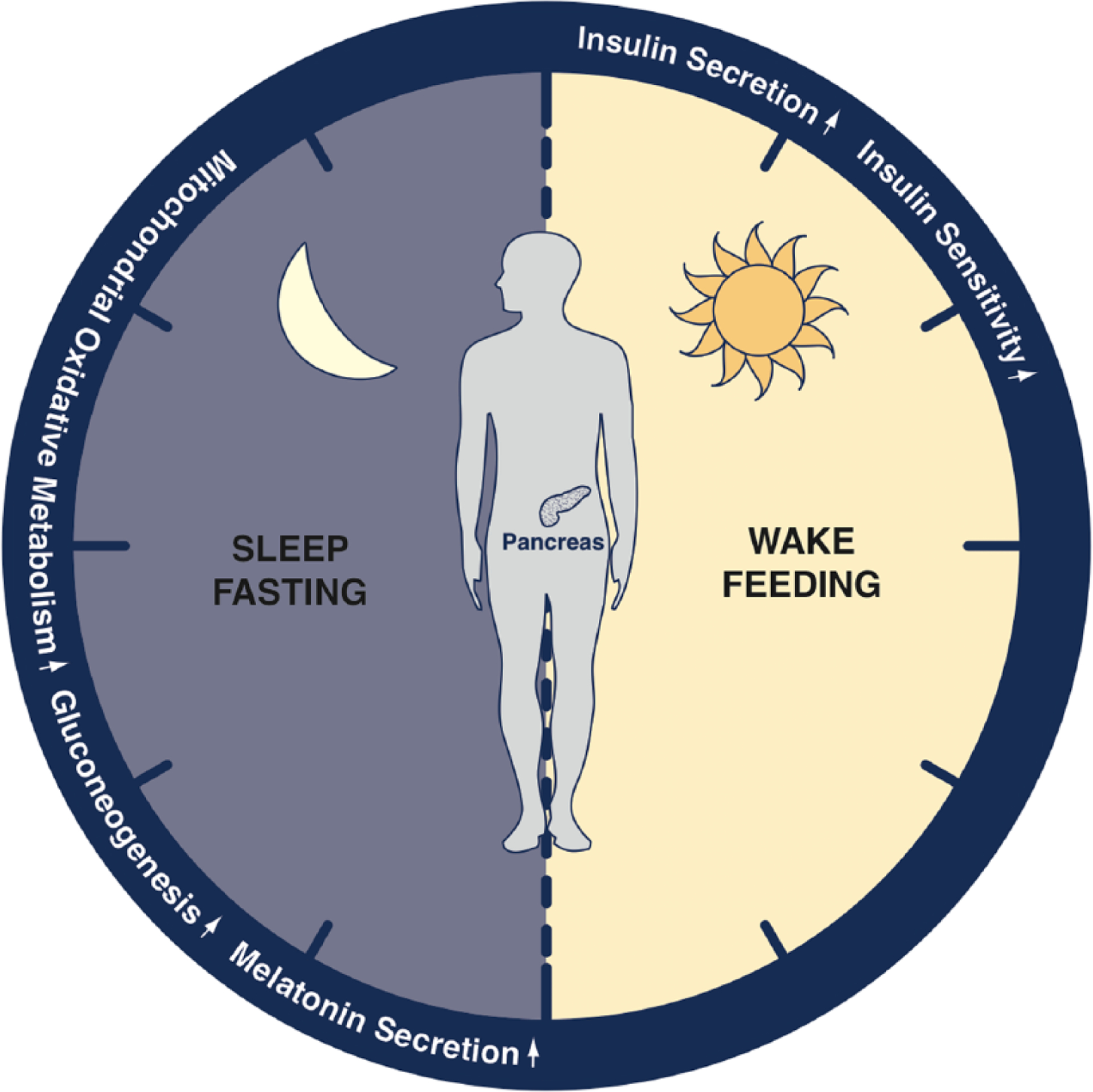
Regulation of glucose homeostasis by the circadian clock. Blood glucose levels are coordinately regulated across the 24-h light-dark cycle by molecular clocks expressed throughout the body. The cell-autonomous clock within insulin-producing pancreatic beta cells anticipates the start of the active or feeding period by increasing glucose-stimulated insulin secretion, which in turn stimulates the clearance of glucose from the blood. Both insulin secretion and peripheral insulin action decline as the day progresses, while the liver clock increases glucose synthesis to sustain blood glucose levels in anticipation of sleep. Brain-derived melatonin increases in the evening in humans and may sensitize beta cells to secrete insulin in response to glucose in the morning. Molecular clocks distributed within beta cells, brain, and peripheral insulin sensitive tissues coordinate blood glucose levels across the day-night cycle.
In addition to the cell-autonomous effects of the circadian clock within distinct metabolic tissues on glucose homeostasis, islet clocks in vivo are exposed to a variety of systemic signals that in turn contribute to blood glucose homeostasis in the intact animal. For example, the beta cell clock is influenced by time-of-day-dependent variations in levels of the circulating GLP-1 hormone. GLP-1 is secreted in response to nutrient ingestion by enteroendocrine L-cells in the intestine and potentiates glucose-stimulated insulin secretion in beta cells (Mojsov et al., 1987). Recent studies have demonstrated that GLP-1 secretion stimulated by administering an oral glucose load exhibits circadian rhythmicity in rats and that rhythmic expression of mRNAs encoding the circadian PAR-bZIP transcription factor thyrotroph embryonic factor (TEF) and extracellular signal-regulated kinase modulator pleiotropic regulatory locus-1 (PRL-1) contributes to circadian GLP-1 secretion in L-cells (Gil-Lozano et al., 2014). Interestingly, maximal glucose-stimulated GLP-1 secretion and GLP-1-induced insulin secretion in rats occurred at ZT22, close to the transition from the dark-to-light period when beta cells are most sensitive to glucose in mice (Gil-Lozano et al., 2014; Marcheva et al., 2010). Rhythmic circulating GLP-1 levels are therefore an additional layer of systemic circadian control that acts synergistically with the pancreatic clock to regulate blood insulin and glucose levels throughout the day (Fig. 2).
