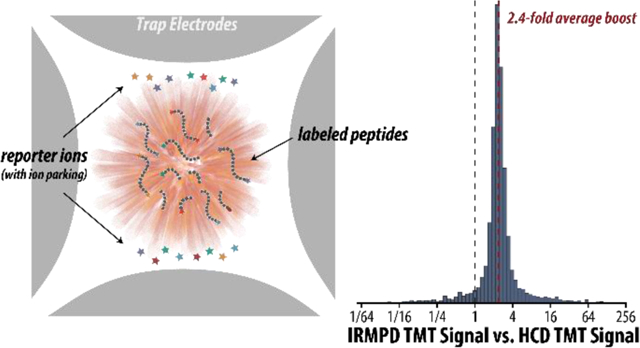
Abstract
Isobaric tagging facilitates multiplexed experiments that can determine sequences and relative amounts of peptides in biological samples using tandem mass spectrometry (MSn). Limited reporter ion generation limits quantitative accuracy and precision. As reporter ions are susceptible to unintended fragmentation and scattering by high-energy collisions, we activated peptides with IR photons and prevented successive dissociation of generated reporter ions with ion parking, which altogether boosted reporter ion yield by up to 55%. Even so, unintended co-isolation of contaminating peaks in MS2 experiments distorts reporter ion intensities and can distort quantitative information. MS3 experiments address contamination by generating reporter ions via collisional activation (HCD) of one or more peptide product ions rather than the isolated peptide precursor ion. Because HCD performance is related to m/z, activation of multiple synchronously isolated product ions generates less than optimal reporter ion intensities. In this work, we show that using infrared multiphoton dissociation (IRMPD), which is not dependent on m/z, to generate reporter ions from ten synchronously isolated peptide product ions results in a 2.4-fold increase in reporter ion intensities, significantly enhancing the sensitivity and dynamic range of quantitation via isobaric tagging.
Graphical Abstract
INTRODUCTION
By affording the ability to multi-plex protein analysis, isobaric tags, e.g., Tandem Mass Tags (TMT) [1], iTRAQ reagents [2], and N,N-dimethyl leucine (DiLeu) tags [3], provide a high-throughput approach to quantitation using mass spectrometry [4]. During tandem mass spectrometry, peptide cations labeled with isobaric reagents fragment to produce reporter ions of distinct mass-to-charge (m/z) values. These reporter ions facilitate simultaneous analysis of multiple samples, i.e., quantitative multiplexing [5,6]. In practice, however, limited reporter ion signal-to-noise (S/N) can hamper quantitative accuracy and precision. A method to boost the efficiency of reporter ion generation is especially attractive for quantitation of low abundance peptides where precursor ion flux is limited.
Besides limited reporter ion signal from low abundance peptide precursors, quantitative performance in isobaric tagging is often distorted by co-isolation of contaminating ions [7,8]. If the isolated m/z range of interest is purified through charge reduction ion/ion reactions [7] or fragmentation [8] the resulting product ions can be isolated and fragmented to produce reporter ions with reduced contamination and ratio distortion. In the latter case, multiple b- and y-type fragments are synchronously isolated via a MultiNotch isolation waveform prior to MS3 activation to boost reporter ion intensities, relative to the standard MS3 scan type [9].
Although the MultiNotch approach can mitigate ratio distortion due to contaminating ions, it introduces some challenges. Typically, higher-energy collision dissociation (HCD), i.e., beam-type collisional dissociation, is used to fragment peptides and to produce low molecular weight reporter ions [10]. HCD of individual ions is very fast and efficient; however, its dependence on precursor m/z limits reporter ion production efficiency when applied concurrently to several synchronously isolated peptide fragments. In other words, when co-fragmenting multiple product ions spanning a range of m/z values, one must compromise the collision energy setting as each fragment’s most optimal energy will be different. Lower energies are needed for low m/z ions to reduce scattering and fragmentation of the desired reporter ions, and higher energies are needed to efficiently fragment high m/z ions [11–13].
Infrared multiphoton dissociation (IRMPD) has emerged in the past three decades as a robust and effective method for peptide and protein dissociation [14,15]. Whereas HCD is dependent on precursor m/z and charge state through the implementation of a voltage offset [16–18], IRMPD is dependent on the precursor’s number of aromatic and amide chromophores, as well as charge state [19,20]. We supposed that a combination of IRMPD and ion parking [21] would maximize reporter ion generation from tagged peptides by eliminating the m/z dependence inherent in HCD. Herein, we test this hypothesis and demonstrate that IRMPD produces up to 55% more reporter ion signal than HCD of individual peptide fragments and thereby enhances relative quantitation in a multiplexed experiment of TMT-labeled peptides. We further present the compounded benefit of IRMPD when multiple fragment ions are synchronously isolated, as in MultiNotch MS3-based quantitation of TMT-tagged tryptic peptides, which produces up to an average of 139% more reporter ion signal than HCD.
EXPERIMENTAL SECTION
Materials
Ovalbumin (OVA) peptide (323 – 329) with sequence ISQAVHAAHAEINEAGR (Fisher Scientific) was labeled with the 10-plex TMT-129 and TMT-130 reagents. Desalted and dried down peptides were brought up in 200 mM (TEAB). Labeling reagents were brought to room temperature and 0.8 mg of each TMT reagent was resuspended in 100% ACN. OVA peptide was combined with TMT-129 or 130 reagent (4000:1 tag/peptide) and incubated for 4 hours at room temperature. The labeling reaction was quenched with 0.8 μL of 50% hydroxylamine, dried down, and desalted. The labeled peptide was diluted to 5 μM in 50% methanol and 0.2% formic acid in water and for electrospray mass spectrometry. Pierce TMT11plex Yeast Digest Standard (Thermo Fisher Scientific) was reconstituted in 0.2% formic acid to a final concentration of 0.25 μg/μL and 375 ng peptides were injected onto the column.
