Abstract
Enzymes in the ergosterol-biosynthetic pathway are the targets of a number of antifungal agents including azoles, allylamines, and morpholines. In order to understand the response of Saccharomyces cerevisiae to perturbations in the ergosterol pathway, genome-wide transcript profiles following exposure to a number of antifungal agents targeting ergosterol biosynthesis (clotrimazole, fluconazole, itraconazole, ketoconazole, voriconazole, terbinafine, and amorolfine) were obtained. These profiles were compared to the transcript profiles of strains containing deletions of one of the late-stage ergosterol genes: ERG2, ERG5, or ERG6. A total of 234 genes were identified as responsive, including the majority of genes from the ergosterol pathway. Expression of several responsive genes, including ERG25, YER067W, and YNL300W, was also monitored by PCR over time following exposure to ketoconazole. The kinetics of transcriptional response support the conditions selected for the microarray experiment. In addition to ergosterol-biosynthetic genes, 36 mitochondrial genes and a number of other genes with roles related to ergosterol function were responsive, as were a number of genes responsive to oxidative stress. Transcriptional changes related to heme biosynthesis were observed in cells treated with chemical agents, suggesting an additional effect of exposure to these compounds. The expression profile in response to a novel imidazole, PNU-144248E, was also determined. The concordance of responsive genes suggests that this compound has the same mode of action as other azoles. Thus, genome-wide transcript profiles can be used to predict the mode of action of a chemical agent as well as to characterize expression changes in response to perturbation of a metabolic pathway.
Opportunistic fungal infections have become a life-threatening problem for individuals with compromised immune systems, and azoles represent a significant portion of drugs used to treat systemic infection by fungal commensal organisms. Fluconazole has emerged as the primary therapy for the treatment of oropharyngeal candidiasis in human immunodeficiency virus-infected patients (38, 39). Azoles such as fluconazole inhibit lanosterol 14α-demethylase (Erg11p), an enzyme in the ergosterol-biosynthetic pathway in yeast (54). The nitrogens in the azole ring form a complex with the heme iron component of the cytochrome group, resulting in the inhibition of the enzyme (54).
Widespread treatment with azoles has led to clinical resistance. Isolation of resistant strains has led to the intensive study of the molecular mechanisms by which the organism can compensate to permit growth in the presence of these drugs (39). Various clinical isolates of azole-resistant Candida albicans contain point mutations in the gene encoding Erg11p (26, 51) or have increased expression of this gene (50), leading to decreased susceptibility to azole drugs. In addition to altering the target gene, many clinical isolates of C. albicans upregulate multidrug-resistant pumps belonging to the ATP-binding cassette (ABC) transporter and major facilitator families in response to azole exposure, resulting in lower susceptibility due to increased efflux (41, 50).
Ergosterol is an essential component of yeast plasma membranes which affects membrane fluidity, permeability, and the activity of membrane-bound enzymes (8, 9, 36). In Saccharomyces cerevisiae, ergosterol is also a major component of secretory vesicles and has an important role in mitochondrial respiration (9, 36, 55). Ergosterol has been predicted to play a role in oxygen sensing (44), defined by the well-characterized sparking function of this sterol (31, 40). Genes in the ergosterol pathway exhibit transcriptional regulation in response to mutations in other ERG genes and resulting sterol limitation (2, 23, 29). When overexpressed, genes such as CYB5, COX3, and RPL27 contribute to altered sensitivity to azoles (27, 47). Thus, analysis of the genome-wide transcriptional changes in response to ergosterol perturbation may reveal novel mechanisms for resistance, or possibly even new sites for chemical intervention, in addition to increasing understanding of the cellular response to perturbations of ergosterol biosynthesis.
This study finds a convergent pattern of gene expression between drug-treated cells and cells with genetic alterations in the same targeted pathway. From these data a set of genes has been identified that may be predictive of an “ergosterol response.” Several of these genes do not respond to other treatments studied (data not shown), rendering them useful as surrogate expression markers for monitoring cellular responses to agents which perturb ergosterol biosynthesis. Other responsive genes have been identified that offer insight into the relation of ergosterol biosynthesis to important physiological changes in the cell, such as mitochondrial function. In addition, this study defines a method by which genome-wide transcriptional changes in yeast can be analyzed following exposure to a drug with an uncharacterized mode of action.
MATERIALS AND METHODS
Antifungal agents.
Clotrimazole, biotin-11-CTP, and biotin-16-UTP were obtained from Sigma Chemical Company; ketoconazole was from Biomol Research Labs, Inc., itraconazole was from Accurate Chemical and Scientific Corp., Westbury, N.Y., and terbinafine was from TCI, Tokyo, Japan. Amorolfine, fluconazole, voriconazole, and PNU-144248E were synthesized at Pharmacia & Upjohn.
Yeast strains.
S. cerevisiae BY4743 (a/α his3Δ/his3Δ leu2Δ/leu2Δ +/lys2Δ met15Δ/+ ura3Δ/ura3Δ) and strains 30568 (erg6Δ/erg6Δ in a BY4743 background), 30590 (erg5Δ/erg5Δ in a BY4743 background), and 30788 (erg2Δ/erg2Δ in a BY4743 background) were used. All were obtained from Research Genetics (www.resgen.com).
MIC determinations.
A 10-ml culture of YPD medium (1% Difco yeast extract, 2% Difco peptone, 2% glucose) was inoculated from a colony and grown overnight at 30°C to saturation. The culture was then diluted to an optical density at 600 nm (OD600) of 0.1, and 50 μl was used to inoculate a 96-well U-bottom culture plate (Costar, Corning, N.Y.) containing 50 μl of twofold serially diluted test compounds (starting at a final concentration of 100 μM in 100 μl). The culture was allowed to grow in the presence of drug for 24 h at 30°C in a SpectraMax Plus (Molecular Dynamics Corp., Sunnyvale, Calif.), and the MIC was determined as the drug concentration in the first well with no growth.
Cell culture and drug exposure.
A 40-ml culture of YPD medium was inoculated from a colony and grown overnight at 30°C and 140 rpm to saturation (∼1 × 108 to 2 × 108 cells/ml). The culture was diluted 10-fold and allowed to recover from stationary phase for 2 h. In the case of drug-treated cells, drug was then added to each culture at a concentration equivalent to 0.5 times the MIC (concentrations used were 0.6 μM clotrimazole, 25 μM fluconazole, 0.6 μM itraconazole, 4 μM ketoconazole, 0.6 μM PNU-144248E, 0.19 μM voriconazole, 0.1 μM amorolfine, and 50 μM terbinafine), and the cultures were incubated for 90 min. Strains containing mutations were diluted to 2 × 106 cells/ml and harvested at late-logarithmic phase (∼5 × 107 cells/ml). Strains 30568 and 30788 were twofold less dense than BY4743 and 30590; the culture volume of these strains was doubled in order to equalize the total number of cells in the preparation. Cells were pelleted at 1,500 rpm at 20°C for 5 min in an IEC (Needham, Mass.) MP4R centrifuge. The pellets were washed with 1 ml of water at 22°C that had previously been treated with diethylpyrocarbonate (DEPC) to inactivate RNases. Cell pellets were placed on ice, and RNA was extracted immediately to minimize change in the expression profile.
RNA preparation and hybridization to Affymetrix microarrays.
RNA preparation and hybridization to Affymetrix (Santa Clara, Calif.) DNA microarrays were performed as described by Wodicka et al. (52). Briefly, cells were harvested, washed with water, and lysed quickly, and RNA was extracted using hot acidic phenol (30). Poly(A)+ RNA was isolated using an Oligotex mRNA kit from Qiagen Inc. (Valencia, Calif.). Double-stranded cDNA was synthesized using the Superscript Choice system (Gibco BRL, Gaithersburg, Md.) and labeled with biotin-11-CTP and biotin-16-UTP with the T7 Megascript System (Ambion Inc., Austin, Tex.) for hybridization to the microarrays. Eleven micrograms of the resulting cRNA was used to probe the four arrays comprising the yeast genome, following the method recommended by Affymetrix Inc. and described by Wodicka et al. (52).
