Abstract
The nucleolus is the largest sub-nuclear domain, serving primarily as the place for ribosome biogenesis. A delicately regulated function of the nucleolus is vital to the cell not only for maintaining proper protein synthesis but is also tightly associated with responses to different types of cellular stresses. Recently, several long non-coding RNAs (lncRNAs) were found to be part of the regulatory network that modulate nucleolar functions. Several of these lncRNAs are encoded in the ribosomal DNA (rDNA) repeats or are transcribed from the genomic regions that are located near the nucleolus organizer regions (NORs). In this review, we first discuss the current understanding of the sequence of the NORs and variations between different NORs. We then focus on the NOR-derived lncRNAs in mammalian cells and their functions in rRNA transcription and the organization of nucleolar structure under different cellular conditions. The identification of these lncRNAs reveals great potential of the NORs in harboring novel genes involved in the regulation of nucleolar functions.
Introduction
As the most prominent structure in almost every single eukaryotic interphase nucleus, the nucleolus has long been known to act as the factory of ribosome biogenesis, including the transcription of the rRNA gene by RNA Polymerase I (Pol I) to the processing of precursor rRNA (pre-rRNA) and the assembly of ribosomal proteins. Additionally, nucleoli play vital roles in other biological processes such as organizing the 3D-genome architecture, acting as a protein reservoir, and sensing environmental stresses (Bersaglieri and Santoro 2019; Boisvert et al. 2007; Boulon et al. 2010; Frottin et al. 2019; Iarovaia et al. 2019; Quinodoz et al. 2018). Thus, proper regulation of nucleolar functions is crucial for normal cellular metabolism and requires a complex molecular network.
The past several decades have witnessed an explosion in our knowledge of long non-coding RNA (lncRNA) genes. lncRNAs are defined as RNAs that are longer than 200 nucleotides (nt) and are not translated into any proteins. Far from their initial impression as ‘genomic junk’ or ‘transcription noise,’ lncRNAs have been proved to play essential roles in all aspects of biological processes, including regulating gene expression at the transcriptional and post-transcriptional levels, chromatin organization, and cellular condensate formation (Guh et al. 2020; Guo et al. 2021; Kopp and Mendell 2018; MacDonald and Mann 2020; Yao et al. 2019). The molecular functions of lncRNAs are often exerted through interactions between the lncRNA and proteins or nucleic acids. This ability of lncRNAs to interact with a wide range of molecules allows them to play versatile roles, such as recruiters, scaffolds, blockers, sponges, etc. (Engreitz et al. 2016; Gil and Ulitsky 2020; Statello et al. 2021; Sun et al. 2018; Thomson and Dinger 2016). Many lncRNAs are key components of specific subcellular domains, and thus, their functions are tightly associated with the functions of the domains (Bridges et al. 2021; Chen 2016). For example, MALAT1 localizes to nuclear speckles and regulates transcription and pre-mRNA splicing (Sun et al. 2018). NEAT1 is localized to paraspeckles, where it is required for the integrity of paraspeckles and functions in pre-mRNA processing (Clemson et al. 2009; Fox et al. 2018; Wang et al. 2020). Several lncRNA species have been reported to localize to the nucleolus and regulate on nucleolar functions. Some of the nucleolar lncRNAs are transcribed outside of the nucleolus and are then directed to nucleoli depending on either the snoRNA-like elements in their sequences (Li et al. 2018; Xing et al. 2017) or specific interactions with certain nucleolar proteins (Wang et al. 2021). Another important mode of lncRNA function is to act locally, regulating the expression of nearby genes or influencing the organization of the surrounding chromatin environment (Gil and Ulitsky 2020; Kopp and Mendell 2018; Statello et al. 2021). Indeed, several lncRNAs are transcribed within the nucleolus and regulate rRNA biogenesis or organize nucleolus structure.
DNA within the nucleolus: nucleolus organizer regions (NORs)
Nucleoli are organized surrounding NORs, which are composed of genomic loci containing rDNA repeats. In humans, regions within the p-arms of the five acrocentric chromosomes Chr13, Chr14, Chr15, Chr21, and Chr22, are devoted to NORs (Fig. 1A). Knowledge about the genomic contents of NORs and the surrounding region within the p-arms (Fig. 1B) remains incomplete. Sequences beyond the rDNA repeats on the NORs are largely unannotated due to inadequate of current sequencing and contig assembly methods for highly repetitive regions (McStay 2016). Recent studies revealed that rRNA genes are not the only active gene elements that are transcribed from these regions (Floutsakou et al. 2013; van Sluis et al. 2019). However, lack of sequence information prevents a systematic discovery of potential novel genes.
Fig. 1.
