Abstract
A 9.8-kb DNA region from the oleandomycin gene cluster in Streptomyces antibioticus was cloned. Sequence analysis revealed the presence of 8 open reading frames encoding different enzyme activities involved in the biosynthesis of one of the two 2,6-deoxysugars attached to the oleandomycin aglycone: l-oleandrose (the oleW, oleV, oleL, and oleU genes) and d-desosamine (the oleNI and oleT genes), or of both (the oleS and oleE genes). A Streptomyces albus strain harboring the oleG2 glycosyltransferase gene integrated into the chromosome was constructed. This strain was transformed with two different plasmid constructs (pOLV and pOLE) containing a set of genes proposed to be required for the biosynthesis of dTDP-l-olivose and dTDP-l-oleandrose, respectively. Incubation of these recombinant strains with the erythromycin aglycon (erythronolide B) gave rise to two new glycosylated compounds, identified as l-3-O-olivosyl- and l-3-O-oleandrosyl-erythronolide B, indicating that pOLV and pOLE encode all enzyme activities required for the biosynthesis of these two 2,6-dideoxysugars. A pathway is proposed for the biosynthesis of these two deoxysugars in S. antibioticus.
The macrolide group of antibiotics comprises a series of compounds mainly active against gram-positive bacteria with clinical application in the treatment of bacterial infections. Oleandomycin (Fig. 1A) is a 14-unit macrolide synthesized by Streptomyces antibioticus. Structurally it contains a macrolactone ring (oleandolide) which is synthesized by the assembly of one starter acetyl coenzyme A (CoA) and six extender methylmalonyl CoA units. To this aglycone, two 6-deoxysugars (l-oleandrose and d-desosamine) are later attached and, probably as late steps in biosynthesis, C-8 hydroxylation and epoxide formation occur after the transfer of the two sugar residues onto the macrolactone. Previous studies have characterized genes involved in the biosynthesis of the polyketide moiety of oleandomycin, genes involved in oleandomycin resistance and secretion, and genes involved in sugar biosynthesis and glycosylation. A large gene encoding the third subunit of the oleandomycin type I polyketide synthase (PKS) has been cloned and sequenced, showing an unusual coding sequence for Streptomyces genes (54). This gene product would catalyze the last two condensation cycles in the biosynthesis of the macrolactone. Two oleandomycin resistance genes (oleB and oleC) have been also cloned and characterized, both encoding ATP-binding cassette (ABC) transporters (34, 44). It has been demonstrated that oleB, an ABC transporter located within the oleandomycin gene cluster, can secrete an inactive glycosylated oleandomycin intermediate (34). Two glycosyltransferase genes (oleI and oleD) and a β-glycosidase (oleR) gene have been identified (20, 43) and shown to participate in modifying oleandomycin intermediates to produce a final inactive intracellular glycosylated derivative (42, 58) that is reactivated after secretion (41). In addition, a gene (oleP) encoding a cytochrome P450 monooxygenase has also been described; it may be responsible for the introduction of the epoxide group characteristic of oleandomycin as one of the latest steps in oleandomycin biosynthesis (45). Recently, five additional genes involved in sugar biosynthesis (oleP1, oleM1, and oleY) and sugar transfer (oleG1 and oleG2) have been reported (35). A proof for the involvement of this chromosomal region in oleandomycin biosynthesis was obtained by insertional inactivation of the oleG1 gene, resulting in a nonproducing mutant (35).
FIG. 1.
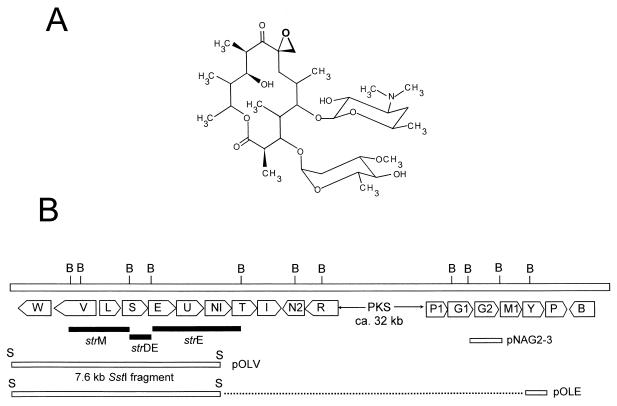
(A) Structure of oleandomycin. (B) Schematic representation of the oleandomycin gene cluster. Solid bars, DNA regions hybridizing against the strDEM genes from the streptomycin pathway; open bars, locations of the 7.6-kb SstI fragment and the PCR-amplified oleY gene. B, BamHI; S, SstI; PKS, PKS genes.