Hybridization signal detection and analysis.
Affymetrix microarrays consist of oligonucleotides synthesized in situ. Each open reading frame (ORF) has a number of corresponding oligomers on the microarray, termed a probe set. A probe set consists of a series of probe pairs: oligomers designed to match the ORF sequence and, for each, a corresponding mismatch oligomer designed to serve as a hybridization control. The Affymetrix Genechip algorithm computes the hybridization signal, termed average difference intensity (ADI), for each probe set, and this value was used for analysis. ADI values range from below zero to more than 15,000; values below 1 were changed to 1 prior to analysis.
Experimental (E) ADI values were compared on a gene-by-gene basis to the ADI value in the untreated control (termed the baseline value [BL], the arithmetic average of ADI values for that ORF derived from six replicate data sets of BY4743 growing in mid-log phase in YPD medium). To compensate for differences between chips in a data set, the ADI and the E/BL ratio for each ORF were each divided by the geometric mean of the E/BL ratios for that chip. Data sets were compared visually using Spotfire Pro 3.0 (Spotfire Corp., Cambridge, Mass.) and numerically using MS Excel.
Quantitative PCR.
Total RNA was extracted as described above and used as a substrate for quantitative reverse transcription-PCR (qRT-PCR). qRT-PCRs were performed using the Taqman Gold PCR kit and analyzed using a Prism 7700 (Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif.). Threshold values are calculated from the standard deviation of the background signal; Ct (cycles to reach threshold) is the cycle number in which the reaction fluorescence surpassed the threshold value. Experimental values were compared to results from a second PCR carried out in the same tube, primed from TEF1, a translation elongation factor.
Probe-primer sets were identified using Primer Express (PE Applied Biosystems). Sequences of primers and fluorescently labeled probes were as follows: for ERG25, 5′-CCAAGCAAGCACCTACTCACAA, 5′-CCAGTATTTCTCCATGAAATTCAATTG, and 6FAM-TTGCAAAATGTCGCCCATTACCAACC-TAMRA; for YNL300W, 5′-CCCATGAGATCAGCACATACGT, 5′-ACCCATGATGGCACCCATA, and 6FAM-CGCTGCCGTTAAAGGCTCCGTTG-TAMRA; and for YER067, 5′-GCCGTTAGGAAACCTGAGCTT, 5′-CTCACCGTAATGCCAGGTGAT, and 6FAM-TTATCAAATGTCTCTTCCTTCGATTTTTCGTGAA-TAMRA (FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine). Primers and probes used for TEF1 were 5′-GTAGAGTTGAAACCGGTGTCATCA, 5′-AACGGACTTGACTTCAGTGGTAACA, and VIC-CAGGTATGGTTGTTACTTTTGCCCCAGCTG-TAMRA (VIC was obtained from PE Biosystems).
RESULTS AND DISCUSSION
Experimental design and methodology.
Following the complete sequence determination of the S. cerevisiae genome, Affymetrix DNA microarrays have emerged as a powerful tool for examining the simultaneous expression pattern of more than 6,000 yeast genes (10, 14, 21, 33, 52). Because of the scope of each data set obtained from a microarray hybridization, treatments used in this study were limited to a single concentration at a single time point. Rich medium (YPD) was used so as not to limit the growth rate, and one doubling time (90 min) was selected for the duration of exposure to agent at 0.5 times the MIC. Cells were exposed to agents and harvested during the mid-logarithmic-growth phase.
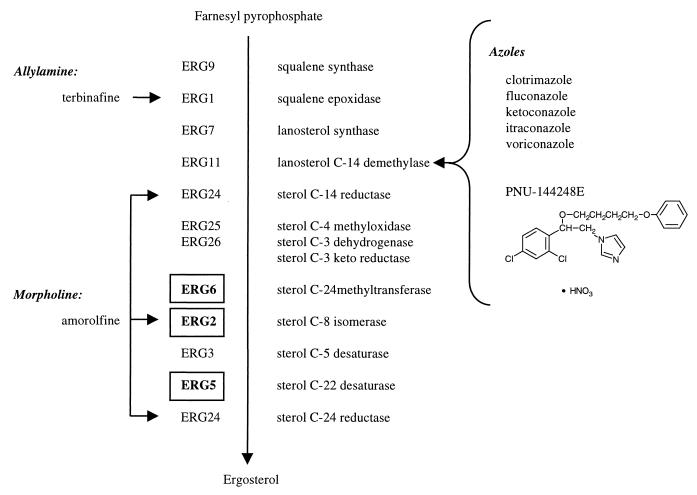
RNA was prepared following exposure of the parent strain, BY4743, to one of five previously characterized azoles (clotrimazole, fluconazole, itraconazole, ketoconazole, and voriconazole), a novel imidazole, PNU-144248E (shown in Fig. 1), an allylamine (terbinafine), or a morpholine (amorolfine) as described in Materials and Methods. In addition, RNA was prepared from three strains each bearing a homozygous deletion of ERG2, ERG5, or ERG6 and from two untreated control cultures. Figure 1 shows the genes involved in the biosynthesis of ergosterol from farnesyl pyrophosphate in S. cerevisiae.
FIG. 1.
Treatments used in the study and their relationships to ergosterol biosynthesis. Gene names are as listed in reference 9). Genes in deletion strains are boldfaced and boxed. Arrows point to the sites of action of the antifungal agents. The structure of PNU-144248E is shown under the azoles.
Identification of gene expression patterns.
Each Affymetrix yeast genome set represents 6,593 ORFs, including 172 control genes. Thus, the 11 data sets from the treatments comprising this study represent 72,523 data points. Hybridization intensity values are expressed in ADI units, as described in Materials and Methods. Following normalization to account for variations in chip intensity (described in Materials and Methods), a filter was applied requiring the experimental ADI value to be above 50 ADI units, thereby limiting the data to 29,428 points. Use of a second filter requiring the baseline ADI value to be greater than 50 ADI units further limited the data to 20,697 points. The ADI values used in the filters were chosen after examination of the background ADI value calculated by the GeneChip software and the ADI values for selected unexpressed genes (e.g., haploid-specific genes in diploid cells [data not shown]); with both approaches, a value of up to ∼25 ADI units corresponded to no signal.
Genes for which ADI ratios changed beyond 1 standard deviation were considered to be responsive to the treatment. By this criterion, 1,154 genes responded with increased mRNA levels in at least one treatment; 1,358 genes responded with decreased mRNA levels relative to the baseline. To distinguish gene responses to perturbations in ergosterol biosynthesis from other transcriptional changes, genes responding in at least 5 of the 11 experimental treatments were considered in the subsequent analysis. The intention of selecting by these criteria is to identify genes that have a convergent pattern of expression across many individual treatments, which may be indicative of a common response.
A total of 156 genes showed significant increases in transcript levels in five or more treatments, and 78 showed significantly decreased transcript levels in five or more treatments. These were annotated using the biological roles assigned by the Yeast Protein Database (YPD) (18). The number and characteristics of the responsive genes grouped according to biological role are shown in Table 1. The category with the largest number of responses (hits) is “unknown” group of 53 genes. Next most abundant are the 36 responsive mitochondrial genes, followed by 22 genes involved in biosynthesis of lipids, fatty acids, and sterols. This group includes nine genes in the ergosterol pathway. The category “other related genes” refers to genes that are related to ergosterol perturbation by other experimental results; these genes are described in Tables 2 and 3 and Fig. 2 and 3. The category “other genes” includes responsive genes each of which was the sole representative of a particular biological pathway, and whose relationship to ergosterol perturbation could not be discerned.