Figure 2.
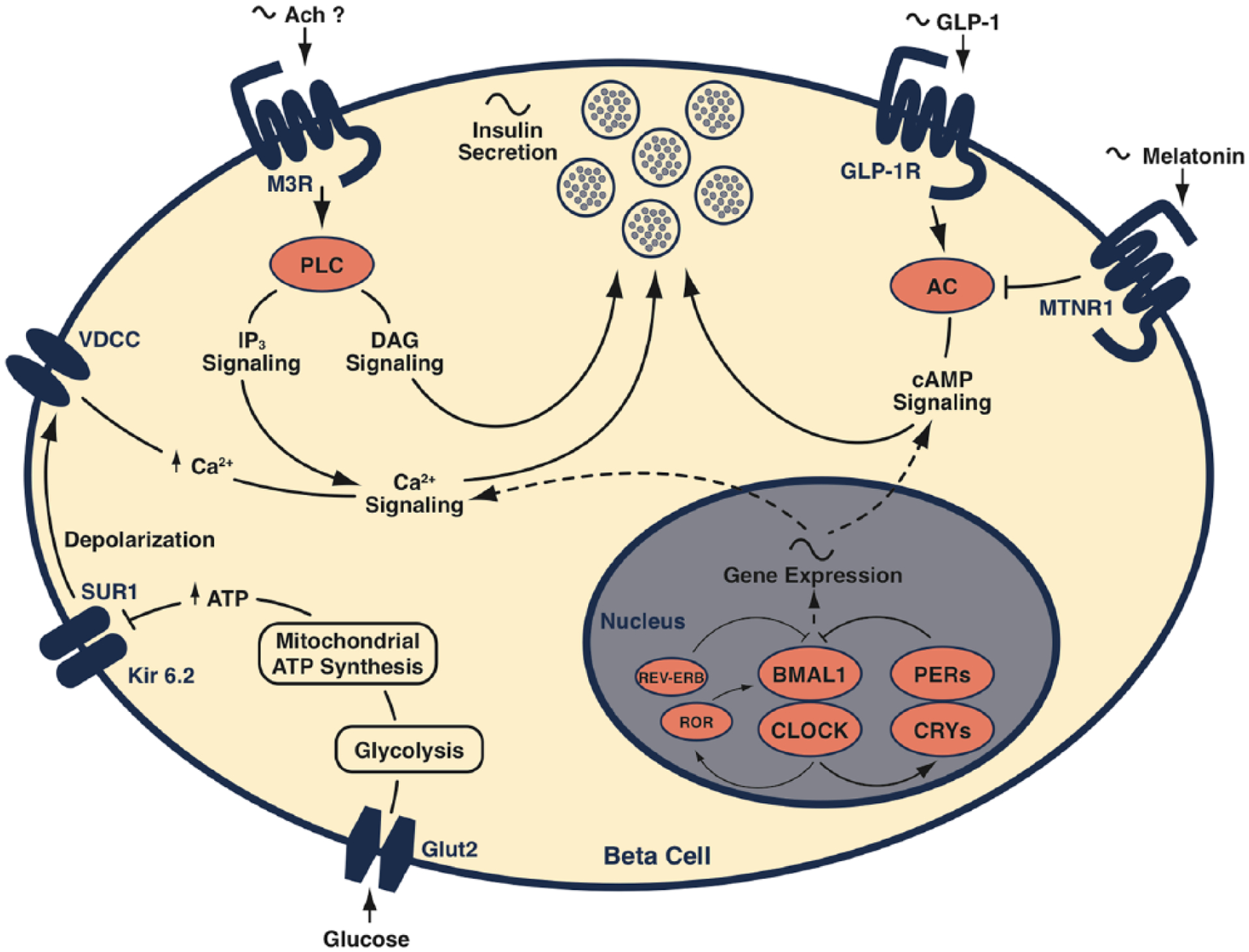
Molecular clock in the beta cell regulates insulin secretion together with circulating systemic factors. The circadian system regulates insulin secretion by driving rhythmic transcription of gene networks involved in glucose-, cAMP-, and Ca2+-stimulated exocytosis. Circulating levels of GLP-1 and melatonin modulate insulin secretion by augmenting or reducing intracellular cAMP levels, respectively. Acetylcholine-stimulated DAG synthesis bypasses clock-controlled signaling pathways to promote insulin secretion, possibly linking brain-derived cholinergic signals to the temporal control of beta cell function.
Clocks throughout the body are also entrained and modulated by extracellular signals such as temperature fluctuations (Buhr et al., 2010; Saini et al., 2012; Morf et al., 2012), serum factors (Balsalobre et al., 1998; Balsalobre et al., 2000b; Gerber et al., 2013), hormones (Le Minh et al., 2001; Chaves et al., 2014), and feeding (Damiola et al., 2000; Kohsaka et al., 2007; Hatori et al., 2012), and exposure to environmental conditions associated with disrupted circadian rhythms has also been shown to affect the pancreatic clock. For example, chronic exposure of mice to constant light (LL) for 10 weeks, which causes a total loss of SCN-driven behavioral rhythms, dampened the amplitude and altered the phase and synchrony of circadian Per1-LUC bioluminescence in isolated islets (Qian et al., 2013). Islets isolated from these LL-exposed rats displayed impaired insulin secretion in response to glucose, but not GLP-1 or tolbutamide, a secretagogue that acts by chemically closing K+ATP channels (Qian et al., 2013), suggesting shared regulation of some, but not all, pathways by the islet cell–autonomous clock and systemic circadian signals perturbed in LL. Similarly, a study testing the impact of simulated shift work on beta cell function found that a circadian disruption (CD) paradigm where mice were exposed to serial 6-h advances in the light-dark cycle every 4 days for 8 weeks inhibited glucose-stimulated insulin secretion and insulin-stimulated glucose clearance (Lee et al., 2013). Beta cell–specific Bmal1 knockout mice displayed more severely impaired insulin secretion when subjected to this CD protocol (Lee et al., 2013), suggesting that proper expression of clock genes in the beta cell might help to prevent hyperglycemia even in the setting of systemic circadian disruption in shift work.