Mass Spectrometry
All MS, MS/MS and MS3 experiments were performed on a quadrupole-Orbitrap-quadrupolar linear ion trap (QLT) MS system (Orbitrap Fusion Lumos, Thermo Fisher Scientific, San Jose, CA, USA) that was modified to include a Firestar Ti-40 Synrad 40 W CO2 continuous wave laser (Mukilteo, WA, USA) which allowed for the infrared photoactivation of peptide precursors within the QLT. The implementations of an infrared laser on this Tribrid MS system have been described previously [22,23]. Adjustments to the instrument Lua control code allowed for implementation of broadband ion parking during IRMPD experiments [24]. The range of frequencies were calculated for the range of reporter ion m/z (126–131) and applied with equal amplitudes using the built-in waveform generator.
Peptide standards were infused at 3–5 μL/min using a 100 mL Hamilton GasTight Valco syringe (Reno, NV) and a Chemyx Fusion 101 syringe pump (Stafford, TX). OVA peptide precursors were introduced to the mass spectrometer via heated electrospray ionization (H-ESI) at 3.5 kV relative to ground with a source temperature of 40 °C and an inlet capillary temperature of 275 °C. The ion funnel radiofrequency (RF) was held at 60% and the source was open to atmosphere for OVA experiments. A reverse-phase column was made in-house with 1.7 μm diameter, 130 A pore size ethylene bridged hybrid C18 particles (Waters) to a length of 30 cm. A high-pressure packing station, capable of reaching pressures of 30,000 psi was used, as described elsewhere [25].
Figure 1 summarizes the MS3 HCD and IRMPD experiments. MS/MS and MS3 scans were conducted in the Orbitrap at a resolving power of 30,000 at 200 m/z. MS/MS scans used HCD with a normalized collisional energy (nce) of 30. MultiNotch MS3 scans were performed with synchronous precursor selection (SPS). For the high-throughput analysis of many tryptic peptides, data-dependent MS2 scans were followed by back-to-back HCD and IRMPD MS3 scans of the same MultiNotch isolation precursors was performed, allowing direct comparisons of methodologies. A similar back-to-back data collection strategy was implemented for the optimization of HCD collision energy (MS3), IRMPD laser power, and number of b- and y- type fragments used for SPS. For IRMPD experiments, precursor cations were activated in the middle section of the high-pressure cell of the quadrupolar linear ion trap. Ion parking limited reporter ion exposure to the laser beam thus preventing noticeable signal depletion on the time frame of the experiment. The manufacturer specifies the laser used in these experiments as a 40 W laser and IRMPD reactions ranged from 5 to 50% power. HCD normalized collision energy was varied from 10 to 110 nce. For targeted analysis on the effect of specific peptides and fragments on reporter ion generation, an MSn experimental workflow was written into the instrument’s Lua code to enable real-time data collection of targeted b- and y-type fragment ions for subsequent MS3 activation and facile iteration of method parameter optimization.
Figure 1. Overview of peptide identification and TMT quantitation with MultiNotch IRMPD MS3.
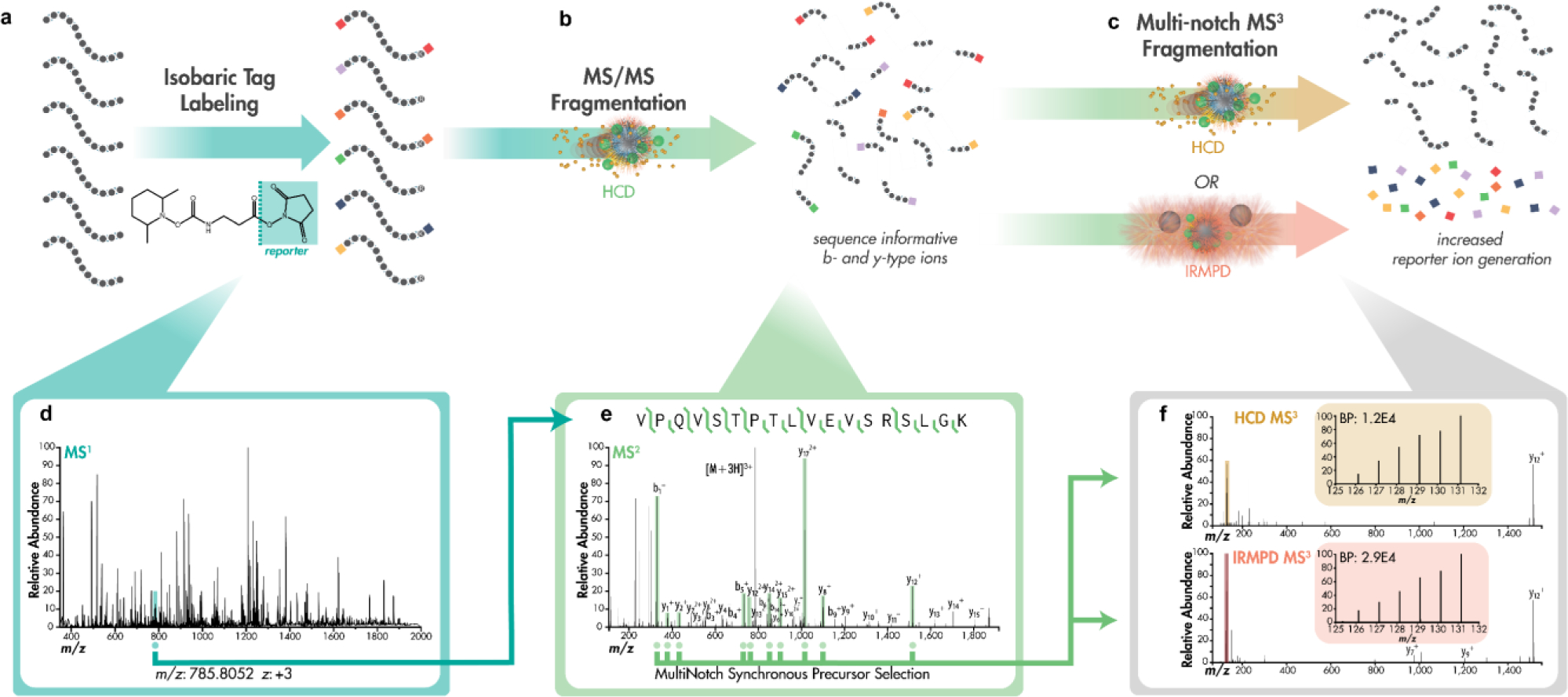
a) Stable isobaric labeling of peptides allows for quantitative mass spectrometry-based proteomics experiments in which differentially tagged peptides appear indistinguishable in MS1 scans. b) Fragmentation by lower energy HCD produces sequence informative b- and y-type ions and limited reporter ions. c) MultiNotch Synchronous Precursor Selection allows for isolation and fragmentation of high signal fragment ions by either higher energy HCD or IRMPD. d) A MS1 survey scan reveals precursor targets for e) data dependent MS2 acquisition with lower energy HCD. f) Synchronous selection of the top ten most abundant fragment ions allows for either higher energy or IRMPD to increase reporter ion generation.