Here we report the identification of eight additional genes involved in the biosynthesis of d-desosamine and l-oleandrose. A pathway is proposed for the biosynthesis of these two 6-deoxysugars by S. antibioticus, and roles are suggested for the different sugar-biosynthetic genes, including those reported here as well as others reported previously (35, 43). We also report the construction of two plasmids capable of promoting the biosynthesis of dTDP-l-olivose and dTDP-l-oleandrose, and evidence is presented for the transfer of these two dTDP-activated sugars to the erythromycin aglycone (erythronolide B) by the oleandrosyl glycosyltransferase (oleG2).
MATERIALS AND METHODS
Microorganisms, culture conditions, and vectors.
S. antibioticus ATCC 11891, an oleandomycin producer, was used as a donor of chromosomal DNA. Growth in liquid medium was carried out at 30°C in Trypticase soy broth (TSB; Oxoid). Streptomyces albus J1074 (ilv-1 sal-2 R− M−) (6) was used as a host for biotransformation experiments. Escherichia coli XL1-Blue (5) was used as a host for subcloning. Plasmids are listed in Table 1. pIJ2925 (23) and pUC18 (63) were used as vectors for subcloning experiments and DNA sequencing. pKCE is an integrative plasmid vector derived from pKC796 (24) in which the erythromycin resistance gene (ermE) was cloned as a 1.7-kb BglII fragment into the BamHI site of this vector (E. Fernández, unpublished data). pIAGO is a multicopy replicative bifunctional (Streptomyces-E. coli) plasmid derived from pWHM3 (57) that contains the promoter of the erythromycin resistance gene (ermE) from Saccharopolyspora erythraea as a 0.28-kb KpnI-BamHI fragment and that can be selected by resistance to thiostrepton in Streptomyces and to ampicillin in E. coli. pIAGO was used for gene expression in Streptomyces. pEM4 was used for subcloning and gene expression in Streptomyces (43). When antibiotic selection of transformants was needed, 25 μg of thiostrepton/ml, 25 μg of apramycin/ml, or 200 μg of erythromycin/ml was used.
TABLE 1.
Plasmid constructs used in this study
| Plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| pIJ2925 | pUC derivative | 23 |
| pKCE | pKC796 containing the erythromycin resistance gene | E. Fernández, unpublished data |
| pIAGO | pWHM3 containing a 0.28-kb KpnI-BamHI fragment including the erythromycin resistance promoter | This study |
| pEM4 | pWHM4 containing the erythromycin resistance promoter | 43 |
| pUC2G | pUC18 with a 5.8-kb BglII fragment from cosAB35 containing the 3′ end of the third subunit of the oleandomycin PKS gene, oleP1, oleG1, and oleG2 | This study |
| pOLV | pIAGO containing a 7.6-kb SstI fragment including the oleW, oleV, oleL, oleS, oleE, and oleU genes and the 5′ end of oleNI | This study |
| pOLE | pOLV containing a 1.1-kb PCR fragment including the oleY gene | This study |
| pNAG2-1 | pEM4 with a 1.4-kb BamHI-EcoRI fragment from pUC2G | This study |
| pNAG2-2 | pIJ2925 with a 1.7-kb HindIII-EcoRI fragment from pNAG2-1 | This study |
| pNAG2-3 | pKCE with a 1.7-kb BglII fragment from pNAG2-2 | This study |
DNA manipulation and sequencing.