CIRCADIAN CONTROL OF PANCREATIC ISLET FUNCTION IN HUMANS
Both epidemiologic and population genetics studies have shown strong associations between alterations in circadian clock function in shift workers and heritable genetic variants (single nucleotide polymorphisms) with the risk of hyperglycemia and diabetes in humans (Knutsson, 2003; Dupuis et al., 2010). The effect of recurrent disruptions of the day-night cycle that occur in shift work can be simulated in the laboratory by exposing humans to 28-h days, where the usual 12-h:12-h light-dark cycle is extended to 14-h:14-h, resulting in days that are 4 h longer than usual. In this paradigm, humans experience a complete 12-h inversion of the external light-dark cycle after 3 successive 28-h days, and circadian clock–controlled biomarkers such as blood cortisol levels display a pattern that is antiphasic to its usual relationship with the light-dark cycle (Scheer et al., 2009). While misaligned, humans displayed significantly increased glucose and insulin levels after ingesting food at regularly scheduled mealtimes, which for some subjects reached levels seen in prediabetes (Scheer et al., 2009; Morris et al., 2015), indicating that circadian misalignment caused reductions in both insulin sensitivity (increasing the demand for insulin secretion) and beta cell compensation for elevated glucose levels (insufficient insulin to lower blood glucose).
An additional link between circadian function and human glucose metabolism has come from genome-wide association studies (GWAS) testing for heritable genetic variation associated with fasting glucose levels and diabetes incidence in human populations, which have identified associations between polymorphisms in close proximity to genes involved in circadian rhythm regulation to human beta cell function. Specifically, single nucleotide variants mapping to the circadian clock repressor CRY2 (Dupuis et al., 2010) and a receptor for the circadian hormone melatonin MTNR1B (Bouatia-Naji et al., 2009; Dupuis et al., 2010) are associated with elevated fasting glucose levels in multiple human populations. Genetic variants in BMAL1 and CRY2 have also been linked with development of type 2 diabetes (Woon et al., 2007; Hu et al., 2010). In addition, allelic variants of CRY2 have been associated with a decrease in 2 different oral glucose tolerance test–based disposition indices in middle-aged Danish people due to decreased insulin sensitivity (Boesgaard et al., 2010). While CRY2 and MTNR1B are expressed in human pancreatic islets, they are also broadly expressed in other tissues involved in glycemic control (Dupuis et al., 2010). However, humans carrying the MTNR1B variant associated with diabetes and hyperglycemia have been found to display reduced insulin secretion independently of their level of glycemic control (Fadista et al., 2014; Staiger et al., 2008), suggestive of a role of MTNR1B in pancreatic beta cell function. Genomic analyses further substantiated a role of the MTNR1B variant in human islet cells, where MTNR1B mRNA expression was increased due to enhanced binding of key beta cell transcription factors FOXA2 and NEUROD1 to a transcriptional enhancer of the MTNR1B gene (Gaulton et al., 2015). Importantly, melatonin, the endogenous ligand for the MTNR1B receptor, is synthesized and released into circulation at night by endocrine cells in the pineal gland in response to rhythmic signals originating in the SCN (Mulder et al., 2009). Melatonin acutely inhibits glucose-, KCl-, and cAMP-stimulated insulin secretion in rodent islets and beta cells when administered in conjunction with stimulating concentrations of glucose and the cAMP agonist forskolin (Peschke et al., 2002), most likely by coupling to inhibitory Gi proteins that limit the synthesis of the insulinotropic molecules cAMP and cGMP (von Gall et al., 2002). The inhibition of insulin secretion by melatonin is consistent with the decline in human insulin secretory capacity in the evening (Morris et al., 2015) and with impaired beta cell function in humans with increased sensitivity to melatonin signaling. However, the kinetics of melatonin are complex, and prolonged treatment of rodent beta cells with melatonin for 12 h, mimicking the endogenous exposure in vivo, also sensitizes cells to forskolin- and GLP-1-stimulated insulin secretion (Kemp et al., 2002), enhances transcription of cAMP response element binding protein (CREB) target genes (Nishiyama and Hirai, 2014), and protects islets from stress-induced damage (Costes et al., 2015). Moreover, rare coding variants that inhibit MTNR1B function have been found to associate with type 2 diabetes risk in human populations (Bonnefond et al., 2012), and reduced rates of nighttime melatonin secretion are associated with human diabetes (McMullan et al., 2013), suggesting that while melatonin appears to acutely suppress insulin secretion, it may also exert long-lasting effects that promote secretory function and beta cell survival. In the future, it will be important to understand how circadian transcription factors are influenced by diabetes-associated genomic variants since they are enriched in enhancer elements that regulate gene transcription in islets (Gaulton et al., 2010; Pasquali et al., 2014) and may sensitize individuals to the adverse effects of circadian disruption.