Data Analysis
Mass spectra and chromatogram information was accessed in the vendor’s post-acquisition software (XCalibur Qual Browser, version 4.0). No microscans were performed. Peak lists and intensities from XTRACT were input into the Interactive Peptide Spectral Annotator (IPSA) for sequencing and fragmentation efficiency metrics [26]. All fragment matches were made within a 10-ppm tolerance. A Python script was written using RawQuant [27] to directly extract precursor, fragment, and reporter ions and their corresponding intensities (in terms of signal-to-noise). Additionally, code was written in R to plot parameter optimization and corresponding statistics. The OMSSA algorithm and COMPASS software suite were used for searching and processing data [28,29]. Peptides were searched with a 25-ppm tolerance around the monoisotopic precursor and a 10-ppm tolerance on fragment ion masses. Search results were filtered to a 1% unique peptide FDR based on expectation value (E-value) and ppm error using COMPASS [30,31]. See the Supporting Information for more detailed methods.
RESULTS AND DISCUSSION
To assess the potential benefit of IRMPD for reporter ion generation from TMT-labeled peptide fragments, we labeled OVA (323 – 329, ISQAVHAAHAEINEAGR) with the N129 TMT reagent and subjected it to HCD and IRMPD. That is, the singly-labeled, triply charged (m/z 668.7) precursor was isolated (2 Th) and dissociated with 30% normalized collisional energy (nce). Because the TMT tag is attached to the N-terminus, we identified the ten most abundant b-type ions resulting from this dissociation and subjected each of them to a range of HCD offset voltages (separately). Figure 2a presents the amount of reporter ion signal generated at different HCD offset voltages. The inset illustrates a correlation between optimal HCD offset voltage for reporter ion generation and b-type ion m/z. This correlation reveals that higher mass and lower charge ions require a steeper voltage gradient to achieve the same degree of fragmentation. While this outcome is expected based upon theory [11–13], the trend highlights a shortcoming in using HCD to generate reporter ion from co-isolated peptide fragments spanning an m/z range [8]. Although the HCD offset voltage chosen might be optimized for one of the co-isolated fragments, it will be suboptimal for the others, thus generating less reporter signal than theoretically possible.
Figure 2.
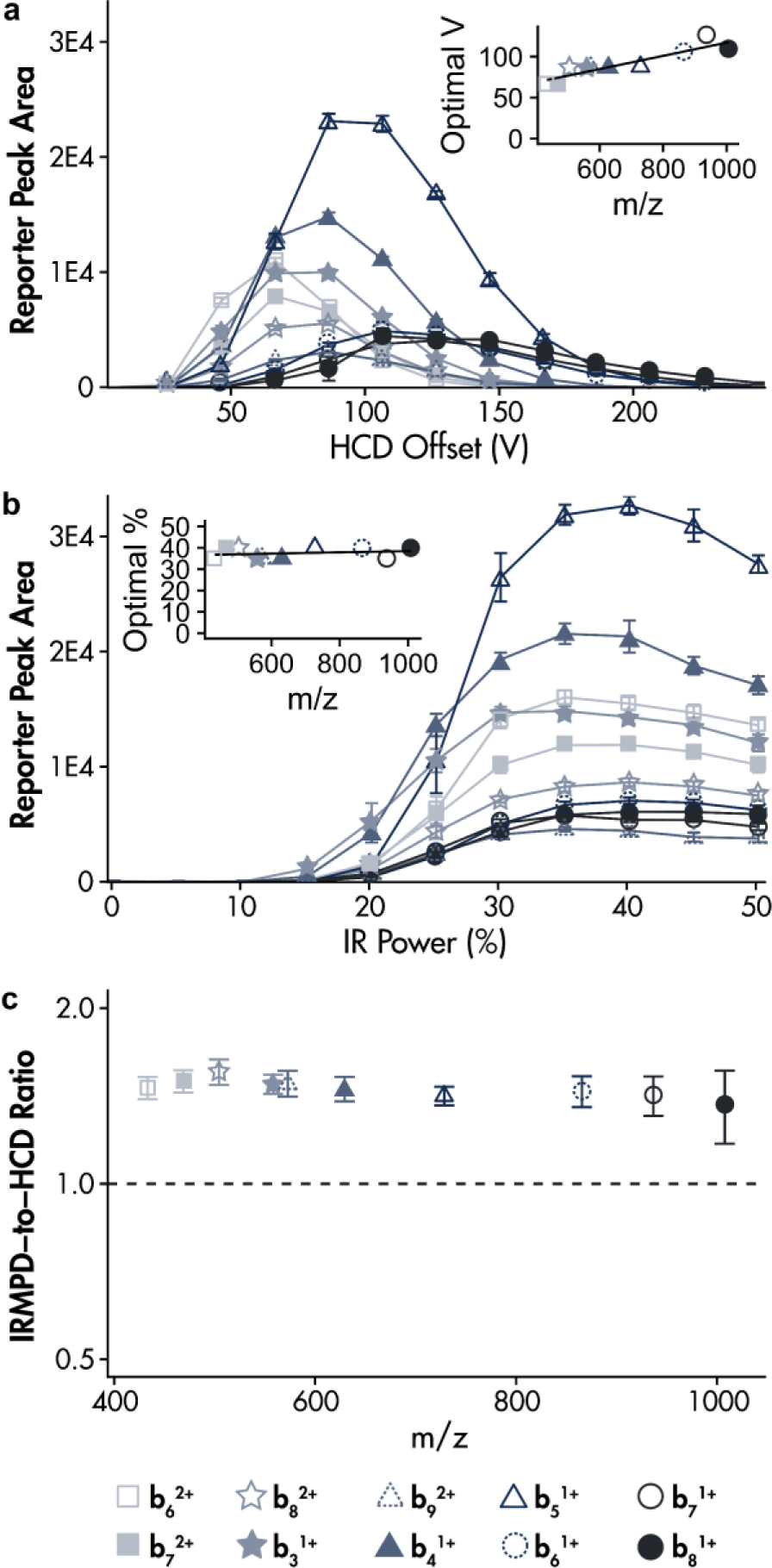
TMT reporter (130N) signal generated from the ten most abundant b-type ions resulting from 30% HCD of [OVA + 130N + 3H]3+ at various (a) HCD offset voltages and (b) IR laser powers. A 20 V parking waveform was used to preserve the TMT reporter (q = 0.8) during IRMPD. Insets illustrate that (a) the optimal HCD offset for reporter generation varies with precursor b ion m/z and (b) the optimal IR power for reporter generation is constant with respect to precursor b ion m/z. (c) Percent increase of TMT reporter generated at optimal IR power (see (b) inset) over reporter generated at optimal HCD voltage (see (a) inset) for the ten b-type ions.