TABLE 1.
Genes responding in five or more treatments
| Biological rolea | No. of genesb | No. of hitsc | Hit/gene ratio | No. of drug hitsd | Drug hit/ gene ratio | No. of KO hitse | KO hit/ gene ratio | No. (%) of genes responding to saturated culture | % of genes responding to:
|
Direction of changef | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Drug treatment | KO | ||||||||||
| Unknownfg | |||||||||||
| Up | 26 | 154 | 5.92 | 124 | 4.77 | 30 | 1.15 | 0 (0) | 80 | 20 | Up |
| Up/sat'd | 8 | 52 | 6.50 | 35 | 4.38 | 17 | 2.13 | 8 (100) | 67 | 33 | Up |
| Down | 11 | 67 | 6.09 | 56 | 5.09 | 11 | 1.00 | 0 (0) | 84 | 16 | Down |
| Down/sat'd | 8 | 50 | 6.25 | 41 | 5.13 | 9 | 1.13 | 8 (100) | 82 | 18 | Down |
| Mitochondrial | 32 | 194 | 6.06 | 145 | 4.53 | 49 | 1.53 | 23 (72) | 75 | 25 | Up |
| 4 | 25 | 6.25 | 20 | 5.00 | 5 | 1.25 | 1 (25) | 80 | 20 | Down | |
| Lipid/fatty acid/sterol | 16 | 101 | 6.31 | 63 | 3.94 | 38 | 2.38 | 1 (6) | 62 | 38 | Up |
| 6 | 37 | 6.16 | 28 | 4.67 | 9 | 1.50 | 2 (33) | 76 | 24 | Down | |
| Protein synthesis and processing | 15 | 90 | 6.00 | 73 | 4.87 | 17 | 1.13 | 0 (0) | 81 | 19 | Up |
| 9 | 54 | 6.00 | 42 | 4.67 | 12 | 1.33 | 6 (67) | 78 | 22 | Down | |
| Stress | 15 | 90 | 6.00 | 59 | 3.93 | 31 | 2.07 | 5 (33) | 66 | 34 | Up |
| 5 | 31 | 6.20 | 25 | 5.00 | 6 | 1.20 | 4 (80) | 81 | 19 | Down | |
| Membrane associated | 8 | 45 | 5.62 | 39 | 4.88 | 6 | 0.75 | 0 (0) | 87 | 13 | Up |
| 8 | 50 | 6.25 | 36 | 4.50 | 14 | 1.75 | 3 (38) | 72 | 28 | Down | |
| Amino acid biosynthesis | 9 | 58 | 6.44 | 43 | 4.78 | 15 | 1.67 | 7 (78) | 74 | 26 | Up |
| Cell cycle control | 7 | 38 | 5.43 | 31 | 4.43 | 7 | 1.00 | 0 (0) | 82 | 18 | Up |
| Chromatin associated | 6 | 35 | 5.83 | 29 | 4.83 | 6 | 1.00 | 0 (0) | 83 | 17 | Up |
| Carbohydrate metabolism | 6 | 35 | 5.83 | 24 | 4.00 | 11 | 1.83 | 5 (83) | 69 | 31 | Up |
| Vesicle associated | 5 | 29 | 5.80 | 23 | 4.00 | 6 | 1.20 | 0 (0) | 79 | 21 | Down |
| Cell wall biosynthesis | 5 | 33 | 6.60 | 26 | 5.20 | 7 | 1.40 | 3 (60) | 79 | 21 | Up |
| Nucleotide metabolism | 5 | 28 | 5.60 | 19 | 3.80 | 9 | 1.80 | 3 (60) | 68 | 32 | Down |
| Other related genes | 4 | 24 | 6.00 | 19 | 4.75 | 5 | 1.25 | 1 (25) | 79 | 21 | Down |
| 3 | 17 | 5.67 | 11 | 3.67 | 6 | 2.00 | 1 (33) | 65 | 35 | Up | |
| Other genes | 12 | 66 | 5.50 | 58 | 4.83 | 8 | 0.67 | 2 (17) | 88 | 12 | Down |
Obtained from the YPD (18).
Number of responsive genes in each functional class.
No. of hits, number of total treatments in which the genes responded.
No. of drug hits, number of drug treatments in which the genes responded.
No. of KO hits, number of genetic disruptions (knockouts [KO]) to which the genes responded.
Up, increased transcript levels; down, decreased transcript levels.
Up/sat'd and Down/sat'd, increased and decreased transcript levels, respectively, in response to saturated culture.
TABLE 2.
Genes encoding products associated with cell envelope structure or function
| Function of gene product(s) | Gene(s)a |
|---|---|
| Involved in protein glycosylation | ALG52,4, ALG3, ALG9, ALG63,3, ALG81,4, ALG10, transferase (PMT family; PMT23,2) CWH41, GLS21,5, MNS1 |
| O-glycosylated proteins | AGA1, AGA2, BAR1, CTR1, CTS12,3, CWP1, CWP2, DAN1, FET3, FLO1, FLO5, FLO9, FLO10, FUS1, GAS1, GIT1, HKR1, HSP150, KEX2, KNH1, KRE13,4, KRE92,3, MID2, MSB2, PEX15, PGM1, PGM2, PIR1, PIR3, PRB11,4, SAG1, SEC20, SED1, SED4, SLG1, SRO4, SSR1, STA1, STA2, TIP12,5, TIR13,3, TIR2, YLR110C |
| Responsive membrane proteinsb | BAP22,3, BAP30,6, DIP53,4, FET41,7, HNM13,5, HXT10,5, HXT32,4, PHO870,5, RCS11,4, SMF13,4, YDR373W1,4, YGR138C0,5, YKL146W1,4, YNL065W0,6, YNL321W2,3, YOR161C2,3, YOR271C2,4 |
| GPI-anchored proteins | AGa1, AGA1, CWP1, CWP2, EGT21,4, FLO1, FLO5, FLO9, GAS1, ICWP, KRE13,4, PRY3, SED1, TIP12,5, TIR13,3, TIR2, YAP3, YCR089W, YDR055W0,5, YDR134C, YDR534C, YEL040W, YER150W0,5, YGR189C, YJR151C, YLR110C, YNL300W3,7, YOR009W, YOR214C |
| Involved in synthesis of β-1,6-glucan | CWH53, FKS2, KNR4, HKR1, KRE13,4, KRE5, KRE6, KRE92,3, KRE11, SKN1 |
| Osmotic-stress related | |
| Part of osmotic-stress response signal pathway | HOG12,3 |
| Transcription factor with a role in salt tolerance | CIN51,6 |
| Induced by osmotic stress | DDR483,4, DDR2, GRE1, GRE23,4, GRE3, HOT1, PTP2, PTP3, SIP18, SKN7, SKK2, SSK1, STE11, SLN1, SHO1, YPD1 |
| Responsive proteins involved in secretionb | VID240,7, RET23,4, SEC170,5, COP12,5, LST83,4, NHX10,5, YGL054C0,6, YHR138C0,5 |
Boldfaced genes were responsive in the study; boldfaced underlined genes had decreased transcript levels. In each superscript, the first number is the number of genetic perturbations (out of a total of three) to which the gene responded and the second is the number of drug treatments to which the gene responded. Lists of genes were obtained from the YPD (18) and from references 4, 9, 15, 16, 17, and 19.
Only responsive genes are shown.
TABLE 3.