While genetic studies provide clues concerning the link between human diabetes and the circadian system, studies in human islets further support the idea that the clock plays an important role in metabolic function. For example, analyses of RNA expression from human cadaveric islets reveal self-sustained oscillations in the transcription of core circadian genes and mRNAs encoding key factors involved in beta cell health (Saini et al., 2016; Perelis et al., 2015), indicating that the rhythmic regulation of transcription is conserved from rodents to humans. Live-cell imaging studies of dispersed human islets expressing a Bmal1 promoter fragment driving luciferase (Bmal1-Luc) have further revealed that both beta and nonbeta cells displayed cell-autonomous Bmal1-Luc rhythms, indicating the presence of a functional clock in multiple islet cell types in humans (Pulimeno et al., 2013). Similar to findings in rodent islets, human islets displayed circadian patterns in basal insulin secretion when continuously perifused, and inhibition of the circadian clock by siRNA-mediated silencing of the CLOCK gene abolished these secretory rhythms and reduced insulin secretion (Saini et al., 2016). Further, RNA-seq analyses in synchronized human islets, or in human islet cultures with CLOCK inhibition, demonstrate that the clock controls networks of genes that regulate stimulus-induced exocytosis, including effectors of muscarinic Gq coupled receptor signaling, cAMP signaling, insulin vesicle fusion, and glucose sensing (Perelis et al., 2015; Saini et al., 2016), similar to findings in rodent islets. Interestingly, gene expression studies comparing the transcriptomes of human islets isolated from normoglycemic compared with type 2 diabetic donors have found significant reductions in the expression of several basic helix-loop-helix Per-Arnt-Sim (bHLH-PAS) transcription factors, including the core clock component BMAL1 (Gunton et al., 2005), as well as CRY2, PER2, and PER3 (Stamenkovic et al., 2012), in the diabetic state. In addition, CRY2 and PER3 expression levels were highly correlated with donor glycated hemoglobin levels, providing an added link between the regulation of circadian gene transcription and glucose control in humans (Stamenkovic et al., 2012).
CONCLUSIONS AND FUTURE DIRECTIONS
Collectively, in vivo and in vitro studies in both rodents and humans reveal that multiple circadian control mechanisms modulate the secretion of insulin across the day-night cycle. Cell-autonomous expression of CLOCK/BMAL1 in beta cells drives genome-wide cycles in the transcription of mRNAs regulating the formation, trafficking, and exocytosis of insulin-containing vesicles that prime cells to maximally secrete insulin at specific windows of time. Additional layers of regulation establish the rhythmicity of glucose homeostasis in the intact animal, including variation in hepatic transcription cycles and in the gastrointestinal tract where incretin hormones are produced (such as GLP-1), as well as the brain-driven melatonin, neuroendocrine, and behavioral cycles. While still incompletely known, white adipose, brown adipose, and skeletal muscle clocks also contribute to energy balance and in turn likely affect glucose homeostasis (Paschos et al., 2012; Gerhart-Hines et al., 2013; Schroder et al., 2015; Hodge et al., 2015). Thus, the cell-autonomous circadian pattern of insulin secretion is one piece of a complex puzzle that emerges in uncovering mechanisms underlying the temporal dynamics of glucose homeostasis throughout the day. Disruption of external circadian entrainment by light cycle manipulation or simulated shift work has been shown to inhibit clock function and insulin secretion in the beta cell (Lee et al., 2013; Qian et al., 2013) and as such may account in part for subtypes of diabetes in these individuals. Additional mechanisms of circadian disruption in shift workers may involve alterations in autonomic nervous system signals or feeding-derived factors that affect islet function. However, the relative contribution of the brain versus peripheral clocks, as well as the repertoire of secreted factors that confer timing cues to the beta cell, remains largely unknown and constitutes an area of active investigation. For example, Per2 transcriptional oscillations in isolated islets can be entrained by mimetics of circulating and physiologic conditions, including the cAMP agonist forskolin (which mimics downstream GLP-1 signaling), the glucocorticoid agonist dexamethasone, and simulated body temperature cycles (Marcheva et al., 2010; Perelis et al., 2015; Pulimeno et al., 2013; Saini et al., 2016), and therefore alterations in the circadian pattern of circulating GLP-1, glucocorticoid hormones, and body temperature rhythms (Balsalobre et al., 2000a; Balsalobre et al., 2000b; Schibler et al., 2015) may represent mechanisms that contribute to the dysregulation of the islet clock during shift work. Further, the observation that mimetics of cholinergic signaling bypass the insulin secretory defects in Bmal1−/− islets presents the intriguing possibility that under some circumstances, the autonomic nervous system may override impairment of the cell-autonomous clock to promote insulin secretion. Indeed, studies have long noted the existence of a preabsorptive “cephalic phase” of insulin secretion that is highly conserved across animal species and regulated by brain-derived signals transmitted by cholinergic vagal innervation (Power and Schulkin, 2008; Powley and Berthoud, 1985). In addition to providing insight into new potential mechanisms for circadian regulation of the beta cell, the observation that acetylcholine receptor activity and downstream DAG signaling restores insulin secretion suggests that this pathway may be a therapeutic target to enhance insulin secretion in diabetic humans with a circadian component to their disease.