Whereas optimal HCD offset voltage varies greatly with fragment m/z (60 to 90 V for these fragments), Figure 2b shows that optimal IRMPD energy for reporter ion generation from these same b-type product ions is independent of fragment m/z, with maximal reporter output at 35–40% laser power. Based on several optimization studies (see Figure S1, supplemental information), we applied 20 V of a dipolar parking waveform with the reporter ion at a q value of 0.8 to preserve the reporter during 5 ms of IR activation within the high-pressure trap. Our choice of parking conditions prioritized high q values to maximize the m/z range of the ion trap and low irradiation times to minimize the IRMPD time penalty. From this result, we conclude that IRMPD of a group of co-isolated peptide fragments could more efficiently produce reporter signal than HCD.
In addition to maximizing reporter ion generation for a group of co-isolated fragments, Figure 2c illustrates that IRMPD increases reporter signal from a single fragment. The percent increase of reporter signal generated via IRMPD with the optimal IR power from Fig. 2b over HCD with the optimal offset voltage from Fig. 2a ranges from 35% to 55% for these fragments using our IRMPD and parking setup.
To obtain data on a diverse set of peptides we performed analyses on a TMT11plex yeast digest standard. A method (vide supra) ran back-to-back scans applying either HCD or IRMPD to synchronously selected peptide fragments generated via HCD. In total we collected data from 15,850 precursors, resulting in 5,622 identified TMT-labeled peptides. Figure 3 presents the general trends in total reporter ion intensity when applying a range of HCD nce (Fig. 3a) and IR laser power (Fig. 3b) to ten synchronously selected fragments. When applied to an SPS MS3 scan, the HCD nce is calculated with respect to an intensity-weighted average mass of the synchronously selected peptide fragments. On average, the optimal HCD collision energy was 50 nce, and the optimal IR power was 40% using 5 ms irradiation. Note that the intensity weighted average m/z determines the normalized collision energy for HCD experiments using synchronously selected precursors to address the issue shown by Figure 2a. In the case of HCD activation of single precursors, the maximal TMT reporter intensity occurs at the same HCD nce for all peptides resulting from the linear relationship between precursor m/z and optimal HCD offset voltage. When activating multiple precursors, however, there is some deviation in optimal HCD nce depending on how a single HCD offset voltage is calculated from multiple values of m/z.