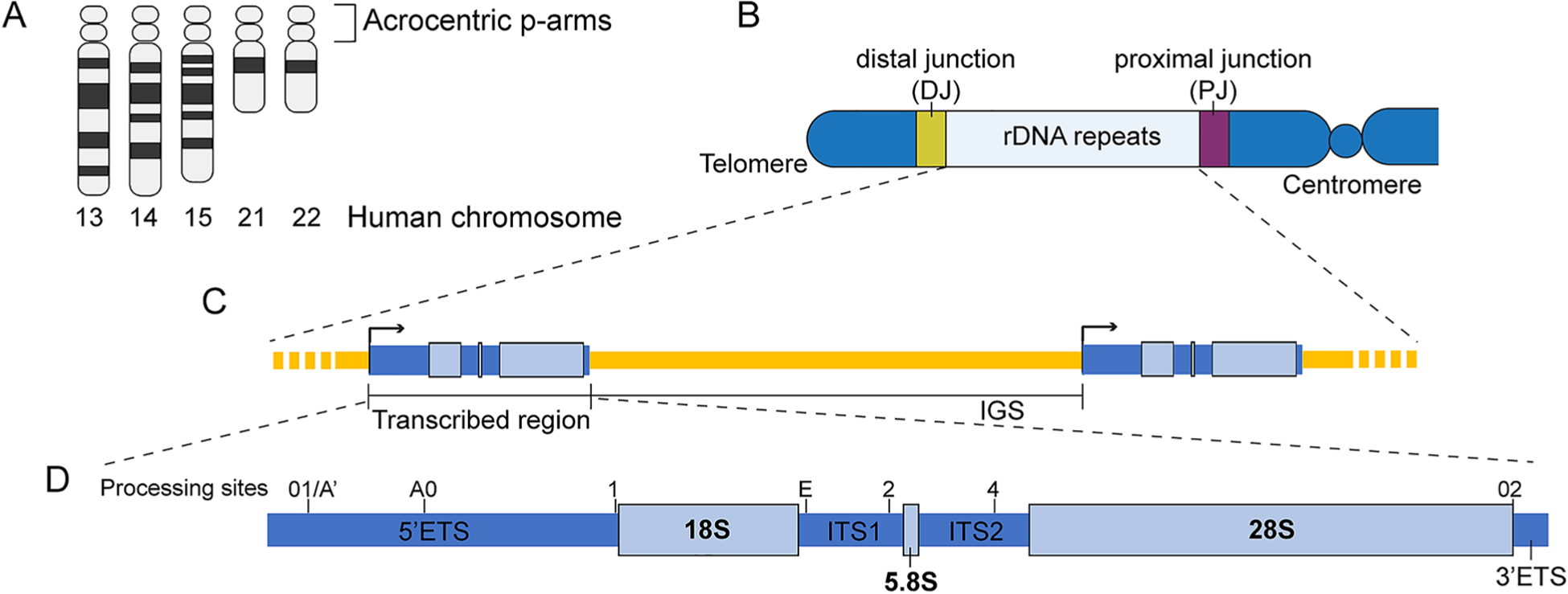
NOR arrangement. A A schematic of the five acrocentric chromosomes in human cells. B A schematic of the organization of the acrocentric p-arm. C A schematic showing that rDNA genes form a head-to-tail tandem array on the acrocentric p-arms. D A schematic of the rDNA transcribed region in human cells. Major pre-rRNA processing sites are indicated. IGS intergenic spacer, ETS external transcribed spacer, ITS internal transcribed spacer
Variations within the rDNA repeat
A significant part of the NOR is made up of the rDNA repeats arranged in a head-to-tail tandem array (Fig. 1C), each of which contains the transcribed region that produces full-length pre-rRNA and the intergenic spacer (IGS) region. Besides the sequence of the 18S, 5.8S, and 28S, the transcribed region also contains several spacer sequences, including 5′ and 3′ external transcribed spacers (ETSs) and two internal transcribed spacers (ITS1 between 18S and 5.8S and ITS2 between 5.8S and 28S) (Fig. 1D). These regions are removed from pre-rRNA during rRNA synthesis. Interestingly, although the sequences of the mature rRNAs are highly conserved across species (Arnheim et al. 1980; Sinclair and Brown 1971), spacer regions have rapidly diverged. Although some studies have suggested several structurally conserved motifs within the ITS1 and ITS2 regions, their sequences and lengths vary extensively among species. In fact, sequences of the transcribed spacers are widely used for the identification of fungi species (Chen et al. 2001; Fujita et al. 2001; Schoch et al. 2012). Additionally, differences in the structures and lengths of the IGS were found not only across species but also between individuals of a single species (Arnheim et al. 1980; Reeder et al. 1976; Smirnov et al. 2016). The size of the IGS ranges from ~ 2 kbp in yeast to ~ 30 kbp in humans. This sequence evolution, particularly seen in the expansion of size, suggests that regulatory elements and even functional genes may be emerging within these spacer regions during evolution.
While the differences within transcribed spacers largely contribute to the efficiency and accuracy of the varied steps of pre-rRNA processing in different species, the significance of evolution of the IGS is not fully understood. The human IGS contains many different kinds of repeats, including simple repeats, microsatellites, retrotransposon-derived repeats, etc. (Gonzalez and Sylvester 1995; Smirnov et al. 2016). Among these types of repeats, Alu elements are predominant, constituting ~ 18% of the IGS (Gonzalez and Sylvester 1997). Interestingly, most of these Alu repeats are “old,” meaning that they transposed into the IGS at a very early time during evolution (Gonzalez and Sylvester 1997). Their relatively clustered distribution suggests that these “disrupted” regions might not be of regulatory importance to rDNA expression or other bioprocesses. The profound differences in IGS length occasionally seen between individuals are believed to be due to their potential to be recombination hotspots. A recent study found that a ~ 2 kbp region is perfectly duplicated in the rDNA sample collected from Chr21, and this region is not included in the current rDNA reference sequence (Kim et al. 2018). Another interesting feature of the IGS repeats is that long polypyrimidine repeats of [TC]n are found throughout the sequence, possibly serving as sites for protein binding or triple-stranded DNA formation (Sakai et al. 1995). Additionally, several lncRNAs are reported to be transcribed from different [TC] n-enriched regions and engage in sequestering certain proteins. These lncRNAs will be discussed below.
It has also been reported that the human population harbors thousands of rRNA variants (Kuo et al. 1996; Parks et al. 2018). The vast majority of the variations between variants consists of single-nucleotide polymorphisms (SNPs), which tend to occur in low-complexity regions. Some of the differences are found within functional regions of mature rRNAs, such as regions responsible for rRNA-protein binding. However, there have not been sufficient functional studies to determine whether these SNPs influence rRNA processing and/or functions. Interestingly, there are conserved variants between human and mouse, and many of these variants show tissue-specific expression in mice (Parks et al. 2018; Tseng et al. 2008). Bulk sequencing methods do not give a definitive view of the expression pattern of human rRNA variants, and thus, it is difficult to speculate their specific roles based on available analyses. In fact, oocyte-specific 5S rRNA variants have long been known in Xenopus and in several types of fish (Locati et al. 2017a, b; Wegnez et al. 1972). Recently, Locati et al. revealed that in zebrafish, the four types of rRNAs went through a transition from maternal-specific variants to somatic-specific ones (Locati et al. 2017a). The biological significance of having a distinct set of rRNAs during the early development stage is not clear. Maternal-specific rRNAs were computationally predicted to show high mRNA binding affinity (Locati et al. 2017b). A similar model seems to be observed in the case of bacteria, where rRNA variants are involved in stress-induced translational alteration by targeting different mRNAs (Kurylo et al. 2018; Song et al. 2019). It would be interesting to test whether such a mechanism exists in eukaryotic organisms.
Sequence beyond the rDNA arrays: distal and proximal junctions
Due to the highly repetitive and heterochromatic features of the NORs, the sequences flanking the rDNA arrays have long been missing from the human genome assembly. The attempts to identify the sequences adjacent to rDNA repeats are ongoing. Earlier studies using clones generated by restriction digestions managed to identify some short sequences extended beyond the rDNA repeats, usually no more than 10 kbp at a time. Thanks to the development of artificial chromosome systems such as YACs and BACs as well as the next-generation sequencing methods, several mega-base pairs of sequences have been annotated (Floutsakou et al. 2013; van Sluis et al. 2019). Despite the striking variability in the rDNA copy numbers present in each of the NORs, the sequences immediately adjacent to rDNA arrays showed an unusually high similarity across the five acrocentric chromosomes (Floutsakou et al. 2013).