Conversely, extensive evidence shows that circadian clocks are sensitive to nutrient excess and are dysregulated in obesity and diabetes. For example, genetic rodent models for obesity and metabolic dysregulation, such as the leptin-deficient ob/ob mice (Lepob/ob), the leptin receptor-deficient db/db mice (Leprdb/db), and the Zucker obese rats, all exhibit disruptions in circadian behavior, including feeding rhythms, diurnal locomotor activity rhythms, and sleep (Laposky et al., 2006; Laposky et al., 2008; Megirian et al., 1998; Mistlberger et al., 1998). Interestingly, changes in peripheral clock gene expression in Lepob/ob mice precede metabolic abnormalities (Ando et al., 2011), suggesting that altered clock function may contribute to metabolic decline. Furthermore, diabetic Leprdb/db mice overexpressing Cry1 in the liver exhibit lower blood glucose levels and improved insulin sensitivity, suggesting that compounds that enhance circadian gene expression may provide therapeutic benefit to individuals with type 2 diabetes (Zhang et al., 2010). Similarly, a recent study found mitigating effects of the small molecule Nobiletin on metabolic disorders in Leprdb/db mice by enhancing the amplitude of circadian rhythms (He et al., 2016). Studies using high-fat diet feeding, which causes obesity and diabetes as well as dysregulation of behavioral, molecular, and metabolic circadian rhythms in mice (Kohsaka et al., 2007; Eckel-Mahan et al., 2013), have shown that restoring the feeding rhythm by restricting feeding time to the “correct” time of day protects mice from developing pathologies associated with diabetes (Arble et al., 2009; Hatori et al., 2012). Moreover, clock expression specifically in the beta cell may exert a protective role during high-fat feeding, since beta cell–specific RipCreER;Bmal1flx/flx mice display decreased compensatory beta cell proliferation and increased glucose levels (Rakshit et al., 2016). It has become clear that the circadian clock is sensitive to metabolic and hormonal changes that occur as consequences of nutrient excess, and future studies are needed to determine whether clock-controlled signaling in the beta cell might be a therapeutic target to improve glucose control in diabetes.
Recent advances in genome-wide transcriptional and cis-regulatory mapping of circadian regulation not only have identified the repertoire of clock-controlled genes within each tissue but also have revealed tissue-specific differences in clock control of enhancer landscapes in distinct tissues. However, many outstanding questions remain regarding the mechanisms and consequences of circadian transcription factor (TF) regulation in beta cells. First, the observation that CLOCK/BMAL1 regulate circadian islet genes by binding to enhancers co-occupied by developmental TFs such as PDX1 suggests that there may be reciprocal regulation of cycling genes by other pancreatic TFs in the beta cell. For example, physiological inhibition of PDX1 or other TFs that occurs following high-fat feeding (Reimer and Ahren, 2002) in wild-type mice may impair beta cell function by inhibiting CLOCK/BMAL1 activity. Second, since circadian gene transcription cycles are enabled by the activity of not only the core circadian activators and repressors but also downstream CLOCK/BMAL1-controlled circadian PAR-bZIP TFs (DBP, HLF, TEF, and E4BP4) and nuclear receptors (RORα/β/γ and REV-ERBα/β), it is therefore possible that alterations in the expression or genomic localization of these TFs may direct and modulate circadian gene transcription in the beta cell. For example, Nrf2 is a downstream target of CLOCK/BMAL1 and encodes a TF that regulates the circadian pattern of antioxidant gene expression in beta cells (Lee et al., 2013). Third, information gained by genomic mapping can be used to interrogate how circadian dysregulation and heritable genetic sequence variants associated with human diabetes contribute to disease risk. The discovery of variants mapping to CRY2 and MTNR1B raises the possibility that there may be subpopulations of individuals with increased metabolic vulnerability to circadian disruption. In the future it will be important to identify the genome-wide sites of circadian TF binding in human islets to determine whether the clock regulates enhancers containing disease-associated variants and whether the sequence variants affect circadian TF binding and rhythmic gene transcription.
Collectively, genetic analyses and physiologic studies indicate that the cell-autonomous circadian clock within pancreatic beta cells is a key regulator of mammalian glucose homeostasis and that its dysregulation is strongly associated with human diabetes. Recent drug screening studies have identified small-molecule drugs that enhance circadian clock function and protect mice from metabolic syndrome (He et al., 2016; Bass, 2016), raising the possibility that the circadian system may ultimately represent a therapeutic target to enhance beta cell metabolic maturation and function.
ACKNOWLEDGMENTS
This research was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) grants 2R01DK090625 and R01DK100814, and JDRF 17-2013-511 and 1-INO-2014-178-A-V, and the University of Chicago Diabetes Research and Training Center grant P60DK020595.
Footnotes
CONFLICT OF INTEREST STATEMENT
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: J.B. has financial interest in Reset Therapeutics.