Plasmid DNA preparations, restriction endonuclease digestions, alkaline phosphatase treatments, ligations, and other DNA manipulations were carried out according to standard procedures for E. coli (47) and for Streptomyces (21). Southern hybridization was performed according to standard procedures (21). Sequencing was performed by using the dideoxynucleotide chain termination method (48) and the Cy5 AutoCycle Sequencing Kit (Pharmacia Biotech). Both DNA strands were sequenced with primers supplied in the kits or with internal oligoprimers (17-mer) using an ALF-express automatic DNA sequencer (Pharmacia). Computer-assisted database searching and sequence analyses were carried out using the University of Wisconsin Genetics Computer Group program package (9) and the BLASTP program (1).
Construction of an S. albus strain expressing oleG2.
pUC2G is a pUC18 construct containing a 5.8-kb BglII fragment subcloned into the BamHI site of the vector. The insert in pUC2G contains the 3′ end of the third subunit of the oleandomycin PKS gene and the entire oleP1, oleG1, and oleG2 group of genes. From this construct, a 1.45-kb BamHI-EcoRI fragment carrying the oleG2 gene (the BamHI site is shown in Fig. 1B and the EcoRI site comes from the pUC polylinker) was subcloned into the same sites of pEM4. In this construct (pNAG2-1) the oleG2 gene is now under the transcriptional control of the erythromycin resistance gene promoter (ermE*p). To place appropriate restriction sites at both ends of ermE*p and oleG2, the fragment was rescued from pNAG2-1 as a 1.7-kb HindIII-EcoRI fragment and subcloned into the same sites of pIJ2925, generating pNAG2-2. Then a 1.7-kb BglII fragment was subcloned into the BglII site of the integrative vector pKCE, generating pNAG2-3. This construct contains two divergent ermEp's, one controlling transcription of oleG2 and the other controlling that of ermE. pNAG2-3 was digested with XhoI, blunt ended, and religated in order to inactivate the apramycin resistance gene. This selectable marker was eliminated from this construct in order to have the possibility in the future of introducing into the strain harboring this construct other plasmids containing the apramycin cassette as an antibiotic marker. The final construct was then used to transform S. albus protoplasts, and transformants were selected by their resistance to erythromycin (200 μg/ml). Then erythromycin-resistant colonies were assayed for their sensitivity to 25 μg of apramycin/ml by replica plating. One of these colonies (strain NAG2) was selected as a host for further studies.
Construction of plasmids synthesizing dTDP-l-olivose and dTDP-l-oleandrose.
Two plasmids containing sugar-biosynthetic genes were constructed as follows.
(i) dTDP-l-olivose-synthesizing plasmid.
A 7.6-kb SstI fragment (shown in Fig. 1B) from cosAB61 (containing the oleW, oleV, oleL, oleS, oleE, and oleU genes and the 5′ end of oleNI) was subcloned into the SstI site of pIAGO upstream of ermEp, generating pOLV. This construct contains all the putative oleandrose-biosynthetic genes except the oleY gene, which is located at the other end of the cluster downstream of the polyketide synthase genes (see Fig. 1B). Transcription of these genes in this construct is dependent upon their own promoters.
(ii) dTDP-l-oleandrose-synthesizing plasmid.
The oleY gene was amplified by PCR using the following oligoprimers: 5′ AGAGAAGCTTGCTAGCCCGGACCACGCGAAGGACCTTTCACATG 3′ for the 5′ end of the gene and 5′ AGTCTAGATTAATTAACTAGTTGTCGTTCCAGAACGGCTCCCGGG 3′ for the 3′ end of the gene (HindIII and NheI sites [underlined] were included in the first oligoprimer, and PacI and XbaI sites [underlined] were included in the second, to facilitate subcloning). PCR conditions were as follows. One hundred nanograms of template DNA was mixed with 30 pmol of each primer and 2 U of Vent DNA polymerase (New England Biolabs) in a total reaction volume of 50 μl containing 2 mM each deoxynucleoside triphosphate, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100, and 10% dimethyl sulfoxide (Merck). This reaction mixture was overlaid with 50 μl of mineral oil (Sigma), and the polymerization reaction was performed in a thermocycler (MiniCycler; MJ Research) under the following conditions: an initial denaturation of 3 min at 98°C; 30 cycles of 30 s at 98°C, 1 min at 67°C, and 3 min at 72°C; and a final extension step of 5 min at 72°C. The PCR product was purified and subcloned into pUC18 as a HindIII-XbaI fragment and then rescued as an NheI-XbaI fragment for subcloning into the XbaI site of pOLV in the right orientation and downstream of ermEp. This construct (pOLE) contained all putative dTDP-l-oleandrose-biosynthetic genes.