A short junction clone containing sequences originated from an area between the rDNA repeats and centromere was initially identified in 1995 (Sakai et al. 1995). Based on this sequence, ~ 200 kbp of proximal junction (PJ) was obtained by screening of the BAC library (Floutsakou et al. 2013). Almost the entire PJ sequence is found to be segmentally duplicated (Floutsakou et al. 2013), which is a common feature of centromeric and pericentromeric regions (She et al. 2004). The PJ sequence, thus, seems to possess little distinctiveness in the genome. Indeed, DNA-fluorescence in situ hybridization (DNA-FISH) showed that the PJ probe hybridized to other locations far from any NORs (Floutsakou et al. 2013; van Sluis et al. 2019). This feature of the PJ region makes it hard to characterize any specific regulatory elements and, therefore, hinders further exploration of the PJ sequence. Interestingly, the position of the junction of PJ/rDNA resides in ITS1 of the transcribed region, and its exact position is not uniform among acrocentric chromosomes. However, the significance of this heterogeneity has not been explored. It is of interest to examine whether the 5’ETS/18S and partial ITS1 located immediately adjacent the PJ are functional. A recent study reported that the rDNA copies at both ends of the rDNA arrays tend to accumulate more mutations and, thus, appear likely to be functional pseudogenes (Robicheau et al. 2017). More careful sequencing analyses need be performed to verify this conclusion.
The distal junction (DJ) is more widely studied because its sequence is mostly unique to acrocentric chromosomes. Not surprisingly, the DJ contains numerous repeats, including Alu-derived repeats, large blocks of Centromeric Repeats (CER), and a striking > 100 kbp inverted repeat (Floutsakou et al. 2013; van Sluis and McStay 2019). Unlike the PJ, the rDNA/DJ junction is at a fixed position within the IGS, located ~ 4 kbp upstream of the pre-rRNA transcription start site. By DNA-FISH, the DJ is observed to be anchored in the perinucleolar heterochromatin (PNH). Ectopic DJ inserted in metacentric chromosomes also localized at the PNH (Floutsakou et al. 2013), suggesting that this anchorage is likely be an active targeting event and not a passive one merely due to the close position of the DJ to rDNA within the nucleolus. Proteins such as CCCTC-binding factor (CTCF) are implied to be involved in this anchorage and chromatin organization (Floutsakou et al. 2013). However, the DJs of the silenced NORs are located away from the nucleoli, suggesting that other factors, probably the distinct structure of the silenced NORs counteract the nucleolus-directing function of the DJ. Even though the DJ is embedded in the heterochromatin region, its chromatin is not heterochromatic in nature but rather in a relatively open state showing transcription activity (Floutsakou et al. 2013; van Sluis et al. 2019). Two RNA Pol II-transcribed, fully processed lncRNAs are readily detected from the inverted repeat region of DJ (van Sluis et al. 2019). Details regarding these two RNAs will be discussed in a later section. It is reasonable to believe that more novel transcripts might be produced from the DJ or PJ regions that have the potential to exert important regulatory functions.
The sequence of rDNA repeats present within the NORs on different acrocentric chromosomes is revealed to be homogenous. This is generally believed to be a result of exchanges of rDNA repeats between heterologous chromosomes. The PJ and DJ sequences also show extensive similarity among NOR-containing chromosomes and seem to be conserved among primates (Gonzalez and Sylvester 1997; Sakai et al. 1995; van Sluis et al. 2019). For example, ~ 200 kbp PJ sequence shows a 93.3% inter-chromosomal conservation, with variations predominantly attributed to the Alu repeats and the position of the PJ/rDNA junction (Floutsakou et al. 2013). On the other hand, the DJ sequence shows a higher overall inter-chromosomal identity (~ 99% similarity), exhibiting gradually decreased conservation (~ 90% similarity) preferentially at the far-distal region (toward the telomere) (van Sluis et al. 2019). In this particular study, the authors categorized the DJ contigs into three groups based on the major variations within the DJ sequence (van Sluis et al. 2019). Members in the same group share > 99.5% identity even in the far-distal regions. Two heterologous acrocentric chromosomes can harbor precisely the same indels within their DJ sequences. These observations indicate that inter-chromosomal exchanges on the acrocentric arms are not restricted only to rDNA repeats but rather involves larger chromosomal regions. More strikingly, structural variations within these repeat sequences were readily observed by DNA-FISH not only among human individuals but also between homologous alleles within a single cell (van Sluis et al. 2019). Whether such variations are caused by intrinsic sequence differences between alleles is difficult to determine by the current sequencing technologies due to difficulties in assigning reads to individual acrocentric chromosomes. In addition, the current sampling size is still limited, preventing us from drawing a full picture of the total variations among acrocentric chromosomes in the human population. Nevertheless, it raises an interesting possibility that at least in humans, NORs on different acrocentric chromosomes might be more diverged than what was historically represented. These NORs might harbor specific subsets of genes and/or regulatory elements making them distinct from each other.
Long non-coding RNA and nucleolar functions
A recent study in mouse neural cells revealed an enrichment of several lncRNAs in the nucleoli compared to protein-coding mRNAs (Li et al. 2018). Although a thorough transcriptome analysis of the human nucleolus is not yet performed, there is ample evidence to speculate that human nucleolus also contains a significant number of lncRNAs. In fact, many nucleolus-enriched lncRNA genes have been identified, and the regulatory roles of their encoded lncRNAs in nucleolar functions have been discovered. Several of them are transcribed from the NORs. As indicated above, the IGS region is reported to encode several RNAs which appear to be versatile in their function, having roles not only restricted to the cis regulation of rDNA genes but also influencing the overall organization of the nucleolus under certain cellular conditions (Zentner et al. 2011).