Figure 3.
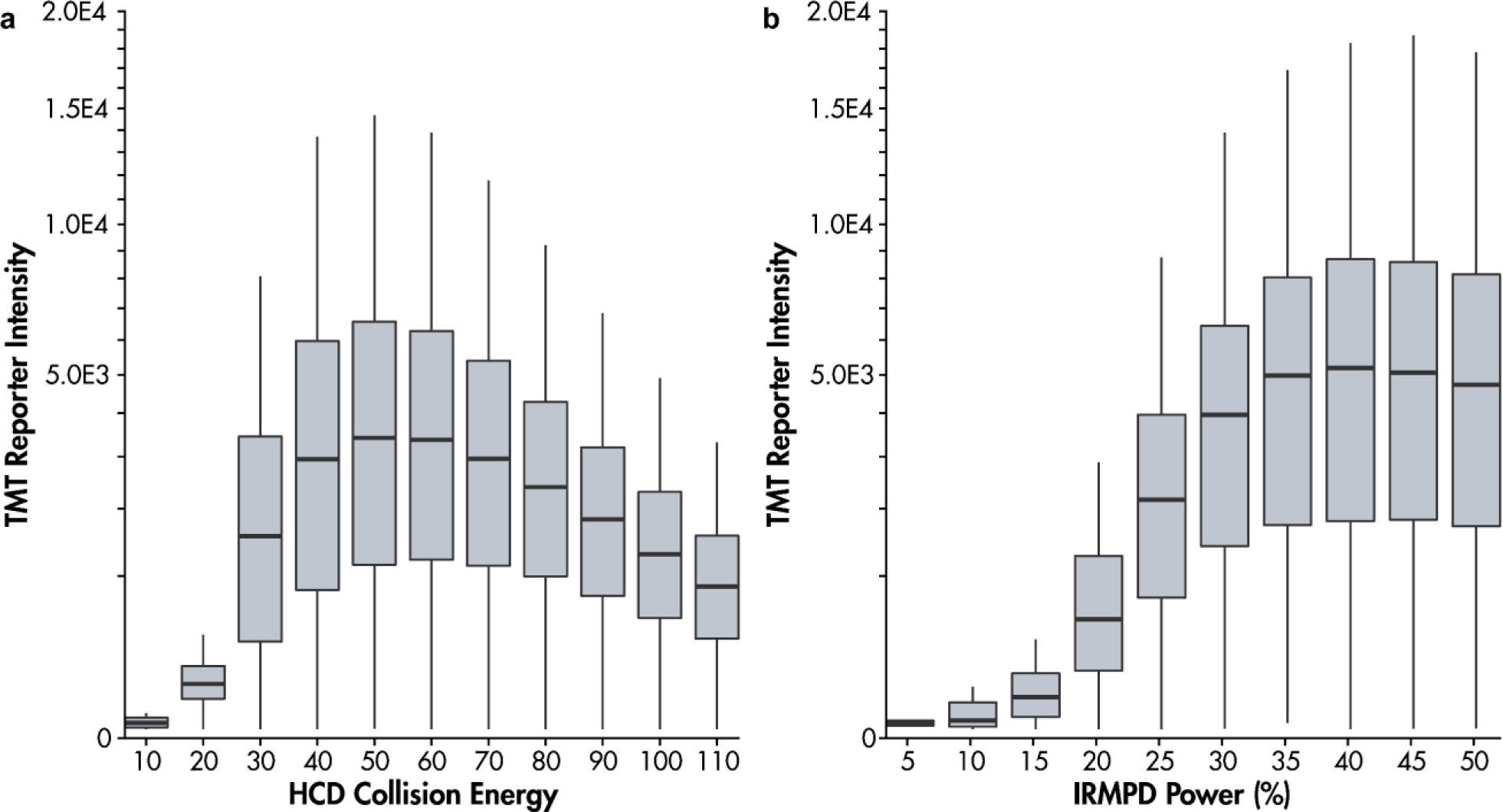
Dependence of TMT reporter intensity on (a) HCD normalized collision energy and (b) IRMPD laser power (fixed at 5 ms irradiation time). Thousands of TMT labeled peptides from the yeast digest standard were fragmented and 10 synchronously selected peptide fragments were activated with HCD and IRMPD in back-to-back scans across energy levels.
The increase in reporter ion intensity due to IRMPD relative to HCD is illustrated across yeast peptide m/z in Figure 4a. Higher m/z precursors tend to yield more reporter ions with IRMPD and/or yield less reporter ions with HCD. Fragmentation of higher m/z precursors likely generate fragments with a wider range of m/z, therefore increasing the penalty on reporter ion generation via HCD. To examine the effect of IRMPD and ion parking on reporter quantitation, the identified peptides were partitioned into intensity-based quartiles and the distribution of measured ratios among TMT reporter ion channels were compared. Specifically, the ratios were determined by calculating the average TMT reporter signal-to-noise of each of the four gene knockout variants and dividing these averages by the average of the other TMT channels (e.g., average intensity of TMT-131C and TMT-131N for WT relative to the average intensity of all other reporter channels, corresponding to Δhis4, Δmet6, and Δura2). Excepting peptides from protein knockouts, all measured ratios should be 1:1. Figure 5 shows all identified peptides ranked by their measured intensities. For each quartile, histograms display the distribution of measured reporter ratios obtained by HCD and IRMPD after applying a TMT reporter ion S/N cut-off of 100. Notably, 50% of the least abundant peptides exceed the S/N cut-off when using IRMPD as opposed to 23% when using HCD. Advancing to higher abundance peptides diminishes this discrepancy; however, IRMPD always results in more peptides exceeding the S/N cut-off of 100.
Figure 4.
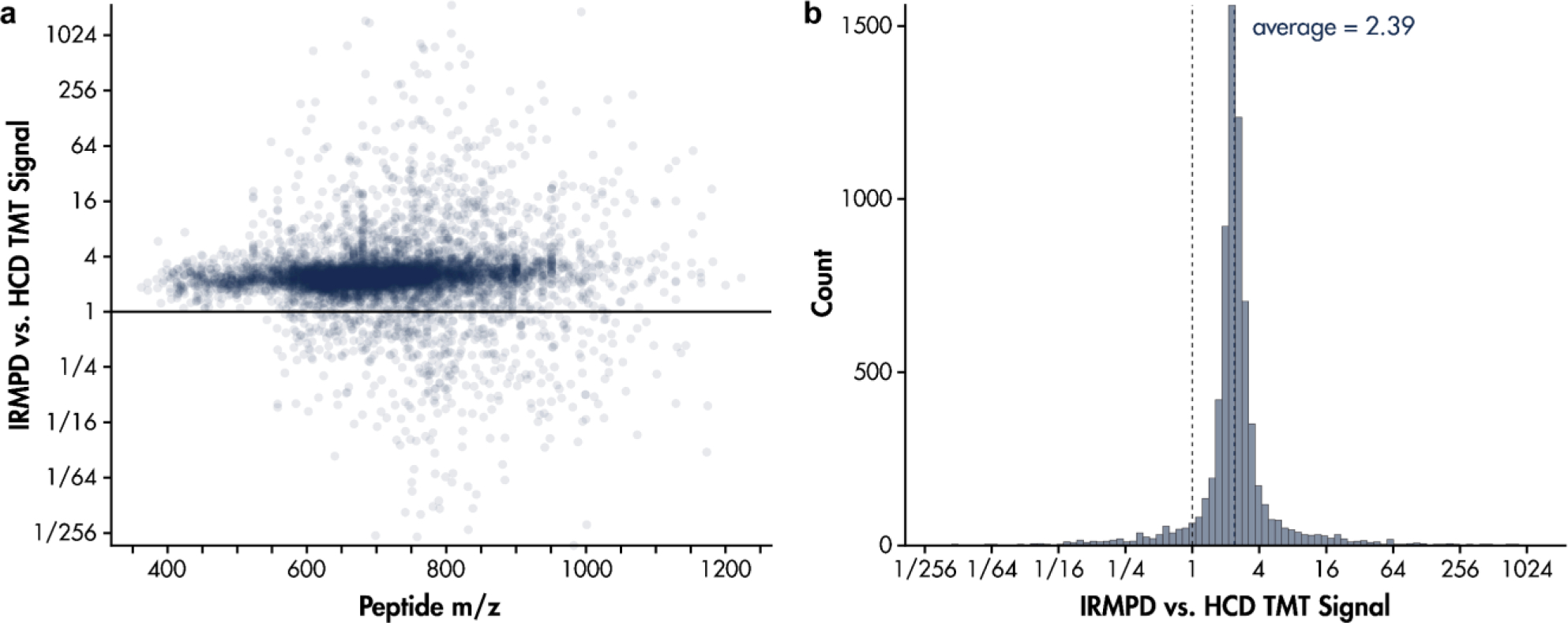
Back-to-back analysis of using IRMPD vs. HCD for generation of reporter ions from the standard TKO yeast standard. Both experiments used HCD (30 nce) to generate fragments from individually isolated peptides and either 50 nce HCD or 40% IR for 5ms to generate TMT reporter from 10 synchronously selected peptide fragments. A 20 V parking waveform was applied during IRMPD to minimize loss of reporter due to photodissociation. (a) Ratio of reporter signal generated via IRMPD over HCD vs. yeast peptide m/z. (b) Histogram of reporter signal increase due to IRMPD over HCD for all peptides.