Biotransformation of erythronolide B.
Spores of S. albus IAGS1 and S. albus IAGS2 were inoculated into TSB containing 2.5 μg of thiostrepton/ml. After incubation for 24 h at 30°C and 250 rpm, the mycelium obtained was used to inoculate plates with 25 ml of solid R5 medium modified as previously described (14), supplemented with 25 μg of thiostrepton/ml and 100 μg of erythronolide B/ml. Four plates were inoculated with each strain so as to obtain confluent growth. After 6 days of incubation at 28°C, solid cultures of each strain were extracted twice with 50 ml of ethyl acetate, and the resulting extracts were evaporated under a vacuum and finally redissolved in 500 μl of methanol.
Chromatographic techniques.
Thin-layer chromatography was performed on precoated silica gel 60 F254 plates using dichloromethane-methanol (90:10, vol/vol) as a solvent. Erythromycin-related compounds were stained by spraying the plates with p-anisaldehyde–sulfuric acid–ethanol (1:1:9, vol/vol/vol), followed by heating at 100°C for about 30 s. High-performance liquid chromatographic (HPLC) analysis was performed using a Symmetry C18 column (Waters) with acetonitrile and water as solvents. Samples were eluted with a gradient from 10 to 50% acetonitrile in 20 min, followed by 10 min under isocratic conditions in 50% acetonitrile. The flow rate was 1 ml/min, and detection was done with a photodiode array detector (Waters), monitoring peaks at 210 nm. New peaks with absorption spectra identical to that of erythronolide B were collected and analyzed by thin-layer chromatography. The new products were purified with several injections using the same column and solvents described for analysis, but mobile-phase conditions were optimized in each case. The new product made by S. albus NAG2(pOLV) was isolated with 30% acetonitrile in water under isocratic conditions. For the derivative corresponding to S. albus NAG2(pOLE), a linear gradient from 25 to 50% acetonitrile in 30 min was used.
Metabolite identification.
HPLC analyses were performed on a GILSON liquid chromatography system equipped with a variable wavelength detector at 215 nm and a Kromasil C18 5-μm stainless steel column (inside diameter, 250 by 4.6 mm) fitted with a Kromasil C18 5-μm precolumn (inside diameter, 50 by 4.6 mm). The mobile phase was acetonitrile–methanol–0.065 M ammonium acetate (pH 6.7) (35:15:50, vol/vol/vol). The flow rate of the mobile phase was 1.0 ml/min, and the column was operated at 30°C. HPLC coupled to mass spectrometry was carried out on a chromatography system (Waters) equipped with a FINNIGAN TSQ 7000 mass spectrometer (full scan range from 0 to 750 average mass units).
Nucleotide sequence accession number.
The 9.8-kb DNA region containing oleW, oleV, oleL, oleS, oleE, oleU, oleNI, and oleT has been submitted to GenBank under accession no. AF055579.
RESULTS
Cloning and sequencing of a chromosomal region of S. antibioticus encoding sugar-biosynthetic genes.
A cosmid library of chromosomal DNA from S. antibioticus (44) was screened by in situ colony hybridization using as probes the strD, strE, and strM genes of Streptomyces griseus, which encode several enzymes catalyzing early stages in the biosynthesis of the sugar moieties in the streptomycin pathway. These genes have been shown to be useful probes for the isolation of gene clusters involved in the biosynthesis of the 6-deoxysugar moieties of different antibiotics (51). Using these genes as probes, we isolated two overlapping clones (cosAB61 and cosAB63) which simultaneously hybridized against the three probes. Using Southern hybridization it was found that the hybridization bands corresponded to three adjacent BamHI fragments of 2.2 (hybridization against the strM probe), 1.1 (strD and strE probes), and 4 (strE probe) kb (Fig. 1B). These two cosmid clones overlapped with several other clones previously isolated in our laboratory that contain the oleandomycin polyketide synthase genes. A 9.8-kb DNA region including the three hybridizing BamHI fragments and surrounding DNA was sequenced. The sequence was analyzed for open reading frames (ORFs) using the CODONPREFERENCE program (9). Eight ORFs were found, of which two were transcribed in one direction (oleW and oleV) and the other six (oleL, oleS, oleE, oleU, oleNI, and oleT) were transcribed divergently from the first two (Fig. 1B). Five of the genes (oleL, oleS, oleE, oleU, and oleNI) showed some overlapping sequences between the stop codon of one gene and the start codon of the next one. This is usually considered an indication of translational coupling (33).