IGS-rRNA and pRNA
In varies species including mouse, rat, Xenopus, and Drosophila, the IGS bears an additional RNA Pol I promoter besides the canonical rRNA promoter (Cassidy et al. 1987; Grimaldi and Di Nocera 1988; Kuhn and Grummt 1987; Labhart and Reeder 1984; Morgan et al. 1983; Paalman et al. 1995; Putnam and Pikaard 1992). In mice, this Pol I promoter is located ~ 2 kbp upstream of the transcription start site of rRNA, producing a group of transcripts referred as IGS-rRNA (Mayer et al. 2006). Although evidence of such a promoter in humans remains to be elucidated (Agrawal and Ganley 2018), the corresponding region on human rDNA indeed produces transcripts, which behave functionally similar to those in mouse (Fig. 2A) (Guetg et al. 2012; Mayer et al. 2008; Shiao et al. 2009). IGS-rRNA itself is not stable and is maintained at very low levels in the cell (Bierhoff et al. 2010; Santoro et al. 2010). However, it is further processed into 150–300 nt mature RNAs, termed promoter-associated RNAs (pRNAs), which are stabilized by associated proteins (Bierhoff et al. 2010; Santoro et al. 2010; Savic et al. 2014). This processing is not efficient in embryonic stem cells (ESCs) and fully established in differentiated cells, leading to a transition from euchromatic to heterochromatic status of a fraction of rDNA genes during differentiation (Savic et al. 2014).
Fig. 2.
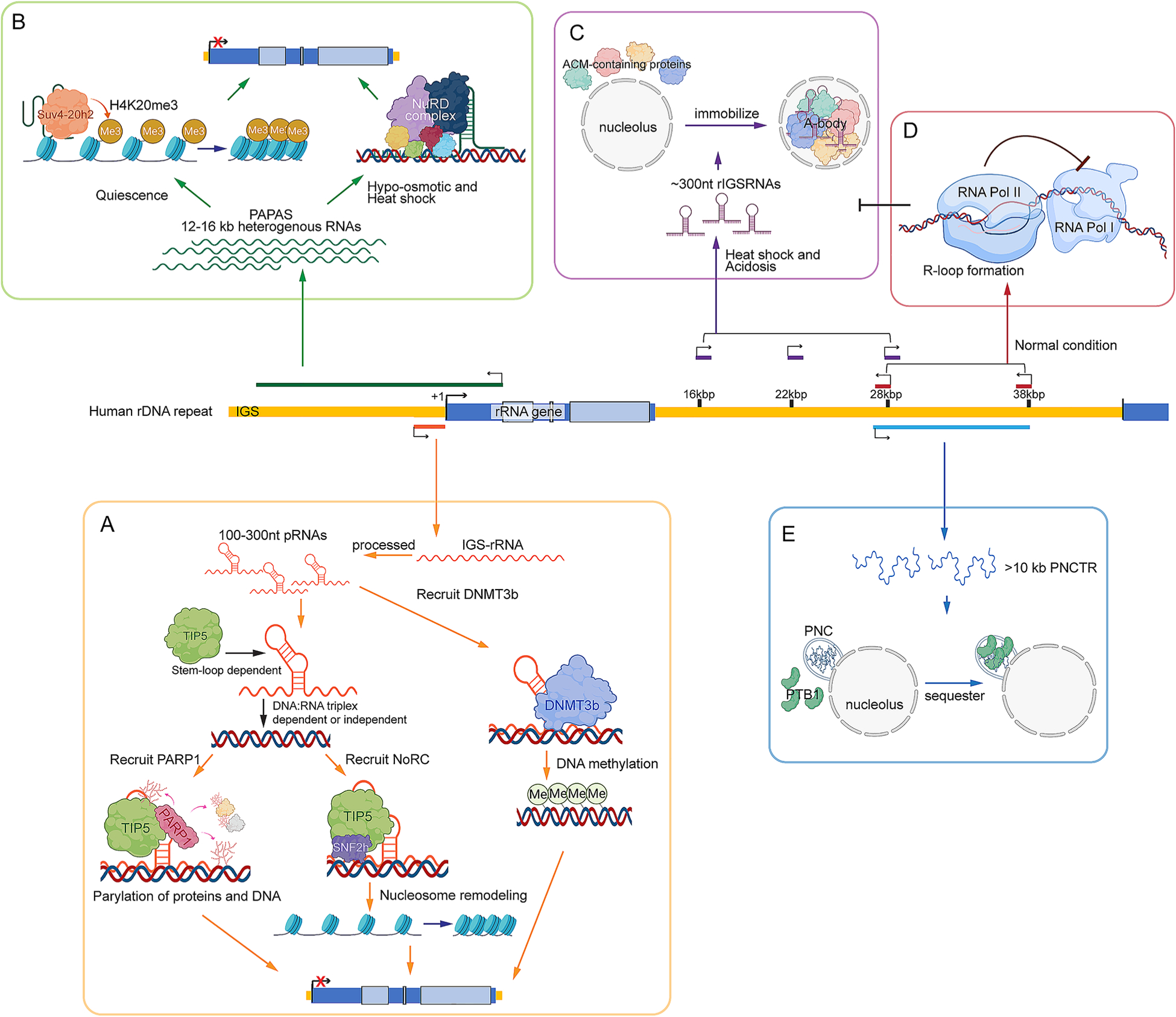
rDNA-derived lncRNAs are involved in the regulation of nucleolar functions. Schematics of the molecular mechanisms under the functions of pRNA (A), PAPAS (B), rIGSRNAs (C), asincRNAs (D), and PNCTR (E). Regions where the rDNA-derived lncRNAs are transcribed from are marked along the human rDNA repeat. + 1 indicates the transcription start site (TSS) of rRNA gene
Studies of pRNAs revealed that this group of ncRNAs functions as key regulators of the nucleolar remodeling complex (NoRC). NoRC is a nucleolar chromatin remodeling complex that consists of the ATPase SNF2h and TIP5 (transcription termination factor 1 (TTF-I)-interacting protein 5) (Strohner et al. 2001). NoRC is recruited to the inactive rDNA promoter and mediates heterochromatin formation via nucleosome sliding (Li et al. 2006) and by recruiting histone modifiers and DNA methyltransferases (Grummt and Langst 2013; Grummt and Pikaard 2003; Guetg et al. 2010; Santoro et al. 2002; Sharifi and Bierhoff 2018; Zhou and Grummt 2005; Zhou et al. 2002). It was found that pRNA modulates NoRC function by directly interacting with TIP5 using a stem-loop structure formed by the region containing rDNA sequences – 127 to – 49 (relative to the transcription start site, TSS, of rRNA) in mouse and – 137 to – 50 in human (Mayer et al. 2008, 2006). Such interaction leads to a conformational change in TIP5, which in turn influences the interaction of TIP5 with other proteins (Mayer et al. 2008). It is also proposed that the 5’ sequence of pRNA, which contains the upstream TTF-I binding site T0 sequence, is responsible for recruiting NoRC to the rDNA promoter by forming a DNA:RNA triplex (Schmitz et al. 2010). However, studies in mouse ESCs suggest that the interaction between pRNAs and TIP5 through the stem-loop structure is sufficient for the recruitment of TIP5 to the nucleolus independent of the formation of DNA:RNA triplex (Savic et al. 2014). On the other hand, the abundance of IGS-rRNA in ESCs compared to differentiated cells impaired the interaction between TIP5 and TTF1, explaining the lack of TIP5 recruitment to rDNA and abolished nucleolar heterochromatin formation in ESCs (Savic et al. 2014). A recent study also suggested that the pRNA-NoRC interaction switches off the ATPase activity of NoRC (Manelyte et al. 2014). Thus, pRNA could act as both a tethering tool for NoRC and also a controller to prevent hyperactivation of the NoRC-mediated nucleosome remodeling activity. In addition, pRNA is also involved in the recruitment of poly (ADP-ribose)-polymerase-1 (PARP1) (Guetg et al. 2012) and DNMT3b (Schmitz et al. 2010) to rDNA, emphasizing its indispensable role in rDNA silencing. The negative regulatory function of pRNA on rRNA expression is in agreement with the increased expression of pRNA in mid-late S phase when NoRC binds to the newly replicated silent rDNA genes (Li et al. 2005; Santoro et al. 2010).