REFERENCES
- Ando H, Kumazaki M, Motosugi Y, Ushijima K, Maekawa T, Ishikawa E, and Fujimura A (2011) Impairment of peripheral circadian clocks precedes metabolic abnormalities in ob/ob mice. Endocrinology 152:1347–1354. [DOI] [PubMed] [Google Scholar]
- Aparicio NJ, Puchulu FE, Gagliardino JJ, Ruiz M, Llorens JM, Ruiz J, Lamas A, and De Miguel R (1974) Circadian variation of the blood glucose, plasma insulin and human growth hormone levels in response to an oral glucose load in normal subjects. Diabetes 23:132–137. [DOI] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, and Turek FW (2009) Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 17:2100–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, and Schibler U (2000a) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289:2344–2347. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, and Schibler U (1998) A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93:929–937. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Marcacci L, and Schibler U (2000b) Multiple signaling pathways elicit circadian gene expression in cultured Rat-1 fibroblasts. Curr Biol 10:1291–1294. [DOI] [PubMed] [Google Scholar]
- Bartness TJ, Song CK, and Demas GE (2001) SCN efferents to peripheral tissues: implications for biological rhythms. J Biol Rhythms 16:196–204. [DOI] [PubMed] [Google Scholar]
- Bass J (2012) Circadian topology of metabolism. Nature 491:348–356. [DOI] [PubMed] [Google Scholar]
- Bass J (2016) Targeting time in metabolic therapeutics. Cell Metab 23:575–577. [DOI] [PubMed] [Google Scholar]
- Boesgaard TW, Grarup N, Jorgensen T, Borch-Johnsen K, Meta-Analysis of Glucose and Insulin-Related Trait Consortium (MAGIC), Hansen T, and Pedersen O (2010) Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia 53:1647–1655. [DOI] [PubMed] [Google Scholar]
- Bonnefond A, Clement N, Fawcett K, Yengo L, Vaillant E, Guillaume JL, Dechaume A, Payne F, Roussel R, Czernichow S, et al. (2012) Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet 44:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, et al. (2009) A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 41:89–94. [DOI] [PubMed] [Google Scholar]
- Bowen AJ and Reeves RL (1967) Diurnal variation in glucose tolerance. Arch Intern Med 119:261–264. [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, and Takahashi JS (2010) Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330:379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buijs RM, Chun SJ, Niijima A, Romijn HJ, and Nagai K (2001) Parasympathetic and sympathetic control of the pancreas: a role for the suprachiasmatic nucleus and other hypothalamic centers that are involved in the regulation of food intake. J Comp Neurol 431:405–423. [DOI] [PubMed] [Google Scholar]
- Buijs RM, la Fleur SE, Wortel J, Van Heyningen C, Zuiddam L, Mettenleiter TC, Kalsbeek A, Nagai K, and Niijima A (2003) The suprachiasmatic nucleus balances sympathetic and parasympathetic output to peripheral organs through separate preautonomic neurons. J Comp Neurol 464:36–48. [DOI] [PubMed] [Google Scholar]
- Carroll KF and Nestel PJ (1973) Diurnal variation in glucose tolerance and in insulin secretion in man. Diabetes 22:333–348. [DOI] [PubMed] [Google Scholar]
- Chaves I, van der Horst GT, Schellevis R, Nijman RM, Koerkamp MG, Holstege FC, Smidt MP, and Hoekman MF (2014) Insulin-FOXO3 signaling modulates circadian rhythms via regulation of clock transcription. Curr Biol 24:1248–1255. [DOI] [PubMed] [Google Scholar]
- Coomans CP, van den Berg SA, Lucassen EA, Houben T, Pronk AC, van der Spek RD, Kalsbeek A, Biermasz NR, Willems van Dijk K, Romijn JA, et al. (2013) The suprachiasmatic nucleus controls circadian energy metabolism and hepatic insulin sensitivity. Diabetes 62:1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes S, Boss M, Thomas AP, and Matveyenko AV (2015) Activation of melatonin signaling promotes beta-cell survival and function. Mol Endocrinol 29:682–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107:21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, and Schibler U (2000) Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev 14:2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, and Albrecht U (2010) The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72:517–549. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, and Sassone-Corsi P (2013) Reprogramming of the circadian clock by nutritional challenge. Cell 155:1464–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, Storm P, Osmark P, Ladenvall C, Prasad RB, et al. (2014) Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A 111:13924–13929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, and Lazar MA (2014) Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159:1140–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardino JJ, Hernandez RE, and Rebolledo OR (1984) Chronobiological aspects of blood glucose regulation: a new scope for the study of diabetes mellitus. Chronobiologia 11:357–379. [PubMed] [Google Scholar]
- Gaulton KJ, Ferreira T, Lee Y, Raimondo A, Magi R, Reschen ME, Mahajan A, Locke A, Rayner NW, Robertson N, et al. (2015) Genetic fine mapping and genomic annotation defines causal mechanisms at type 2 diabetes susceptibility loci. Nat Genet 47:1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, Panhuis TM, Mieczkowski P, Secchi A, Bosco D, et al. (2010) A map of open chromatin in human pancreatic islets. Nat Genet 42:255–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A, Esnault C, Aubert G, Treisman R, Pralong F, and Schibler U (2013) Blood-borne circadian signal stimulates daily oscillations in actin dynamics and SRF activity. Cell 152:492–503. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, et al. (2013) The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature 503:410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Lozano M, Mingomataj EL, Wu WK, Ridout SA, and Brubaker PL (2014) Circadian secretion of the intestinal hormone GLP-1 by the rodent L cell. Diabetes 63:3674–3685. [DOI] [PubMed] [Google Scholar]
- Gunton JE, Kulkarni RN, Yim S, Okada T, Hawthorne WJ, Tseng YH, Roberson RS, Ricordi C, O’Connell PJ, Gonzalez FJ, et al. (2005) Loss of ARNT/HIF1beta mediates altered gene expression and pancreatic-islet dysfunction in human type 2 diabetes. Cell 122:337–349. [DOI] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. (2012) Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab 15:848–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Nohara K, Park N, Park YS, Guillory B, Zhao Z, Garcia JM, Koike N, Lee CC, Takahashi JS, et al. (2016) The small molecule nobiletin targets the molecular oscillator to enhance circadian rhythms and protect against metabolic syndrome. Cell Metab 23:610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge BA, Wen Y, Riley LA, Zhang X, England JH, Harfmann BD, Schroder EA, and Esser KA (2015) The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skelet Muscle 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C, Zhang R, Wang C, Wang J, Ma X, Hou X, Lu J, Yu W, Jiang F, Bao Y, et al. (2010) Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS One 5:e15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hong HK, Chong JL, Indacochea AA, Lee SS, Han M, Takahashi JS, and Hogenesch JB (2012) Brain-specific rescue of Clock reveals system-driven transcriptional rhythms in peripheral tissue. PLoS Genet 8:e1002835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett RJ and Keen H (1969) Diurnal variation of oral glucose tolerance: a possible pointer to the evolution of diabetes mellitus. Br Med J 2:341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp DM, Ubeda M, and Habener JF (2002) Identification and functional characterization of melatonin Mel 1a receptors in pancreatic beta cells: potential role in incretin-mediated cell function by sensitization of cAMP signaling. Mol Cell Endocrinol 191:157–166. [DOI] [PubMed] [Google Scholar]