Deduced functions of the gene products. (i) oleW.
The oleW gene is located at one end of the cluster upstream of the PKS genes. It appears to start at an ATG codon 19 nucleotides downstream from the stop codon of oleV, with a putative ribosomal-binding site 5 nucleotides upstream of the initiation codon (GAAGGGA). The gene spans 987 nucleotides, and downstream of its TGA codon there is a noncoding region with a length of approximately 600 nucleotides, containing DNA sequences that could form secondary structures that could act as transcriptional terminators. The deduced product of oleW is a protein of 327 amino acid residues (Mr, 35,834) closely related to LanT from the landomycin gene cluster of Streptomyces cyanogenus (50.8% identity) (59), Gra-orf26 from the granaticin biosynthetic gene cluster of Streptomyces violaceoruber (49.1% identity) (22), an oxidoreductase from the rifamycin pathway of Amycolatopsis mediterranei (48.9% identity) (2), and RdmF from the rhodomycin biosynthetic pathway of Streptomyces purpurascens (48.9% identity) (32). It has recently been shown that Gra-orf26 (together with Gra-orf27) participates in the 2-deoxygenation of the oleandrose molecule of granaticin (13). Based on the similarity between OleW and Gra-orf26 (Fig. 2), OleW could be a 3-ketoreductase functioning in combination with OleV to achieve the C-2 deoxygenation step of the oleandrose moiety of oleandomycin.
FIG. 2.
Alignment of OleW and related proteins. LanT, landomycin biosynthetic pathway of S. cyanogenus (59); Oxidored, oxidoreductase from the rifamycin pathway of A. mediterranei (2); GraORF26, granaticin biosynthetic pathway of S. violaceoruber (22); RdmF, rhodomycin biosynthetic pathway of S. purpurascens (32). Amino acids that are identical in at least three of the five proteins compared are shown on a solid background, and conservative substituted amino acids are shown on a shaded background.
(ii) oleV.
The oleV gene is 1,425 nucleotides long and starts with an ATG codon located 335 nucleotides upstream of the initiation codon of oleL and diverging from it. A peculiarity of the oleV gene is the presence in its sequence of a TTA codon. The TTA codon has been proposed to be involved in the regulation of differentiation and secondary metabolism in Streptomyces (25). The deduced protein product of oleV (a protein of 474 amino acid residues and an estimated Mr of 53,149) shows strong sequence similarity to DnmT from the daunosamine pathway in Streptomyces peucetius (52.8% identity) (49), Gra-orf27 from the granaticin pathway in S. violaceoruber (50.7% identity) (22), SnoH from the nogalamycin pathway in Streptomyces nogalater (51.5% identity) (64), ORF3 from Streptomyces sp. strain C5 (48.2% identity) (11), and EryBVI from the erythromycin pathway in S. erythraea (51.1% identity) (16, 53). Based on these similarities and on the proposed function for the above-mentioned Gra-orf27 protein (13), a function as a 2,3-dehydratase can be assigned to the OleV protein in l-oleandrose biosynthesis in S. antibioticus.
(iii) oleL.