Genes encoding mitochondrial proteins
| Gene product(s) | Gene(s)a |
|---|---|
| Electron transport complexes, inner mitochondrial membrane: | |
| Ubiquinol cytochrome c reductase complex III | COB, CYT12,4, COR12,3, QCR2, QCR61,4, QCR7, QCR8, QCR9, QCR10, RIP11,5 |
| Cytochrome c (anaerobic isoform) | CYC71,5 |
| Cytochrome c oxidase | COX1, COX2, COX3, COX43,4, COX5A2,4, COX5B1,8, COX6, COX7, COX80,5, COX9, COX12, COX133,4 |
| ATP synthase | ATP13,5, ATP2, ATP3, ATP4, ATP5, ATP6, ATP7, ATP8, ATP9, ATP140,6, ATP153,4, ATP160,5, ATP17, ATP20, INH10,6, TIM11 |
| ADP/ATP carrier protein | AAC1, AAC31,4, PET93,3, YPL134C1,4, YPR021C |
| Elements of the TCA cycle | CIT1, ACO12,4, IDH1, IDH2, KGD11,5, KGD2, LPD1, LSC1, LSC2, SDH12,4, SDH2, SDH3, SDH4, FUM12,4, MDH1 |
| Other responding mitochondrial proteins | ARG5,60,5, ARG71,6, COT13,5, GPD23,4, IDP10,5, ILV52,3, ILV63,5, MRP10,6, PFK270,6, SHM11,4, SOM10,6, SOD22,4, TIM233,3, YAH12,3, YIL154C0,6 |
Boldface, underlining, and superscripts are as explained in footnote a to Table 2. Lists of genes were obtained from the ExPASy/Boehringer Mannheim metabolic pathways (http://expasy.hcuge.ch/cgi-bin/search-biochem-index) and from the YPD (18).
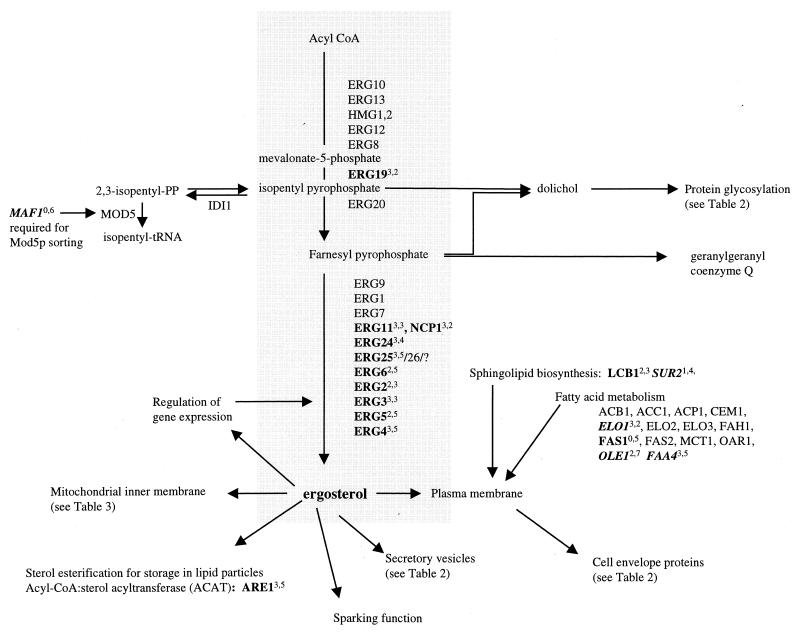
FIG. 2.
Genes involved in the biosynthesis of ergosterol and membrane components. Boldfaced genes were responsive in the study; boldface italics indicate genes with decreased transcript levels. Superscripts indicate the number of treatments to which the gene responded. The first number in each superscript is the number of genetic perturbations (out of a total of three) which elicited a response, and the second number is the number of drug treatments (out of a total of eight) which elicited a response. Lists of genes were obtained from the YPD (18) and reference 9.
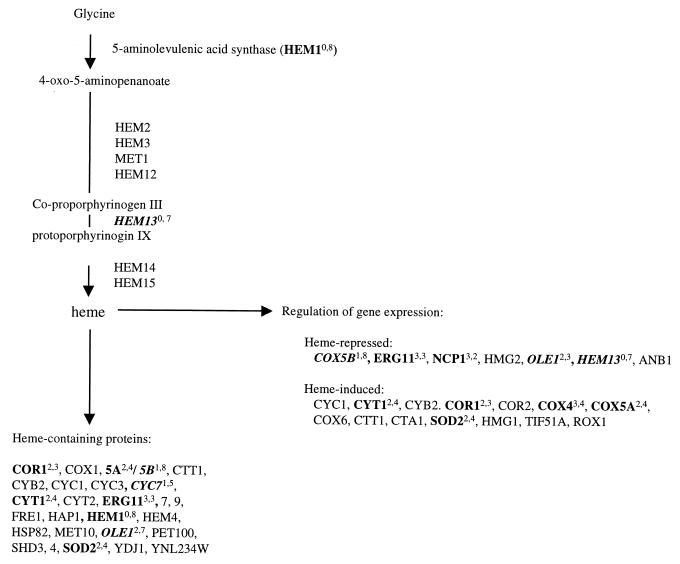
FIG. 3.
Genes involved in the biosynthesis and utilization of heme. Boldface, italics, and superscripts are as described for Fig. 2. Lists of genes were obtained from the YPD (18).
Genes were also categorized by the relative number of responses due to chemical versus genetic perturbation. The percentage of the response that was due to chemical perturbation ranged from highs of 87 to 88%, in the case of membrane-associated proteins, to a low of 62% for the class of lipid-, fatty acid-, or sterol-related genes. Analysis using a k-means algorithm did not reveal transcriptional patterns associated with particular treatment classes (data not shown). Interestingly, the ergosterol genes were highly responsive to genetic disruption of ERG2, ERG5, and ERG6. The 10 responsive ergosterol genes had increased transcript levels in all three mutant strains (Fig. 2) with the exception of the particular gene disrupted in each strain.
As a means to assess the specificity of the ergosterol response identified in this study, the behavior of these responsive genes was examined following the transition from logarithmic growth to saturation. Table 1 shows the number of responsive genes in each category whose transcript levels change as the cells enter saturated growth and undergo the diauxic shift (10, 18; G. F. Bammert and J. M. Fostel, unpublished data). The proportion of responsive genes which are altered in response to saturated growth varies from 0% of genes in several pathways to highs of 78 and 83% of ergosterol-responsive genes related to amino acid and carbohydrate metabolism, respectively. This suggests that some groups of transcripts may be responding to alterations in the metabolic rate rather than to ergosterol perturbation directly.
Responsive genes in the ergosterol pathway.
Figure 2 shows the genes involved in the biosynthesis of ergosterol from acyl coenzyme A (acyl-CoA), highlighting some of the responsive genes identified above. Most notably, nine genes in the ergosterol pathway responded with increased transcript levels to the conditions used in this study. The ergosterol pathway represented the highest proportion of responsive genes identified, in agreement with previous studies showing that this pathway is the target of azoles and is responsive to modulations of the ergosterol level.
The biosynthesis of ergosterol involves the coordination of many factors by which the cell regulates the synthesis of this essential component. ERG19 is reported to contribute to the regulation of flux through the mevalonate pathway (3) and is increased in response to perturbations here. ERG3 expression increases following treatment with antifungal agents (43) and in strains with mutations in ERG2, ERG5, and ERG6 (2). These data are supported by the observations reported here: ERG3 expression increased following drug treatment and in the three ERG deletion strains. Additionally, expression of NCP1, which encodes NADP-cytochrome P450 reductase and the electron donor for squalene epoxidase, lanosterol 14α-demethylase, and sterol C-22 desaturase (45), increases fivefold in a strain constitutively overexpressing ERG11 (48), consistent with coordinate increases in the transcript levels of these two genes in this study.