- Knutsson A (2003) Health disorders of shift workers. Occup Med (Lond) 53:103–108. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, and Bass J (2007) High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab 6:414–421. [DOI] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, and Schibler U (2007) System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Fleur SE, Kalsbeek A, Wortel J, and Buijs RM (2000) Polysynaptic neural pathways between the hypothalamus, including the suprachiasmatic nucleus, and the liver. Brain Res 871:50–56. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, and Evans RM (2011) Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480:552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, and Weitz CJ (2008) Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A 105:15172–15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky AD, Bradley MA, Williams DL, Bass J, and Turek FW (2008) Sleep-wake regulation is altered in leptin-resistant (db/db) genetically obese and diabetic mice. Am J Physiol Regul Integr Comp Physiol 295:R2059–R2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laposky AD, Shelton J, Bass J, Dugovic C, Perrino N, and Turek FW (2006) Altered sleep regulation in leptin-deficient mice. Am J Physiol Regul Integr Comp Physiol 290:R894–R903. [DOI] [PubMed] [Google Scholar]
- Le Minh N, Damiola F, Tronche F, Schutz G, and Schibler U (2001) Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J 20:7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kim MS, Li R, Liu VY, Fu L, Moore DD, Ma K, and Yechoor VK (2011) Loss of Bmal1 leads to uncoupling and impaired glucose-stimulated insulin secretion in beta-cells. Islets 3:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moulik M, Fang Z, Saha P, Zou F, Xu Y, Nelson DL, Ma K, Moore DD, and Yechoor VK (2013) Bmal1 and beta-cell clock are required for adaptation to circadian disruption, and their loss of function leads to oxidative stress-induced beta-cell failure in mice. Mol Cell Biol 33:2327–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. (2010) Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466:627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullan CJ, Schernhammer ES, Rimm EB, Hu FB, and Forman JP (2013) Melatonin secretion and the incidence of type 2 diabetes. JAMA 309:1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megirian D, Dmochowski J, and Farkas GA (1998) Mechanism controlling sleep organization of the obese Zucker rats. J Appl Physiol (1985) 84:253–256. [DOI] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, and Rosbash M (2012) Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife 1:e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milev NB and Reddy AB (2015) Circadian redox oscillations and metabolism. Trends Endocrinol Metab 26:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistlberger RE, Lukman H, and Nadeau BG (1998) Circadian rhythms in the Zucker obese rat: assessment and intervention. Appetite 30:255–267. [DOI] [PubMed] [Google Scholar]
- Mojsov S, Weir GC, and Habener JF (1987) Insulinotropin: glucagon-like peptide I (7–37) co-encoded in the glucagon gene is a potent stimulator of insulin release in the perfused rat pancreas. J Clin Invest 79:616–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, and Schibler U (2012) Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 338:379–383. [DOI] [PubMed] [Google Scholar]
- Morris CJ, Yang JN, Garcia JI, Myers S, Bozzi I, Wang W, Buxton OM, Shea SA, and Scheer FA (2015) Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci U S A 112:E2225–E2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder H, Nagorny CL, Lyssenko V, and Groop L (2009) Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia 52:1240–1249. [DOI] [PubMed] [Google Scholar]
- Muoio DM and Newgard CB (2008) Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev Mol Cell Biol 9:193–205. [DOI] [PubMed] [Google Scholar]
- Nishiyama K and Hirai K (2014) The melatonin agonist ramelteon induces duration-dependent clock gene expression through cAMP signaling in pancreatic INS-1 beta-cells. PLoS One 9:e102073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, and Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109:307–320. [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, and Takahashi JS (2014) Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, et al. (2012) Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med 18:1768–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodriguez-Segui SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Moran I, Gomez-Marin C, van de Bunt M, et al. (2014) Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat Genet 46:136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek CB, Affinati AH, Ramsey KM, Kuo HY, Yu W, Sena LA, Ilkayeva O, Marcheva B, Kobayashi Y, Omura C, et al. (2013) Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342:1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peek CB, Ramsey KM, Levine DC, Marcheva B, Perelis M, and Bass J (2015) Circadian regulation of cellular physiology. Methods Enzymol 552:165–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelis M, Marcheva B, Ramsey KM, Schipma MJ, Hutchison AL, Taguchi A, Peek CB, Hong H, Huang W, Omura C, et al. (2015) Pancreatic beta cell enhancers regulate rhythmic transcription of genes controlling insulin secretion. Science 350:aac4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke E, Muhlbauer E, Musshoff U, Csernus VJ, Chankiewitz E, and Peschke D (2002) Receptor (MT(1)) mediated influence of melatonin on cAMP concentration and insulin secretion of rat insulinoma cells INS-1. J Pineal Res 33:63–71. [DOI] [PubMed] [Google Scholar]
- Peschke E and Peschke D (1998) Evidence for a circadian rhythm of insulin release from perifused rat pancreatic islets. Diabetologia 41:1085–1092. [DOI] [PubMed] [Google Scholar]
- Polonsky KS, Given BD, and Van Cauter E (1988) Twenty-four-hour profiles and pulsatile patterns of insulin secretion in normal and obese subjects. J Clin Invest 81:442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power ML and Schulkin J (2008) Anticipatory physiological regulation in feeding biology: cephalic phase responses. Appetite 50:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley TL and Berthoud HR (1985) Diet and cephalic phase insulin responses. Am J Clin Nutr 42:991–1002. [DOI] [PubMed] [Google Scholar]