There are still questions remaining about the regulation of IGS-rRNA and pRNA. According to an earlier study, pRNA consists of a group of RNA species of different sizes (Mayer et al. 2006). However, the underlying mechanism that controls the differential processing of an IGS-rRNA precursor transcript into pRNA of varying lengths is yet to be understood. It is not certain whether all of the pRNA transcripts of varying lengths execute similar functions. In addition, the regulatory mechanisms underlying the differential processing of pRNA during differentiation remain to be determined. It would be interesting to elucidate whether the pRNA processing is modulated under other circumstances (such as cellular senescence) when the silencing of rRNA genes needs to be reprogrammed. Recently, DHX9 (DExH-Box Helicase 9) was shown to be involved in the processing of IGS-rRNA to pRNA (Leone et al. 2017). However, it was observed that the ATP-helicase activity of DHX9 was not required for pRNA maturation. Considering the RNA-independent interaction between TIP5 and DHX9, it is unclear whether DHX9 acts upstream or downstream in the pRNA processing pathway. Second, the fact that pRNA is generated from a subfraction of hypomethylated active rDNA repeats but directs the maintenance of the repressive status of heterochromatic rDNA repeats suggests that pRNA inhibits the transcription of rDNA repeats in trans (Santoro et al. 2010). It would be interesting to see what epigenetic marks or other chromatic features of the newly replicated silent rDNA repeats direct the recruitment of pRNA to them in trans.
PAPAS
While pRNA is key to the establishment of a relatively stable environment to repress rRNA transcription, another rDNA-derived lncRNA, promoter and pre-rRNA antisense (PAPAS), is involved in the transient repression of rRNA transcription upon specific environmental stresses. PAPAS represents a heterogenous pool of 12–16 kb long transcripts that are transcribed from the antisense strand of rDNA by RNA Pol II, covering a region from the pre-rRNA coding region to the rRNA promoter in the IGS (Fig. 2B) (Bierhoff et al. 2014, 2010). Rather than being synthesized from a defined promoter, PAPAS is transcribed from multiple start sites by RNA polymerase II (Bierhoff et al. 2010). In dividing cells, PAPAS is maintained at lower level even compared to pRNA (Bierhoff et al. 2010). The expression of PAPAS is induced in quiescent cells and also by different types of stresses, including hypotonic stress and heat shock (Bierhoff et al. 2014; Zhao et al. 2016a, b, 2018). All of these conditions are known to partially and transiently shut down rRNA transcription. However, the molecular mechanisms underlying the negative regulatory function of PAPAS on rRNA transcription vary under different conditions. During quiescence, PAPAS mediates the interaction between Suv4-20h2 and UBF-positive rDNA promoters, leading to trimethylation of H4K20 and subsequent chromatin compaction resulting in the repression of rRNA transcription (Bierhoff et al. 2014). Upon stresses such as hypo-osmotic stress and heat shock, Suv4-20h2 is largely degraded through Nedd4/Rsp5p-mediated proteolysis (Zhao et al. 2016a, b). In such instances, PAPAS exerts its repressive function on rRNA transcription by recruiting nucleosome remodeling and deacetylation complex (NuRD) to the rDNA regulatory elements (Zhao et al. 2016a, b, 2018). NuRD is a versatile complex involved in transcriptional repression via chromatin remodeling and histone deacetylation activities (Lai and Wade 2011; Torchy et al. 2015). NuRD is also responsible for the establishment of transcriptionally poised chromatin status at rRNA genes (Xie et al. 2012). Upon heat shock, CHD4, the ATPase subunit of NuRD, is dephosphorylated, which in turn increases its binding affinity with PAPAS (Zhao et al. 2018). In vitro studies show that murine PAPAS recognizes the T-stretch of the enhancer region-located upstream of rRNA TSS in a sequence-specific manner and forms a DNA:RNA triplex. Such interactions, thus, tether NuRD to the target rDNA promoter (Zhao et al. 2018). However, there is no evidence for such a T-stretch of sequence within human rDNA repeats, which raises the question of how PAPAS tethers NuRD in human cells. In addition, there remain several questions regarding whether PAPAS acts in cis or trans. As a low abundant lncRNA that regulates the activity of a gene of numerous copies like rDNA, it is not intuitive to propose that PAPAS functions in trans. On the other hand, for a cis-acting model, it would be interesting to examine whether PAPAS is transcribed from an actively selected array of rDNA repeats and what RNA pol II regulatory factors could be dictating the expression of PAPAS. Nevertheless, the example of PAPAS exemplifies the great potential of lncRNA in regulating nucleolar chromatin structure and gene expression. Similar to PAPAS, quiescence-induced lncRNAs from intracisternal A particle (IAP) retroelements recruit Suv4-20H2 to the IAP elements in order to promote H4K20me3 modification (Bierhoff et al. 2014). These examples highlight common lncRNA-mediated regulatory mechanisms controlling epigenetic modifications resulting in gene repression.