Measurements using promoter fusions in various genetic backgrounds found a number of genes affecting ERG9 expression (23). Two treatments overlapped those reported here: exposure to ketoconazole and expression in an ERG2 disruptant. In both cases ERG9 expression increased (23). While ERG9 did not meet the criterion of change in five or more treatments used here, levels of the ERG9 transcript did increase in four treatments: ERG2 and ERG6 knockouts and exposure to fluconazole or PNU-144248E. The difference in the ketoconazole treatments used in the two studies (18 h the study reported in reference 23 versus 90 min here) may account for the difference, as observed in Fig. 5 for ERG25.
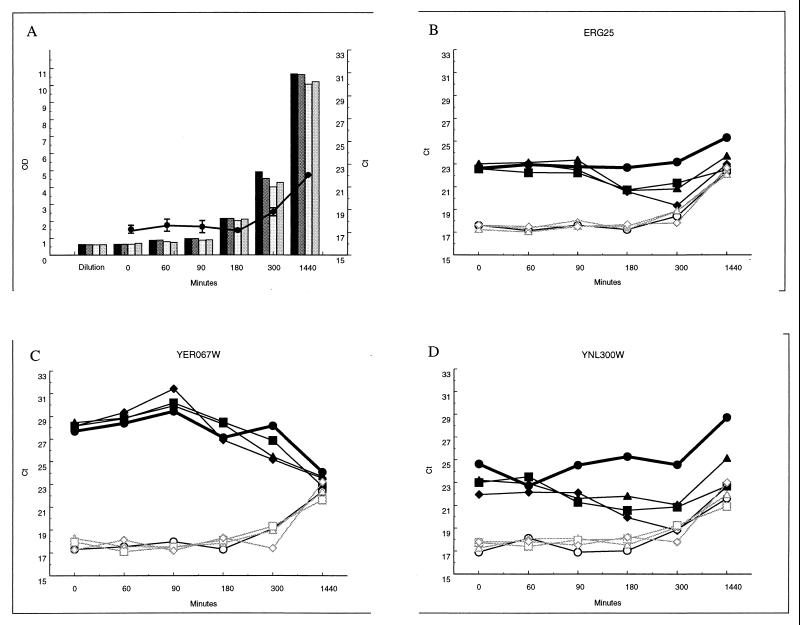
FIG. 5.
Expression time course following exposure to ketoconazole. (A) Culture OD at the times sampled (solid bars, untreated culture; heavily shaded bars, 4 μM ketoconazole; open bars, 8 μM ketoconazole; light shaded bars, 16 μM ketoconazole) and change in TEF1 level as culture goes into saturation (line, average of three measurements of untreated culture in panels B, C, and D; error bars, standard errors). (B through D) Responses of ERG25, YER067W, and YNL300W transcripts (solid symbols) and an internal TEF1 control (open symbols). Culture was left untreated (circles) or treated with ketoconazole at 4 μM (triangles), 8 μM (squares), or 16 μM (diamonds).
Another gene in the pathway, ERG1, did not change significantly in any of the treatments used here, including terbinafine, although Erg1p enzyme activity does increase following terbinafine exposure (28). Erg1p activity responds to oxygen and sterol limitation (34) and appears to be regulated by protein localization or other factors (28). Taken together, these observations suggest that changes in Erg1p activity arise from posttranscriptional regulation.
In a study using promoter fusions as a readout of transcriptional changes, Dimster-Denk et al. (11) observed 19 genes with a change in expression greater than 1.5-fold following 21 h of exposure to fluconazole relative to expression levels in untreated cells. Of these 19 responsive genes, 7 were found to be responsive in this study (ERG2, ERG3, ERG4, ERG5, ERG6, ERG11, and ERG19) and ERG9 expression was changed in four treatments. The other 11 genes had transcript levels too low to be measured reliably, including 5 mating-response genes not expressed in the diploid strain used here (ERG8, ERG12, ARE2, COQ7 [also referred to as CAT5], FAR3, FIG1, HEM14, MFA1, MFA2, MOD5, and STE2). Responses by ERG24 and ERG25 were seen on the microarray but were not reported by Dimster-Denk et al. Thus, the two methods are in agreement on the identification of responsive genes among genes detected by both. The change in transcript level was not always in the same direction in the two studies, however. Decreases in ERG3, ERG4, ERG5, ERG6, and ERG11 transcript levels at 21 h were reported by Dimster-Denk et al. (11), while these genes showed increased transcript levels following 90 min of exposure as measured by microarrays. It is more likely that this reflects differences between the biological responses of these genes at the different exposure times than a discrepancy between the methods used to measure expression.
Other responsive genes related to lipid, fatty acid, and sterol biosynthesis.
In addition to participating in cell membranes, esterified ergosterol is found in lipid particles, which may serve as storage reservoirs or as intermediates in intracellular transport (53, 55). ARE1 is one of the two genes responsible for the esterification of ergosterol, the final step in the pathway leading to ergosteryl, and was responsive in this study. Another responsive gene, CYB5 (encoding cytochrome b5), was identified by an ability to overcome ketoconazole hypersensitivity when overexpressed in an erg11 background (47) and may help protect the organism from azole exposure. ACH1 (encoding acetyl-CoA hydrolase) may be responding to an overproduction of sterol intermediates formed during inhibition of the pathway. FAS1 (encoding fatty-acyl-CoA synthase) deletion strains have reduced levels of ergosterol esters and sphingolipids, indicating a possible role in lipid biosynthesis and metabolism (9). LCB1 encodes serine C-palmitoyltransferase, the first enzyme involved with the biosynthesis of the long-chain base component of sphingolipids. An increase in the level of transcripts of this transferase following perturbation of the ergosterol pathway suggests an interaction between the ergosterol and sphingolipid biosynthetic pathways in yeast.
Transcript levels of several other genes involving lipid, fatty acid, and sterol metabolism decreased in this study. The SUR2 product hydroxylates the sphingoid C-4 of ceramide (15), and the decrease observed in the level of this transcript further suggests an interaction between the sphingolipid and sterol pathways. ELO1 encodes an enzyme responsible for the elongation of fatty acids. A decline in the level of this transcript under the conditions of this study may indicate a compensatory response in cellular fatty acid content to limited sterols following perturbation of the ergosterol pathway. OLE1, encoding δ-9 desaturase, is needed for the formation of unsaturated fatty acids and also shows a decreased transcript level here. OLE1 is repressed by the presence of saturated fatty acids (12); thus, the decline in OLE1 transcript levels may indicate an increase in saturated fatty acid levels, possibly another compensatory response to altered ergosterol. Interestingly, the fatty-acid-responsive repression of OLE1 is mediated through the FAA1 and FAA4 products (7), and the level of FAA4 transcripts decreased here. A connection of fatty acids to perturbation of the ergosterol pathway may suggest a restructuring of the cell membrane in response to reduced ergosterol levels.
Other responsive pathways.
While ergosterol is found throughout the cell membranes, it is most abundant in the plasma membrane and secretory vesicles and is important for mitochondrial respiration (9, 36, 55). Depletion of ergosterol with concomitant accumulation of sterol intermediates can result in alterations in membrane functions, synthesis and activity of membrane-bound enzymes, and mitochondrial activities, as well as in uncoordinated behavior of the yeast cell (36, 49). The changes in transcript pattern reported in Table 3 may reflect these stresses.
It can be predicted that perturbations of ergosterol levels within the cell may affect the functioning of mitochondrial enzymes. This pattern does indeed emerge with the increase in transcript levels of several components of the mitochondrial electron transport system, as shown in Table 3. Transcript levels of four members of the cytochrome c oxidase complex, COX4, COX5A, COX8, and COX13, were increased. Similarly, transcript levels increased for four members of the cytochrome c reductase complex, RIP1, CYT1, QCR6, and COR1, and five genes encoding subunits of ATP synthase, ATP1, ATP14, ATP15, ATP16, and INH1. Other genes involved in energy generation that showed increased transcript levels are listed in Table 3, as are a number of other responsive genes encoding mitochondrial proteins.