- Pulimeno P, Mannic T, Sage D, Giovannoni L, Salmon P, Lemeille S, Giry-Laterriere M, Unser M, Bosco D, Bauer C, et al. (2013) Autonomous and self-sustained circadian oscillators displayed in human islet cells. Diabetologia 56:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Block GD, Colwell CS, and Matveyenko AV (2013) Consequences of exposure to light at night on the pancreatic islet circadian clock and function in rats. Diabetes 62:3469–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakshit K, Hsu TW, and Matveyenko AV (2016) Bmal1 is required for beta cell compensatory expansion, survival and metabolic adaptation to diet-induced obesity in mice. Diabetologia 59:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer MK and Ahren B (2002) Altered beta-cell distribution of pdx-1 and GLUT-2 after a short-term challenge with a high-fat diet in C57BL/6 J mice. Diabetes 51 Suppl 1:S138–S143. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, and Naef F (2011) Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9:e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts HJ (1964) Afternoon glucose tolerance testing: a key to the pathogenesis, early diagnosis and prognosis of diabetogenic hyperinsulinism. J Am Geriatr Soc 12:423–472. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Diaz R, Dando R, Jacques-Silva MC, Fachado A, Molina J, Abdulreda MH, Ricordi C, Roper SD, Berggren PO, and Caicedo A (2011) Alpha cells secrete acetylcholine as a non-neuronal paracrine signal priming beta cell function in humans. Nat Med 17:888–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, and Fitzgerald GA (2004) BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2:e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz de Azua I, Gautam D, Guettier JM, and Wess J (2011) Novel insights into the function of beta-cell M3 muscarinic acetylcholine receptors: therapeutic implications. Trends Endocrinol Metab 22:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad A, Dalla Man C, Nandy DK, Levine JA, Bharucha AE, Rizza RA, Basu R, Carter RE, Cobelli C, Kudva YC, et al. (2012) Diurnal pattern to insulin secretion and insulin action in healthy individuals. Diabetes 61:2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadacca LA, Lamia KA, deLemos AS, Blum B, and Weitz CJ (2011) An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Morf J, Stratmann M, Gos P, and Schibler U (2012) Simulated body temperature rhythms reveal the phase-shifting behavior and plasticity of mammalian circadian oscillators. Genes Dev 26:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Petrenko V, Pulimeno P, Giovannoni L, Berney T, Hebrok M, Howald C, Dermitzakis ET, and Dibner C (2016) A functional circadian clock is required for proper insulin secretion by human pancreatic islet cells. Diabetes Obes Metab 18:355–365. [DOI] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, and Shea SA (2009) Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A 106:4453–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U, Gotic I, Saini C, Gos P, Curie T, Emmenegger Y, Sinturel F, Gosselin P, Gerber A, Fleury-Olela F, et al. (2015) Clock-talk: interactions between central and peripheral circadian oscillators in mammals. Cold Spring Harb Symp Quant Biol 80:223–232. [DOI] [PubMed] [Google Scholar]
- Schroder EA, Harfmann BD, Zhang X, Srikuea R, England JH, Hodge BA, Wen Y, Riley LA, Yu Q, Christie A, et al. (2015) Intrinsic muscle clock is necessary for musculo-skeletal health. J Physiol 593:5387–5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seino S, Shibasaki T, and Minami K (2011) Dynamics of insulin secretion and the clinical implications for obesity and diabetes. J Clin Invest 121:2118–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro ET, Tillil H, Polonsky KS, Fang VS, Rubenstein AH, and Van Cauter E (1988) Oscillations in insulin secretion during constant glucose infusion in normal man: relationship to changes in plasma glucose. J Clin Endocrinol Metab 67:307–314. [DOI] [PubMed] [Google Scholar]
- Staiger H, Machicao F, Schafer SA, Kirchhoff K, Kantartzis K, Guthoff M, Silbernagel G, Stefan N, Haring HU, and Fritsche A (2008) Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine beta-cell function. PLoS One 3:e3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenkovic JA, Olsson AH, Nagorny CL, Malmgren S, Dekker-Nitert M, Ling C, and Mulder H (2012) Regulation of core clock genes in human islets. Metabolism 61:978–985. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Zinkin NT, Stanojevic V, Clarke WL, and Habener JF (1997) Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet 15:106–110. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, and Weitz CJ (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417:78–83. [DOI] [PubMed] [Google Scholar]
- Sturis J, Scheen AJ, Leproult R, Polonsky KS, and van Cauter E (1995) 24-hour glucose profiles during continuous or oscillatory insulin infusion: demonstration of the functional significance of ultradian insulin oscillations. J Clin Invest 95:1464–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. (2005) Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308:1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, Nagano M, Nakahama K, Suzuki Y, Sugano S, et al. (2002) A transcription factor response element for gene expression during circadian night. Nature 418:534–539. [DOI] [PubMed] [Google Scholar]
- Van Cauter E, Polonsky KS, and Scheen AJ (1997) Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr Rev 18:716–738. [DOI] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, and Takahashi JS (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, and Panda S (2012) Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab 16:833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gall C, Stehle JH, and Weaver DR (2002) Mammalian melatonin receptors: molecular biology and signal transduction. Cell Tissue Res 309:151–162. [DOI] [PubMed] [Google Scholar]
- Woon PY, Kaisaki PJ, Braganca J, Bihoreau MT, Levy JC, Farrall M, and Gauguier D (2007) Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc Natl Acad Sci U S A 104:14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Chen L, Grant GR, Paschos G, Song WL, Musiek ES, Lee V, McLoughlin SC, Grosser T, Cotsarelis G, et al. (2016) Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 8:324ra16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. (2010) Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16:1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, and Hogenesch JB (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang Y, Zhou M, Wang S, Hua Z, and Zhang J (2012) Loss of mPer2 increases plasma insulin levels by enhanced glucose-stimulated insulin secretion and impaired insulin clearance in mice. FEBS Lett 586: 1306–1311. [DOI] [PubMed] [Google Scholar]