rIGSRNAs
Human IGS contains numerous [TC]n or [GA]n simple repeats. Interestingly, three regions within the IGS enriched with simple repeats are transcribed by RNA Pol I in response to certain cellular stresses and are further processed into ~ 300 nt lncRNAs (Fig. 2C). These regions are located ~ 16 kbp, ~ 22 kbp, and ~ 28 kbp downstream of the TSS of rRNAs. Therefore, these lncRNAs are named rIGS16, rIGS22, and rIGS28, respectively (Audas et al. 2016, 2012; Wang et al. 2018). Interestingly, different rIGSs are induced by specific cellular stresses; rIGS16 and rIGS22 are induced by heat shock, while rIGS28 is induced only under acidified conditions. These lncRNAs play vital roles in the nucleolus reorganization, specifically by redirecting certain proteins into nucleoli (Audas et al. 2012). Such proteins share similar motifs called amyloid-converting motif (ACM), which contain arginine/histidine clusters (Audas et al. 2016). rIGSRNAs recognize the short cationic ACM and concentrate the ACM-containing proteins in the nucleolus (Wang et al. 2018). Once the seed is established, more proteins aggregate in the nucleolus and are immobilized via amyloidogenesis. Eventually, a reversible amyloid body (A-body) is formed by these ACM-containing proteins at the center of the nucleolus. The discovery of rIGSRNAs and protein amyloidogenesis emphasizes the role of the nucleolus in biological processes besides ribosome biogenesis. It opens a new view that lncRNAs, especially those containing low-complexity sequences, are potentially involved in amyloid formation, a process that is frequently related to diseases such as Alzheimer’s and Huntington’s diseases. In addition, IGS might produce a larger-than-expected pool of transcripts in response to other types of cellular stresses (Audas et al. 2012). One study linked the transcription stress-induced rIGS36 and rIGS39 with the recruitment of PHF6 to the nucleolus and the regulation of rRNA transcription (Todd et al. 2016). Further investigations are required to test whether a general mode of action could be drawn for these different types of stress-induced rIGSRNAs.
sincRNAs and asincRNAs
The IGS region is enriched with active RNA Pol II across the entire sequence (Abraham et al. 2020). This suggests that besides PAPAS, many more RNA Pol II transcripts are produced from the IGS. A recent study focused on antisense intergenic ncRNAs (asincRNAs) that are transcribed by RNA Pol II from the regions ~ 28 kbp and ~ 38 kbp downstream of the rRNA TSS (Fig. 2D). However, in the case of asincRNAs, the act of transcription might be more important for the maintenance of proper nucleolar structure than the actual asincRNA transcripts. The authors observed that the continuous activity of RNA Pol II establishes an R-loop shield, preventing RNA Pol I recruitment to the locus. The continuous transcription from the asincRNA locus under normal conditions, thus, prevents the unwanted RNA pol I-catalyzed synthesis of sense intergenic ncRNAs (sincRNAs) from the same region, including rIGSRNAs (Abraham et al. 2020).
PNCTR
Pyrimidine-rich non-coding transcript (PNCTR) is another lncRNA species transcribed from the ~ 28 kbp IGS region (Fig. 2E). This RNA Pol I transcript (> 10 kb) is much more abundant than rIGSRNAs and shows higher levels in cancer cells compared to normal cells (Yap et al. 2018). The pyrimidine-rich sequence within the PNCTR lncRNA harbors thousands of predicted binding sites of polypyrimidine tract-binding protein 1 (PTB1), an RNA-binding protein (RBP) that regulates pre-mRNA processing. The authors observed that PNCTR sequesters PTB1 to a nuclear domain associated with the nucleolus termed the perinucleolar compartment (PNC), thereby repressing PTB1-mediated pre-mRNA splicing (Yap et al. 2018).
DJ transcripts
As indicated earlier, the DJ sequence located next to NORs harbors two lncRNAs, named disnor 187 and disnor 238 (Floutsakou et al. 2013; van Sluis et al. 2019). These two transcripts are transcribed by RNA Pol II and are processed through RNA splicing and polyadenylation. RNA-FISH experiments using disnor 187 and disnor 238 probes revealed the presence of up to 10 discrete signals at the nucleolar periphery. This localization pattern was similar to that of the DJ DNA-FISH signal, suggesting that these DJ-derived lncRNAs are enriched at their transcription sites (van Sluis et al. 2019). Knocking down of the DJ transcripts resulted in slower cell growth without any obvious cell cycle arrest. In a proportion of the DJ-derived lncRNA-depleted cells, nucleolar segregation was also readily observed, concomitant with defects in RNA Pol I transcription (van Sluis et al. 2019). Future studies would unravel how the DJ-encoded transcripts modulate nucleolus functions.
Perspectives
NOR-derived RNAs symphonize to maintain nucleolus integrity, transiently or stably regulate rRNA gene expression, and help the cells to respond to various cellular stresses. NOR-derived RNA species have the advantage that they are produced directly from the nucleolus so that they could function locally. This is probably the reason why most of them have the very low expression level in the cells. For the lncRNAs that are transcribed from the rDNA repeats, it is intriguing to see how the rDNA is arranged to harbor all these regulatory elements within a rather small region. The evidence so far indicates that these lncRNAs do not have well-defined start sites or endpoints. Based on these, it is tempting to speculate that RNA polymerase units bind throughout the rDNA repeat, and the firing of a subset of them within a certain region of the rDNA repeat in response to cellular stress is regulated by yet-to-be-determined factors. Another thing that we need to keep in mind is that the sequence of the rDNA, except for the coding regions, is generally not well conserved among mammalian species. This raises the question of whether the underlying regulatory mechanisms remain the same in human and mouse even if similar regulatory RNAs are derived in both species. In addition, the recent data from DJ-derived transcripts highlighted the argument that the genomic regions beyond the rDNA repeats may harbor regulatory genes that are yet to be annotated and characterized. With the advances in more accurate long-read sequencing methods, it is reasonable to believe that the boundaries of the known regions flanking the rDNA repeats will be annotated soon (Nurk et al. 2021), and this would result in the identification of novel RNAs that might play essential roles in regulating nucleolar functions.
Acknowledgements
We thank Ms. You Jin Song from Prasanth’s laboratory for her valuable comments. The research in KVP laboratory is supported by funding from National Institute of Health [R01GM132458 and R21AG065748], National Science Foundation [MCB1723008], and Cancer center at Illinois seed grant and Prairie Dragon Paddlers.