Levels of transcripts from four of the five members of the hypoxic gene family (ANB1, COX5b, CYC7, and HEM13) are reduced in response to treatments used in this study (Table 4). Two of these genes, CYC7 and COX5b, encode the hypoxic isoforms of cytochrome c and cytochrome c oxidase. Since their expression levels depend on the level of available oxygen (6, 25), the decrease in transcript levels of these anaerobiosis-induced genes may be occurring in response to increased levels of intracellular oxygen. Another line of evidence to support this hypothesis is the observation that transcripts from three genes involved with oxidative-stress response (GRE2, YDR453C, and SOD2) are increased (Table 4). In the course of normal catalysis, cytochrome c oxidase transfers two electrons to molecular oxygen, and incomplete reactions can result in the formation of reactive oxygen species. Machida et al. (32) have shown that exposure to farnesol results in the generation of reactive oxygen species through an indirect effect on mitochondrial electron transport. Farnesol can be generated from farnesyl pyrophosphate, a precursor to sterols which may accumulate if late stages of ergosterol biosynthesis are inhibited. All these observations are consistent with increased oxidative stress following perturbation of ergosterol biosynthesis.
TABLE 4.
Genes responding to oxygen stress
| Function or response | Gene(s)a |
|---|---|
| Induced by hypoxia | AAC1, ANB1, COX5B1,8, CPR1, CYC71,5, ERG113,3, HEM130,7, HMG2, OLE12,7, ROX1, SUT1 |
| Induced by anaerobiosis | CYB2, ERG113,3, TIP12,5, TIR2, TIR13,3 |
| Pentose phosphate pathway shunt | ZWF13,4 |
| Major oxidant-scavenging enzymes | CCP1, CTT1, TSA1, SOD1, SOD22,4, TRR1, TRX1, TRX2, GLR1, YDR453C2,4, YCL035, AHP1, GRE23,4 |
Perturbation of mitochondrial electron transport could arise from a decrease in ergosterol levels in the inner membrane due to interference with ergosterol biosynthesis or from a direct interaction between the chemical agents used and the mitochondrial enzyme complexes. For example, metal-chelating drugs block reduction at the ubiquinol oxidation site of the cytochrome c complex (5), and azoles inhibit their target, lanosterol demethylase, through an interaction with the heme iron component of the complex (54). Since more than half of the responsive mitochondrial genes are found to respond both to genetic alterations and to drug treatment, it is likely that the effect is mediated through ergosterol biosynthesis and does not arise solely as a direct consequence of drug action.
Interestingly, one drug-specific pattern was observed. Transcript levels of HEM1, the first gene involved in the biosynthesis of heme, increased in the eight drug treatments but not in the three deletions. This suggests a compensatory response to drug treatment not represented in the mutant strains. Transcript levels of HEM13, the rate-limiting step of heme biosynthesis (57), were reduced in response to seven drug treatments and, again, in none of the deletion strains. HEM13 is repressed by heme and oxygen (1, 22), raising the possibility that levels of one or both are elevated following treatment. This is consistent with the hypothesis of increased intracellular oxygen suggested by the responses of anaerobiosis-induced and oxygen stress genes described above. Heme also plays a role in sensing intracellular oxygen levels (57).
Heme plays a central role in sterol synthesis and regulates the transcription of several genes involved with this process (37, 46). The accumulation of 5-aminolevulenic acid, the product of Hem1p, derepresses 3-hydroxy-3-methyl-glytaryl CoA reductase, leading to increased levels of 2,3-oxidosqualene (31). Heme is required for the enzymatic activities of Erg3p (C-5 sterol desaturase) and Erg5p (C-22,23 desaturase) (36). Erg11p also contains heme and shows heme-regulated expression (48). Other genes regulated by heme are listed in Fig. 3. In the present study, levels of transcripts from four heme-induced genes are increased and levels of transcripts from three heme-repressed genes are decreased, consistent with induction of heme-regulated expression, and suggesting elevated intracellular heme levels following treatment, particularly treatment with chemical agents.
Correlation of the expression pattern of a novel azole to other conditions.
Included in this analysis was the expression profile in response to treatment with a compound containing an azole moiety. PNU-144248E contains an imidazole ring, yet is structurally distinct from the other azoles tested. The rationale for including it in this study was to determine if exposure to this compound would result in a pattern of expression similar to that seen in response to treatment using azoles with known biochemical functions, thereby suggesting a similar mode of action. This information could then be used to assess the predictive ability of expression profiles observed in response to an agent with an unknown mode of action. Of the 156 genes with increased transcript levels following ergosterol perturbation, 144, or 92%, also had increased transcript levels in response to treatment with PNU-144248E. All of the ergosterol, lipid, fatty-acid, and sterol metabolism genes and 17 of the 19 genes involved with energy generation were included (the exceptions were QCR6 and COX13). Twenty-nine of the 34 unknowns were also included. From the set of 78 genes with decreased transcript levels following ergosterol perturbations, 40, or 51%, also had decreased transcript levels following treatment with PNU-144248E. These data suggest that PNU-144248E does indeed behave as an azole as measured by cellular responses at the level of gene transcription.
PDR5 transcript levels increased following treatment with PNU-144248E. This gene encodes a member of the multidrug resistance pump family homologous to genes which are upregulated in azole-resistant isolates of C. albicans (41). It might be anticipated that PDR5 transcript levels increase in all of the drug treatments; however, they were unaltered in other treatments used in this study. It is possible that PDR5 requires a longer exposure time for full induction and that PNU-144248E was the only treatment of sufficient potency to elicit a response in the 90 min of exposure used here. PDR5 transcript levels in cells treated with mucidin did not increase until 2 h of exposure (35), and the response to ergosterol perturbation may follow similar kinetics.
Comparison of microarray measurements with PCR measures of transcript levels.
Microarrays represent a new technology for measuring transcript levels. Control experiments indicate that measures using this technique are quantitative; however, these experiments were carried out for a limited subset of the genes on the array (52). It was of interest to compare microarray ADI measurements for responsive genes to measurements of the same RNA preparation using the Taqman quantitative PCR system. This was performed for two genes of interest, ERG25 and YER067 (Fig. 4), one showing an increase in response to treatment and the other a decrease.
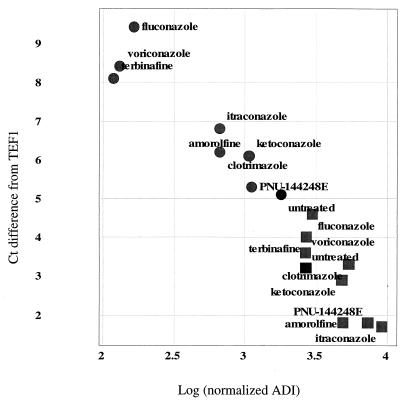
FIG. 4.
Comparison of expression measurements by RT-PCR and microarrays. Each data point represents the measurement of a given gene by each method under a specific treatment, as indicated by the label. Squares, ERG25; circles, YER067W; solid symbols, untreated control. The vertical axis is the change from the TEF1 level; larger numbers signify more cycles needed and thus less starting material in the sample. The horizontal axis indicates the microarray intensity in ADI units, normalized as described in Materials and Methods; ADI units are proportional to transcript levels.
Data from the experiments shown in Fig. 4 were normalized as described in Materials and Methods. Microarray ADI measures are proportional to transcript abundance, while PCR measures of Ct are inversely proportional, i.e., more-abundant transcripts require fewer cycles to achieve detectable levels, and hence the Ct is lower than for less-abundant material. For this reason the correlation seen in Fig. 4 suggests good agreement between measures of transcript levels by these two methods.