Figure 5.
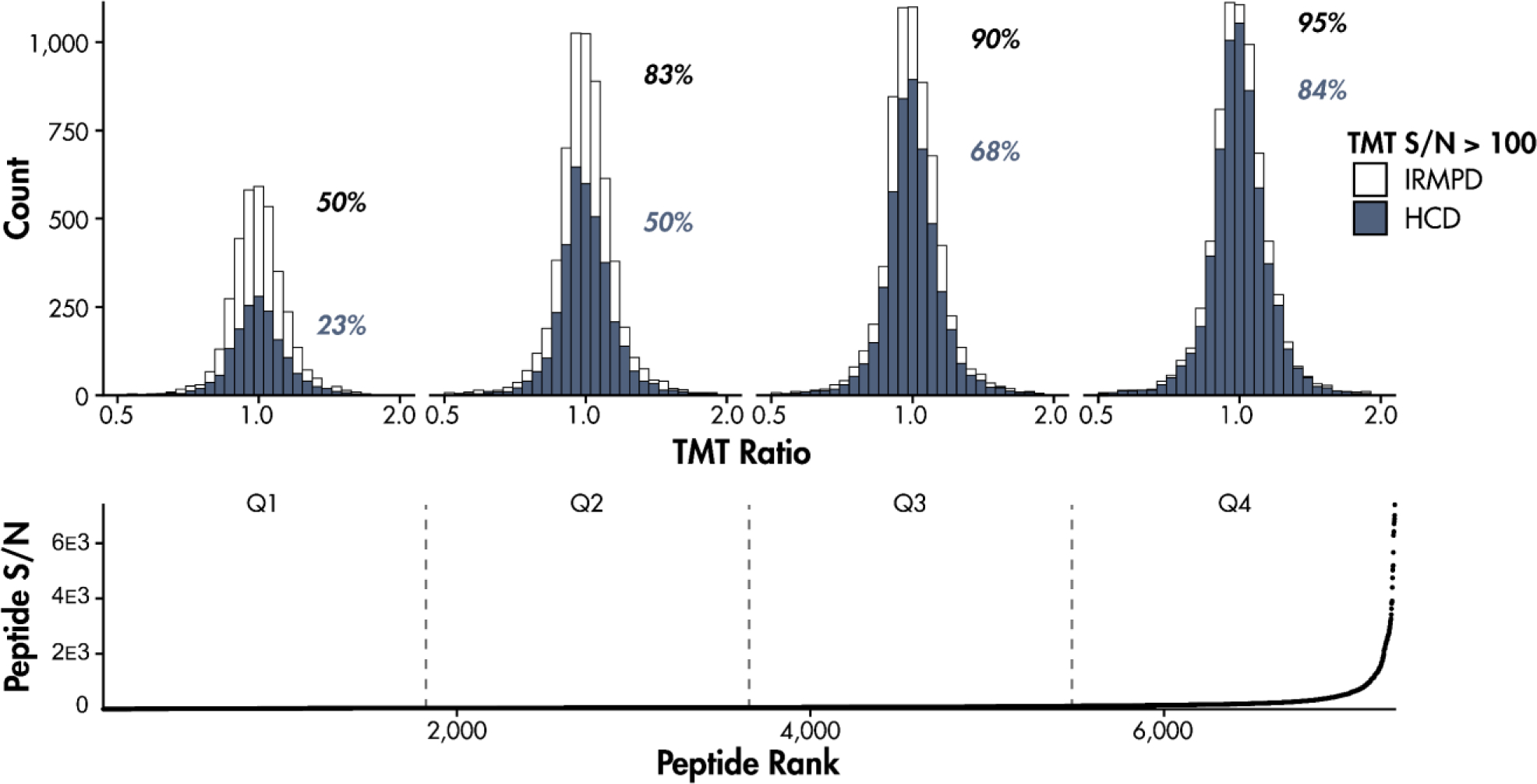
Identified PSMs plotted against their S/N and divided into four quartiles. Each quartile has histograms that illustrate the spread of ratios among TMT reporter ion channels for PSMs with an average TMT S/N greater than 100. Ratios were determined by calculating the average TMT reporter signal-to-noise of each gene knockout variant (WT, Δhis4, Δmet6, and Δura2) and dividing these averages by the average of the other TMT channels. MS3 activation method is indicated by bar color. The percentage of PSMs that exceed the S/N cut-off of 100 using each activation method are shown in each quartile.
Figure S2 shows a different perspective by partitioning the peptides into four quartiles based on TMT reporter ion S/N cut-offs of 100, 250, and 500. Whereas HCD leads to 44% of peptides having TMT SN below 100, IRMPD only produces 20% of peptide with TMT S/N below 100. Conversely, 6% of peptides exceed TMT S/N of 500 using HCD relative to 29% of peptides with IRMPD. This result further shows that IRMPD boosts reporter ion yield without distorting expected TMT ratios as shown by histograms in Figure S2.