Footnotes
Conflict of interest On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, Wang M, Bokros M, Vidya E, Ostrowski LA et al. (2020) Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585:298–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Ganley ARD (2018) The conservation landscape of the human ribosomal RNA gene repeats. PLoS ONE 13:e0207531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheim N, Krystal M, Schmickel R, Wilson G, Ryder O, Zimmer E (1980) Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci USA 77:7323–7327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audas TE, Jacob MD, Lee S (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45:147–157 [DOI] [PubMed] [Google Scholar]
- Audas TE, Audas DE, Jacob MD, Ho JJ, Khacho M, Wang M, Perera JK, Gardiner C, Bennett CA, Head T et al. (2016) Adaptation to stressors by systemic protein amyloidogenesis. Dev Cell 39:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersaglieri C, Santoro R (2019) Genome organization in and around the nucleolus. Cells 8:579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierhoff H, Schmitz K, Maass F, Ye J, Grummt I (2010) Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol 75:357–364 [DOI] [PubMed] [Google Scholar]
- Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I (2014) Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell 54:675–682 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585 [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI (2010) The nucleolus under stress. Mol Cell 40:216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges MC, Daulagala AC, Kourtidis A (2021) LNCcation: lncRNA localization and function. J Cell Biol. 10.1083/jcb.202009045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy BG, Yang-Yen HF, Rothblum LI (1987) Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol 7:2388–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL (2016) Linking long noncoding RNA localization and function. Trends Biochem Sci 41:761–772 [DOI] [PubMed] [Google Scholar]
- Chen YC, Eisner JD, Kattar MM, Rassoulian-Barrett SL, Lafe K, Bui U, Limaye AP, Cookson BT (2001) Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J Clin Microbiol 39:4042–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33:717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Ollikainen N, Guttman M (2016) Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 17:756–770 [DOI] [PubMed] [Google Scholar]
- Floutsakou I, Agrawal S, Nguyen TT, Seoighe C, Ganley AR, McStay B (2013) The shared genomic architecture of human nucleolar organizer regions. Genome Res 23:2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, Bond CS (2018) Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci 43:124–135 [DOI] [PubMed] [Google Scholar]
- Frottin F, Schueder F, Tiwary S, Gupta R, Korner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS (2019) The nucleolus functions as a phase-separated protein quality control compartment. Science 365:342–347 [DOI] [PubMed] [Google Scholar]
- Fujita SI, Senda Y, Nakaguchi S, Hashimoto T (2001) Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J Clin Microbiol 39:3617–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil N, Ulitsky I (2020) Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 21:102–117 [DOI] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE (1995) Complete sequence of the 43-kb human ribosomal DNA repeat: analysis of the intergenic spacer. Genomics 27:320–328 [DOI] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE (1997) Beyond ribosomal DNA: on towards the telomere. Chromosoma 105:431–437 [DOI] [PubMed] [Google Scholar]
- Grimaldi G, Di Nocera PP (1988) Multiple repeated units in Drosophila melanogaster ribosomal DNA spacer stimulate rRNA precursor transcription. Proc Natl Acad Sci USA 85:5502–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Langst G (2013) Epigenetic control of RNA polymerase I transcription in mammalian cells. Biochim Biophys Acta 1829:393–404 [DOI] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS (2003) Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4:641–649 [DOI] [PubMed] [Google Scholar]
- Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R (2010) The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J 29:2135–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Scheifele F, Rosenthal F, Hottiger MO, Santoro R (2012) Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol Cell 45:790–800 [DOI] [PubMed] [Google Scholar]
- Guh CY, Hsieh YH, Chu HP (2020) Functions and properties of nuclear lncRNAs-from systematically mapping the interactomes of lncRNAs. J Biomed Sci 27:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Shi X, Wang X (2021) RNA and liquid-liquid phase separation. Noncoding RNA Res 6:92–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iarovaia OV, Minina EP, Sheval EV, Onichtchouk D, Dokudovskaya S, Razin SV, Vassetzky YS (2019) Nucleolus: a central hub for nuclear functions. Trends Cell Biol 29:647–659 [DOI] [PubMed] [Google Scholar]
- Kim JH, Dilthey AT, Nagaraja R, Lee HS, Koren S, Dudekula D, Wood Iii WH, Piao Y, Ogurtsov AY, Utani K et al. (2018) Variation in human chromosome 21 ribosomal RNA genes characterized by TAR cloning and long-read sequencing. Nucleic Acids Res 46:6712–6725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172:393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Grummt I (1987) A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J 6:3487–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo BA, Gonzalez IL, Gillespie DA, Sylvester JE (1996) Human ribosomal RNA variants from a single individual and their expression in different tissues. Nucleic Acids Res 24:4817–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurylo CM, Parks MM, Juette MF, Zinshteyn B, Altman RB, Thibado JK, Vincent CT, Blanchard SC (2018) Endogenous rRNA sequence variation can regulate stress response gene expression and phenotype. Cell Rep 25:236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P, Reeder RH (1984) Enhancer-like properties of the 60/81 bp elements in the ribosomal gene spacer of Xenopus laevis. Cell 37:285–289 [DOI] [PubMed] [Google Scholar]
- Lai AY, Wade PA (2011) Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer 11:588–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone S, Bar D, Slabber CF, Dalcher D, Santoro R (2017) The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep 18:1248–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang J, Wang M, Li X, Gong H, Tang H, Chen L, Wan L, Liu Q (2018) Activity dependent LoNA regulates translation by coordinating rRNA transcription and methylation. Nat Commun 9:1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I (2005) The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J 24:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Langst G, Grummt I (2006) NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J 25:5735–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locati MD, Pagano JF, Ensink WA, van Olst M, van Leeuwen S, Nehrdich U, Zhu K, Spaink HP, Girard G, Rauwerda H et al. (2017a) Linking maternal and somatic 5S rRNA types with different sequence-specific non-LTR retrotransposons. RNA 23:446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locati MD, Pagano JFB, Girard G, Ensink WA, van Olst M, van Leeuwen S, Nehrdich U, Spaink HP, Rauwerda H, Jonker MJ et al. (2017b) Expression of distinct maternal and somatic 5.8S, 18S, and 28S rRNA types during zebrafish development. RNA 23:1188–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald WA, Mann MRW (2020) Long noncoding RNA functionality in imprinted domain regulation. PLoS Genet 16:e1008930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manelyte L, Strohner R, Gross T, Langst G (2014) Chromatin targeting signals, nucleosome positioning mechanism and non-coding RNA-mediated regulation of the chromatin remodeling complex NoRC. PLoS Genet 10:e1004157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R (2006) Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22:351–361 [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I (2008) The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 9:774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B (2016) Nucleolar organizer regions: genomic “dark matter” requiring illumination. Genes Dev 30:1598–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GT, Reeder RH, Bakken AH (1983) Transcription in cloned spacers of Xenopus laevis ribosomal DNA. Proc Natl Acad Sci USA 80:6490–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A et al. (2021) The complete sequence of a human genome. bioRxiv. 