Changes in transcript levels over time.
A single doubling time was selected for the duration of treatment with chemical agents in this study. It is likely that different transcripts may be responsive at earlier or later times and that the responsive transcripts detected herein may peak at times other than 90 min. In order to assess the response kinetics for genes of interest, measures of transcripts from responsive genes were made following different times of exposure to several of the agents used in this study. ERG25 is a late-stage transcript in the ergosterol pathway and has previously been shown to be responsive to ergosterol levels (29). YER067W and YNL300W are uncharacterized ORFs which respond to ergosterol perturbation but do not change in many other treatments tested (data not shown). YER067W is a ploidy-regulated gene (13). YNL300W is predicted to be a glycosylphosphatidylinositol (GPI)-anchored protein (16) with weak homology to the potential cell wall stress sensor Mid2p (18, 24). Microarray measurements show that both ERG25 and YNL300W transcripts decrease under conditions where the growth rate is slowed, for example, entry into stationary phase, while YER067W transcript levels increase under these conditions (data not shown). This is opposite to the direction of response to ergosterol perturbation, where microarray data show that YER067W levels increase and ERG25 and YNL300W transcripts decrease.
Figure 5 shows the response kinetics for these three genes during a 24-h exposure to ketoconazole. Panel A shows the growth of the cultures used in this experiment both by OD and by the concomitant change in the TEF1 level as the cultures become saturated between 5 and 24 h. Panels B, C, and D show the PCR measures of ERG25, YER067W, and YNL300W RNA, respectively, from cells exposed to ketoconazole at three different doses. All show the TEF1 measure and the experimental transcript. In all cases, TEF1 is relatively unchanged by treatment and serves to normalize slight differences in the total RNA in each reaction tube during logarithmic growth (0, 60, 90, 180, and 300 min). In contrast, exposure to differing concentrations of ketoconazole results in an almost dose-dependent response in the three transcripts measured, and the direction of change is opposite that seen during growth saturation.
One interesting exception is the ERG25 response. The initial microarray measure found a 1.8-fold increase following a 90-min treatment with ketoconazole, and this was confirmed by PCR (Fig. 4). In the time course experiment shown in Fig. 5, however, ERG25 levels were not observed to increase until the 3-h point, when they were 2.5 cycles or approximately 5.6-fold increased relative to levels in untreated cells. This most likely reflects a difference in sensitivity (a 1.8-fold change is within one PCR cycle) or small differences between the cultures used in the two experiments. This experiment illustrates the different strengths of the two methods used. Microarrays reveal the transcript profile at a particular time and state and can be used to identify all transcripts responding under the conditions of the study. Quantitative PCR probes can then be generated to follow the kinetics of expression of transcripts of interest, allowing easy measurement of their response to a variety of treatments.
Conclusions.
Genome-wide transcriptional changes in S. cerevisiae observed in response to treatment of the cells with chemical agents correlate with responses to genetic alterations in the same biosynthetic pathway. A number of responsive genes which encode products with functions impinging on ergosterol biosynthesis or products related to membrane structure and function were identified. This supports the interpretation of expression profiles to define the mode of action of a drug. In addition to changes in transcript level relating directly to ergosterol biosynthesis, expression changes suggestive of interruption of heme biosynthesis and increased intracellular oxygen tension were also observed, indicating additional effects perturbing the ergosterol pathway. The approach used to identify genes responsive to the treatments studied does not rely on prior understanding of the biological effects of the treatments. Thus, it is possible to contemplate applying this method to predict the mode of action of novel agents with antifungal activity. The novel azole PNU-144248E has provided a validation of this method, causing transcriptional changes with a high degree of correlation to those seen following treatment with other azoles with known biological effects. This study has outlined a method to identify genes of interest common to a particular cellular response that will be of utility in the future study of novel antifungal compounds.
ACKNOWLEDGMENTS
We thank many of our colleagues at Pharmacia & Upjohn: Paul May for synthesis of voriconazole, fluconazole, and amorolfine; Sara Morin for identification of PNU-144248E; Mark Johnson for suggesting the method used for data analysis; Ann Berger, Marek Nageic, Don Tong, and Tom Vidmar for numerous helpful discussions; and Cheryl Quinn and Ericka Benson for critical reading of the manuscript. We are grateful for suggestions and guidance regarding quantitative PCRs from Kathryn Becker of PE Applied Biosystems and regarding microarray methodology from Gene Tanimoto and Michael Lelivelt of Affymetrix Inc. We also thank an anonymous reviewer for suggestions on improving the PCR analysis.
REFERENCES
- 1.Amillet J-M, Buisson N, Labbe-Bois R. Characterization of an upstream activation sequence and two Rox1p-responsive sites controlling the induction of the yeast HEM13 gene by oxygen and heme deficiency. J Biol Chem. 1996;271:24425–24432. doi: 10.1074/jbc.271.40.24425. [DOI] [PubMed] [Google Scholar]
- 2.Arthington-Skaggs B A, Crowell D N, Yang H, Sturley S L, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. doi: 10.1016/0014-5793(96)00807-1. [DOI] [PubMed] [Google Scholar]
- 3.Berges T, Guyonnet D, Karst F. The Saccharomyces cerevisiae mevalonate diphosphate decarboxylase is essential for viability, and a single Leu-to-Pro mutation in a conserved sequence leads to thermosensitivity. J Bacteriol. 1997;179:4664–4670. doi: 10.1128/jb.179.15.4664-4670.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boone C, Sommer S S, Hensel A, Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall beta-glucan assembly. J Cell Biol. 1990;110:1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boumans H, van Gaalen M C, Grivell L A, Berden J A. Differential inhibition of the yeast bc1 complex by phenanthrolines and ferroin. Implications for structure and catalytic mechanism. J Biol Chem. 1997;272:16753–16760. doi: 10.1074/jbc.272.27.16753. [DOI] [PubMed] [Google Scholar]
- 6.Burke P V, Raitt D C, Allen L A, Kellogg E A, Poyton R O. Effects of oxygen concentration on the expression of cytochrome c and cytochrome c oxidase genes in yeast. J Biol Chem. 1997;272:14705–14712. doi: 10.1074/jbc.272.23.14705. [DOI] [PubMed] [Google Scholar]
- 7.Choi J-Y, Stuckey J, Hwang S-Y, Martin C E. Regulatory elements that control transcription activation and unsaturated fatty acid-mediated repression of the Saccharomyces cerevisiae OLE1 gene. J Biol Chem. 1996;271:3581–3589. doi: 10.1074/jbc.271.7.3581. [DOI] [PubMed] [Google Scholar]
- 8.Cobon G S, Haslam J M. The effect of altered membrane sterol composition on the temperature dependence of yeast mitochondrial ATPase. Biochem Biophys Res Commun. 1973;52:320–326. doi: 10.1016/0006-291x(73)90990-x. [DOI] [PubMed] [Google Scholar]