To further illustrate benefits of improved reporter ion yield with IRMPD, triplicate LC-MS/MS experiments of TMT 6-plex tagged HAP1 tryptic peptides were quantified at the MS2-level or at the MS3-level with HCD and IRMPD (Figure S3). HCD SPS MS3 allowed for the detection of all six TMT channels for only 68% of its 16,750 PSMs, whereas IRMPD SPS MS3 allowed for detection of all six TMT channels for 80% of its 16,395 PSMs, a 15.2% boost in total quantified PSMs. Although the increased duty cycle of IRMPD experiments reduced total MS3 scans collected by 2%, the boost in reporter ion S/N enabled a greater number of peptides to be quantified relative to the HCD MS3 approach. MS2-based quantitation allowed for quantitation of 85% of all PSMs across all TMT channels, however without the quantitative accuracy of a MS3-based approach (Figure S3).
Although IRMPD produces much more TMT signal than HCD, thereby increasing the number of quantified peptides, it appears that quantitative accuracy and precision are not affected. Figure S4 further illustrates that IRMPD does not reduce interference by increasing TMT signal. The interference-free index (IFI) is defined as
for each protein knockout (his4, met6, and ura2) [32]. Thus, an IFI of 1 indicates no interference. In all three knockouts, IRMPD and HCD produce similar IFI values. This result suggests that quantitative accuracy is limited more by interference reduction through dilution of contaminating ions (e.g., MultiNotch MS3 TMT experiments) rather than improvement of TMT reporter signals.
CONCLUSION
This work builds upon the foundation of TMT-based quantitative proteomics and establishes IRMPD as an ideal activation method for TMT reporter ion generation. By increasing the reporter ion yield, IRMPD boosts the quantitative sensitivity of TMT-based experiments such that a wider dynamic range of analyte concentrations can be quantified. Additionally, higher reporter ion yield increases ion statistics which improves accuracy. This technology should be especially useful for proteomic applications where limited sample starting amounts are available such as enrichments of protein post-translational modification carrying peptide and ultimately single cell applications. In addition to benefitting applications using limited starting material, our observation that IRMPD improves reporter ion generation over HCD more as precursor m/z increases suggests an improvement in quantitation of doubly charged peptides and long peptides from, for example, non-tryptic protease digestions.
Because this approach simply substitutes IRMPD for HCD in an otherwise typical TMT MS3 workflow, it should provide additive benefits to existing isobaric labeling methods and technologies. Increased reporter ion signals should improve quantitative figures of merit for commercial TMT multiplexing reagents that presently offer up to 18-plex [33]. For other isobaric labels, such as iTRAQ [2] and DiLeu [3], reporter ion durability when irradiated will need to be investigated. Although a MS3 approach adequately addresses quantitation issues present in the traditional MS2 TMT approach, the cost has been increased time and reduced reporter ion signals. The release of Orbiter by Schweppe et al. addressed the increased time demand of an MS3 scan by eliminating uninformative MS3 scans and increased the number of quantified peptides two-fold [34].
In this work, we describe the inefficiency of HCD when activating individual peptide fragments and when synchronously activating multiple precursors and demonstrate that IRMPD boosts reporter generation by up to an average of 2.4-fold, without perturbing expected quantitative ratios. With the emergence of real-time database searching, we envision methods that trigger IRMPD scans following non-quantitative HCD scans to boost the number of accurately quantified peptides. These and further improvements have the potential to make isobaric labeling experiments more sensitive and informative, with compounded benefits for experiments utilizing MultiNotch MS3-based quantitation.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Lia Serrano, Chris Mullen, Josh Hinkle, and other members of the Thermo team for helpful discussions. This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (Grant P41GM108538 to J.J.C.) and the National Human Genome Research Institution through a training grant to the Genomic Science Training Program (Grant T32HG002760 to T.M.P.C.).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Expanded materials and methods descriptions. Figure S1, Optimization of IRMPD and parking parameters for improved reporter ion signal. Figure S2, More peptides reveal higher TMT reporter signal using IRMPD. Figure S3, IRMPD generates MS3 spectra with more TMT channels detected in a 6-plex experiment. Figure S4, Interference plots comparing KO peptides between HCD and IRMPD.
Notes
J.J.C. is a consultant for Thermo Fisher Scientific. Raw data files are available online on Chorus (Project ID 1726).
REFERENCES
- [1].Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C, Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS, Anal. Chem. (2003). 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- [2].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ, Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents, Mol. Cell. Proteomics. 3 (2004) 1154–1169. 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- [3].Xiang F, Ye H, Chen R, Fu Q, Li N, N,N-Dimethyl leucines as novel Isobaric tandem mass tags for quantitative proteomics and peptidomics, Anal. Chem. 82 (2010) 2817–2825. 10.1021/ac902778d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Boersema PJ, Aye TT, Van Veen TAB, Heck AJR, Mohammed S, Triplex protein quantification based on stable isotope labeling by peptide dimethylation applied to cell and tissue lysates, Proteomics. 8 (2008) 4624–4632. 10.1002/pmic.200800297. [DOI] [PubMed] [Google Scholar]
- [5].Werner T, Becher I, Sweetman G, Doce C, Savitski MM, Bantscheff M, High-resolution enabled TMT 8-plexing, Anal. Chem. 84 (2012) 7188–7194. 10.1021/ac301553x. [DOI] [PubMed] [Google Scholar]