10.1101/2021.05.26.445798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paalman MH, Henderson SL, Sollner-Webb B (1995) Stimulation of the mouse rRNA gene promoter by a distal spacer promoter. Mol Cell Biol 15:4648–4656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, Blanchard SC (2018) Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv 4:eaao0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Pikaard CS (1992) Cooperative binding of the Xenopus RNA polymerase I transcription factor xUBF to repetitive ribosomal gene enhancers. Mol Cell Biol 12:4970–4980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y et al. (2018) Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174:744–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder RH, Brown DD, Wellauer PK, Dawid IB (1976) Patterns of ribosomal DNA spacer lengths are inherited. J Mol Biol 105:507–516 [DOI] [PubMed] [Google Scholar]
- Robicheau BM, Susko E, Harrigan AM, Snyder M (2017) Ribosomal RNA genes contribute to the formation of Pseudogenes and Junk DNA in the human genome. Genome Biol Evol 9:380–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Ohta T, Minoshima S, Kudoh J, Wang Y, de Jong PJ, Shimizu N (1995) Human ribosomal RNA gene cluster: identification of the proximal end containing a novel tandem repeat sequence. Genomics 26:521–526 [DOI] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I (2002) The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet 32:393–396 [DOI] [PubMed] [Google Scholar]
- Santoro R, Schmitz KM, Sandoval J, Grummt I (2010) Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO Rep 11:52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic N, Bar D, Leone S, Frommel SC, Weber FA, Vollenweider E, Ferrari E, Ziegler U, Kaech A, Shakhova O et al. (2014) lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15:720–734 [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24:2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding C (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi S, Bierhoff H (2018) Regulation of RNA polymerase I transcription in development, disease, and aging. Annu Rev Biochem 87:51–73 [DOI] [PubMed] [Google Scholar]
- She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C et al. (2004) The structure and evolution of centromeric transition regions within the human genome. Nature 430:857–864 [DOI] [PubMed] [Google Scholar]
- Shiao YH, Lupascu ST, Gu YD, Kasprzak W, Hwang CJ, Fields JR, Leighty RM, Quinones O, Shapiro BA, Alvord WG et al. (2009) An intergenic non-coding rRNA correlated with expression of the rRNA and frequency of an rRNA single nucleotide polymorphism in lung cancer cells. PLoS ONE 4:e7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair JH, Brown DD (1971) Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry 10:2761–2769 [DOI] [PubMed] [Google Scholar]
- Smirnov E, Cmarko D, Mazel T, Hornacek M, Raska I (2016) Nucleolar DNA: the host and the guests. Histochem Cell Biol 145:359–372 [DOI] [PubMed] [Google Scholar]
- Song W, Joo M, Yeom JH, Shin E, Lee M, Choi HK, Hwang J, Kim YI, Seo R, Lee JE et al. (2019) Divergent rRNAs as regulators of gene expression at the ribosome level. Nat Microbiol 4:515–526 [DOI] [PubMed] [Google Scholar]
- Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22:96–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I (2001) NoRC–a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J 20:4892–4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Hao Q, Prasanth KV (2018) Nuclear long noncoding RNAs: key regulators of gene expression. Trends Genet 34:142–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17:272–283 [DOI] [PubMed] [Google Scholar]
- Todd MA, Huh MS, Picketts DJ (2016) The sub-nucleolar localization of PHF6 defines its role in rDNA transcription and early processing events. Eur J Hum Genet 24:1453–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchy MP, Hamiche A, Klaholz BP (2015) Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci 72:2491–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H, Chou W, Wang J, Zhang X, Zhang S, Schultz RM (2008) Mouse ribosomal RNA genes contain multiple differentially regulated variants. PLoS ONE 3:e1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sluis M, McStay B (2019) Nucleolar DNA double-strand break responses underpinning rDNA genomic stability. Trends Genet 35:743–753 [DOI] [PubMed] [Google Scholar]
- van Sluis M, Gailin MO, McCarter JGW, Mangan H, Grob A, McStay B (2019) Human NORs, comprising rDNA arrays and functionally conserved distal elements, are located within dynamic chromosomal regions. Genes Dev 33:1688–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tao X, Jacob MD, Bennett CA, Ho JJD, Gonzalgo ML, Audas TE, Lee S (2018) Stress-induced low complexity RNA activates physiological amyloidogenesis. Cell Rep 24:1713–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li K, Huang W (2020) Long non-coding RNA NEAT1-centric gene regulation. Cell Mol Life Sci 77:3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hu X, Song W, Xu H, Xiao Z, Huang R, Bai Q, Zhang F, Chen Y, Liu Y et al. (2021) Mutual dependency between lncRNA LETN and protein NPM1 in controlling the nucleolar structure and functions sustaining cell proliferation. Cell Res 31:664–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegnez M, Monier R, Denis H (1972) Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett 25:13–20 [DOI] [PubMed] [Google Scholar]
- Xie W, Ling T, Zhou Y, Feng W, Zhu Q, Stunnenberg HG, Grummt I, Tao W (2012) The chromatin remodeling complex NuRD establishes the poised state of rRNA genes characterized by bivalent histone modifications and altered nucleosome positions. Proc Natl Acad Sci USA 109:8161–8166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, Dong R, Yang L, Chen LL (2017) SLERT regulates DDX21 rings associated with pol I transcription. Cell 169:664–678 [DOI] [PubMed] [Google Scholar]
- Yao RW, Wang Y, Chen LL (2019) Cellular functions of long noncoding RNAs. Nat Cell Biol 21:542–551 [DOI] [PubMed] [Google Scholar]
- Yap K, Mukhina S, Zhang G, Tan JSC, Ong HS, Makeyev EV (2018) A short tandem repeat-enriched RNA assembles a nuclear compartment to control alternative splicing and promote cell survival. Mol Cell 72:525–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC (2011) Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res 39:4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Dammert MA, Grummt I, Bierhoff H (2016a) lncRNA-induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep 14:1876–1882 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Dammert MA, Hoppe S, Bierhoff H, Grummt I (2016b) Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res 44:8144–8152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Senturk N, Song C, Grummt I (2018) lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev 32:836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Grummt I (2005) The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol 15:1434–1438 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Santoro R, Grummt I (2002) The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J 21:4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]