- 9.Daum G, Lees N D, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids in Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 11.Dimster-Denk D, Rine J, Phillips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the Genome Reporter Matrix. J Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- 12.Fujiwara D, Yoshimoto H, Sone H, Harashima S, Tamai Y. Transcriptional co-regulation of Saccharomyces cerevisiae alcohol acetyltransferase gene, ATF1, and delta-9 fatty acid desaturase gene, OLE1, by unsaturated fatty acids. Yeast. 1998;14:711–721. doi: 10.1002/(SICI)1097-0061(19980615)14:8<711::AID-YEA263>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Galitski T, Saldanha A J, Styles C A, Lander E S, Fink G R. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 14.Gray N S, Wodicka L, Thunnissen A W H, Norman T C, Kwon S, Espinoza F H, Morgan D O, Barnes G, LeClerc S, Meijer L, Kim S H, Lockhart D J, Schultz P G. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;1281:523–528. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 15.Haak D, Gable K, Beeler T, Dunn T. Hydroxylation of Saccharomyces cerevisiae ceramides requires Sup2p and Scs7p. J Biol Chem. 1997;272:29704–29710. doi: 10.1074/jbc.272.47.29704. [DOI] [PubMed] [Google Scholar]
- 16.Hamada K, Terashima H, Arisawa M, Yabuki N, Kitada K. Amino acid residues in the ω-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J Bacteriol. 1999;181:3886–3889. doi: 10.1128/jb.181.13.3886-3889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamada K, Fukuchi S, Arisawa M, Baba M, Kitada K. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:53–59. doi: 10.1007/s004380050706. [DOI] [PubMed] [Google Scholar]
- 18.Hodges P E, McKee A H Z, Davis B P, Payne W E, Garrels J I. Yeast Protein Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue S B, Takewaki N, Takasuka T, Mio T, Adachi M, Fujii Y, Miyamoto C, Arisawa M, Furuichi Y, Watanabe T. Characterization and gene cloning of 1,3-beta-d-glucan synthase from Saccharomyces cerevisiae. Eur J Biochem. 1995;23:845–854. doi: 10.1111/j.1432-1033.1995.tb20770.x. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson D J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 21.Jelinsky S A, Samson L D. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keng T. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2616–2623. doi: 10.1128/mcb.12.6.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy M A, Barbuch R, Bard M. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1445:110–122. doi: 10.1016/s0167-4781(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 24.Ketela T, Green R, Bussey H. Saccharomyces cerevisiae Mid2p is a potential cell wall stress sensor and upstream activator of the PKC1-MPK1 cell integrity pathway. J Bacteriol. 1999;181:3330–3340. doi: 10.1128/jb.181.11.3330-3340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwast K E, Burke P, Staahl B, Poyton R. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc Natl Acad Sci USA. 1999;96:5446–5451. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14alpha-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 27.Launhardt H, Hinnen A, Munder T. Drug-induced phenotypes provide a tool for the functional analysis of yeast genes. Yeast. 1998;14:935–942. doi: 10.1002/(SICI)1097-0061(199807)14:10<935::AID-YEA289>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 28.Leber R, Landl K, Zinser E, Ahorn H, Kohlwein S D, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Kaplan J. Characterization of yeast methyl sterol oxidase (ERG25) and identification of a human homologue. J Biol Chem. 1996;271:16927–16933. doi: 10.1074/jbc.271.28.16927. [DOI] [PubMed] [Google Scholar]
- 30.Lin R-J, Kim D-H, Castanotto D, Westaway S, Rossi J J. RNA preparations from yeast cells. In: Kreig P A, editor. A laboratory guide to RNA isolation and synthesis. New York, N.Y: Wiley-Liss, Inc.; 1996. pp. 43–50. [Google Scholar]
- 31.Lorenz R T, Casey W M, Parks L W. Structural discrimination in the sparking function of sterols in the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:6169–6173. doi: 10.1128/jb.171.11.6169-6173.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Machida K, Tanaka T, Fujita K-I, Taniguchi M. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain. J Bacteriol. 1998;180:4460–4465. doi: 10.1128/jb.180.17.4460-4465.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marton M J, DeRisi J L, Bennett H A, Iyer V R, Meyer M R, Roberts C J, Stoughton R, Burchard J, Slade D, Dai H, Bassett D E, Hartwell L H, Brown P O, Friend S H. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 34.M'baya B, Fegueur M, Servouse M, Karst F. Regulation of squalene synthase and squalene epoxidase activities in Saccharomyces cerevisiae. Lipids. 1989;24:1020–1023. doi: 10.1007/BF02544072. [DOI] [PubMed] [Google Scholar]
- 35.Michalkova-Papajova M, Obernauerova M, Subik J. Role of the PDR gene network in yeast susceptibility to the antifungal antibiotic mucidin. Antimicrob Agents Chemother. 2000;44:418–420. doi: 10.1128/aac.44.2.418-420.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks L W. Metabolism of sterols in yeast. Crit Rev Microbiol. 1978;6:301–341. doi: 10.3109/10408417809090625. [DOI] [PubMed] [Google Scholar]
- 37.Parks L W, Casey W M. Physiological implications of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 38.Revankar S G, Kirkpatrick W R, McAtee R K, Dib O P, Fothergill A W, Redding S W, Rinaldi M G, Patterson T F. Detection and significance of fluconazole resistance in oropharyngeal candidiasis in human immunodeficiency virus-infected patients. J Infect Dis. 1996;174:821–827. doi: 10.1093/infdis/174.4.821. [DOI] [PubMed] [Google Scholar]
- 39.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez R J, Low C, Bottema C D K, Parks L W. Multiple functions for sterols in Saccharomyces cerevisiae. Biochim Biophys Acta. 1985;837:336–343. doi: 10.1016/0005-2760(85)90057-8. [DOI] [PubMed] [Google Scholar]
- 41.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slekar K H, Kosman D J, Culotta V C. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J Biol Chem. 1996;271:28831–28836. doi: 10.1074/jbc.271.46.28831. [DOI] [PubMed] [Google Scholar]
- 43.Smith S J, Crowley J H, Parks L W. Transcriptional regulation by ergosterol in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5427–5432. doi: 10.1128/mcb.16.10.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith S J, Parks L W. Requirement of heme to replace the sparking sterol function in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1997;1345:71–76. doi: 10.1016/s0005-2760(96)00165-8. [DOI] [PubMed] [Google Scholar]
- 45.Sutter T R, Loper J C. Disruption of the Saccharomyces cerevisiae gene for NADPH-cytochrome P450 reductase causes increased sensitivity to ketoconazole. Biochem Biophys Res Commun. 1989;160:1257–1266. doi: 10.1016/s0006-291x(89)80139-1. [DOI] [PubMed] [Google Scholar]
- 46.Thorsness M, Schafer W, D'Ari L, Rine J. Positive and negative transcriptional control by heme of genes encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:5702–5712. doi: 10.1128/mcb.9.12.5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truan G, Epinat J C, Rougeulle C, Cullin C, Pompon D. Cloning and characterization of a yeast cytochrome b5-encoding gene which suppresses ketoconazole hypersensitivity in a NADPH-P-450 reductase-deficient strain. Gene. 1994;142:123–127. doi: 10.1016/0378-1119(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 48.Turi T G, Loper J C. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14 alpha-demethylase (ERG11) J Biol Chem. 1992;267:2046–2056. [PubMed] [Google Scholar]
- 49.Vanden Bossche H. Biochemical targets for antifungal azole derivatives: hypothesis on the mode of action. Curr Top Med Mycol. 1985;1:313–351. doi: 10.1007/978-1-4613-9547-8_12. [DOI] [PubMed] [Google Scholar]
- 50.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;41:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Bark M, Bruner D A, Gleeson A, Deckelbaum R J, Aljinovic G, Pohl T M, Rothstein R, Sturley S L. Sterol esterification in yeast: a two-gene process. Science. 1996;272:1353–1356. doi: 10.1126/science.272.5266.1353. [DOI] [PubMed] [Google Scholar]
- 54.Yoshida Y, Aoyama Y. Interaction of azole antifungal agents with cytochrome P-45014DM purified from Saccharomyces cerevisiae microsomes. Biochem Pharmacol. 1987;36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]
- 55.Zinser E, Paltauf F, Dunn G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zitomer R S, Carrico P, Deckert J. Regulation of hypoxic gene expression in yeast. Kidney Int. 1997;51:507–513. doi: 10.1038/ki.1997.71. [DOI] [PubMed] [Google Scholar]
- 57.Zitomer R S, Lowry C V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]