- [6].Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC, Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags, Anal. Chem. 80 (2008) 2921–2931. 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- [7].Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ, Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging, Nat. Methods. 8 (2011) 933–935. 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ting L, Rad R, Gygi SP, Haas W, MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics, Nat. Methods. 8 (2011) 937–940. 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP, MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes, Anal. Chem. 86 (2014) 7150–7158. 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Savitski MM, Mathieson T, Zinn N, Sweetman G, Doce C, Becher I, Pachl F, Kuster B, Bantscheff M, Measuring and managing ratio compression for accurate iTRAQ/TMT quantification, J. Proteome Res. 12 (2013) 3586–3598. 10.1021/pr400098r. [DOI] [PubMed] [Google Scholar]
- [11].Diedrich JK, Pinto AFM, Yates JR, Energy dependence of HCD on peptide fragmentation: Stepped collisional energy finds the sweet spot, J. Am. Soc. Mass Spectrom. 24 (2013) 1690–1699. 10.1007/s13361-013-0709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dayon L, Pasquarello C, Hoogland C, Sanchez JC, Scherl A, Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags, J. Proteomics. 73 (2010) 769–777. 10.1016/j.jprot.2009.10.015. [DOI] [PubMed] [Google Scholar]
- [13].Scherl A, Shaffer SA, Taylor GK, Hernandez P, Appel RD, Binz PA, Goodlett DR, On the Benefits of Acquiring Peptide Fragment Ions at High Measured Mass Accuracy, J. Am. Soc. Mass Spectrom. 19 (2008) 891–901. 10.1016/j.jasms.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Little DP, Speir JP, Senko MW, Connor PBO, McLafferty FW, Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing, Anal. Chem. 66 (1994) 2809–2815. 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- [15].Crowe MC, Brodbelt JS, Infrared multiphoton dissociation (IRMPD) and collisionally activated dissociation of peptides in a quadrupole ion trap with selective IRMPD of phosphopeptides, J. Am. Soc. Mass Spectrom. 15 (2004) 1581–1592. 10.1016/j.jasms.2004.07.016. [DOI] [PubMed] [Google Scholar]
- [16].Cooks RG, Collision-Induced Dissociation of Polyatomic Ions, in: Collis. Spectrosc, Springer US, 1978: pp. 357–450. 10.1007/978-1-4613-3955-7_8. [DOI] [Google Scholar]
- [17].McLuckey SA, Principles of collisional activation in analytical mass spectrometry, J. Am. Soc. Mass Spectrom. 3 (1992) 599–614. [DOI] [PubMed] [Google Scholar]
- [18].Shukla AK, Futrell JH, Tandem mass spectrometry: Dissociation of ions by collisional activation, J. Mass Spectrom. 35 (2000) 1069–1090. . [DOI] [PubMed] [Google Scholar]
- [19].Madsen JA, Brodbelt JS, Comparison of Infrared Multiphoton Dissociation and Collision-Induced Dissociation of Supercharged Peptides in Ion Traps, J. Am. Soc. Mass Spectrom. 20 (2009) 349–358. 10.1016/j.jasms.2008.10.018. [DOI] [PubMed] [Google Scholar]
- [20].Hunt DF, Shabanowitz J, Yates JR, Mclver RT, Hunter RL, Syka JEP, Amy J, Tandem Quadrupole—Fourier Transform Mass Spectrometry of Oligopeptides, Anal. Chem. 57 (1985) 2728–2733. 10.1021/ac00290a065. [DOI] [PubMed] [Google Scholar]
- [21].Chrisman PA, Pitteri SJ, McLuckey SA, Parallel ion parking of protein mixtures, Anal. Chem. 78 (2006) 310–316. 10.1021/ac0515778. [DOI] [PubMed] [Google Scholar]
- [22].Riley NM, Westphall MS, Hebert AS, Coon JJ, Implementation of activated ion electron transfer dissociation on a quadrupole-Orbitrap-linear ion trap hybrid mass spectrometer, Anal. Chem. 89 (2017) 6358–6366. 10.1021/acs.analchem.7b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Peters-Clarke T, Schauer K, Riley N, Lodge J, Westphall M, Coon J, Optical Fiber-Enabled Photoactivation of Peptides and Proteins, Anal. Chem. 92 (n.d.) 12363–12370. 10.1021/acs.analchem.0c02087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McLuckey SA, Chrisman PA, Pitteri SJ, Parallel ion parking in ion traps, (2006) 19 pp. [DOI] [PubMed] [Google Scholar]
- [25].Shishkova E, Hebert AS, Westphall MS, Coon JJ, Ultra-High Pressure (>30,000 psi) Packing of Capillary Columns Enhancing Depth of Shotgun Proteomic Analyses, Anal. Chem. 90 (2018) 11503–11508. 10.1021/acs.analchem.8b02766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Brademan DR, Riley NM, Kwiecien NW, Coon JJ, Interactive peptide spectral annotator: A versatile web-based tool for proteomic applications, Mol. Cell. Proteomics. 14 (2019) 193–201. http://www.mcponline.org/ (accessed October 2, 2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kovalchik KA, Moggridge S, Chen DDY, Morin GB, Hughes CS, Parsing and Quantification of Raw Orbitrap Mass Spectrometer Data Using RawQuant, J. Proteome Res. 17 (2018) 2237–2247. 10.1021/acs.jproteome.8b00072. [DOI] [PubMed] [Google Scholar]
- [28].Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH, Open mass spectrometry search algorithm, J. Proteome Res. 3 (2004) 958–964. 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- [29].Wenger CD, Phanstiel DH, Lee MV, Bailey DJ, Coon JJ, COMPASS: A suite of pre- and post-search proteomics software tools for OMSSA, Proteomics. (2011). 10.1002/pmic.201000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nesvizhskii AI, Aebersold R, Interpretation of shotgun proteomic data: The protein inference problem, Mol. Cell. Proteomics. 4 (2005) 1419–1440. 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- [31].Elias JE, Gygi SP, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry, Nat. Methods. 4 (2007) 207–214. 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- [32].Paulo JA, O’Connell JD, Gygi SP, A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments, J. Am. Soc. Mass Spectrom. 27 (2016) 1620–1625. 10.1007/s13361-016-1434-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li J, Cai Z, Bomgarden RD, Pike I, Kuhn K, Rogers JC, Roberts TM, Gygi SP, Paulo JA, TMTpro-18plex: The Expanded and Complete Set of TMTpro Reagents for Sample Multiplexing, J. Proteome Res. 20 (2021) 2964–2972. 10.1021/acs.jproteome.1c00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Schweppe DK, Eng JK, Yu Q, Bailey D, Rad R, Navarrete-Perea J, Huttlin EL, Erickson BK, Paulo JA, Gygi SP, Full-Featured, Real-Time Database Searching Platform Enables Fast and Accurate Multiplexed Quantitative Proteomics, J. Proteome Res. 19 (2020) 2026–2034. 10.1021/acs.jproteome.9b00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