The oleL gene starts with an ATG codon and ends in a TGA stop codon. This gene contains in its sequence three TTA codons, two of which, interestingly, are located close to the 5′ end of the gene (3rd and 11th codon positions, respectively), which is in agreement with a potential role in the regulation of secondary metabolism. The oleL gene would code for a polypeptide of 204 amino acids and an estimated Mr of 22,248. Comparison of the deduced product of this gene with proteins in databases showed clear similarities with various dTDP-4-keto-6-deoxyglucose 3,5-epimerases involved in 6-deoxysugar biosynthesis in the streptomycin pathway by Streptomyces glaucescens (50% identity; PIR accession no. S44236), the StrM epimerase from S. griseus, which is also involved in streptomycin biosynthesis (50% identity) (40), the EryBVII protein from the erythromycin pathway from S. erythraea (42.4% identity) (16, 53), and the DnmU protein from the daunorubicin pathway in S. peucetius (42.8% identity) (37). OleL also resembles different RfbC proteins involved in lipopolysaccharide biosynthesis in Mycobacterium tuberculosis (7) and Shigella flexneri (30). On the basis of the above-mentioned similarities, the OleL protein would be responsible for the epimerization occurring at carbons 3 and 5 of the corresponding dTDP intermediate in the biosynthesis of l-oleandrose. Confirmation for this role was obtained by complementing a non-erythromycin-producing mutant from S. erythraea (strain BVII98) defective in 3,5-epimerase activity. Expression of the oleL gene in this mutant under the control of ermEp restored erythromycin production as detected by a bioassay against Bacillus subtilis and thin-layer chromatography using erythromycin A as a standard.
(iv) oleS.
The oleS gene starts at a GTG codon and would end in a TGA codon. It would code for a polypeptide of 356 amino acids and an Mr of 38,109. Its deduced product showed clear similarities with dTDP-d-glucose synthases from streptomycetes involved in the biosynthesis of different antibiotics. The highest scores were with MtmD from the mithramycin pathway in Streptomyces argillaceus (70.2% identity) (28), StrD from the streptomycin pathway in S. griseus (68.2% identity) (40), and DnmL from the daunorubicin pathway in S. peucetius (58.1% identity) (18). All of these enzymes show a well-conserved motif close to the N terminus (LAGGSGTRLRP in OleS) (31). These enzymes are presumed to be involved in the activation of glucose-1-phosphate into dTDP-d-glucose as the earliest step in 6-deoxysugar biosynthesis. In the case of the MtmD synthase (28), such activity has been demonstrated by an in vitro enzymatic assay.
(v) oleE.
The translational starting codon (ATG) of the oleE gene also overlaps the stop codon of the previous gene (oleS). It would code for a polypeptide of 335 amino acids with an estimated Mr of 36,664. Its deduced product showed high similarity with several enzymes that have been proposed to participate in 4,6-dehydration during 6-deoxysugar biosynthesis: MtmE from the mithramycin pathway in S. argillaceus (70.2% identity) (28), StrE from the streptomycin pathway in S. griseus (64.7% identity) (40), Gdh from S. erythraea, a dehydratase which has been shown not to be involved in erythromycin biosynthesis (64.2% identity) (26), and TylA2 from the tylosin pathway in Streptomyces fradiae (63% identity) (31). An interesting feature of all these proteins is the presence of a conserved sequence close to the N terminus which contains a βαβ fold with an NAD+ binding motif, GXGXXG (50, 60). This 4,6-dehydration activity has been demonstrated in vitro for TylA2 (32) and MtmE (29). Therefore, OleE might act as a dTDP-glucose 4,6-dehydratase involved in early steps of the biosynthesis of d-desosamine and l-oleandrose.
(vi) oleU.
oleU is immediately downstream of oleE and also shows an apparent translational coupling with it. The oleU gene would code for a polypeptide of 295 amino acids and an estimated Mr of 32,098. The OleU deduced product displays similarity to a large family of sugar reductases involved in antibiotic biosynthesis: StrL from the streptomycin pathway in S. griseus (57.4% identity) (40), DnmV from the daunorubicin pathway in S. peucetius (27.8% identity) (37), and EryBIV from the erythromycin pathway in S. erythraea (24.8% identity) (16, 53). It also resembles the well-characterized UDP-galactose 4-epimerase encoded by the galE gene of E. coli (3). All the members of this family, including OleU, contain two conserved motifs: one close to the N terminus which is characteristic of a nucleotide-binding site (60) and a second motif (YGRFKRDGE in OleU) in which the tyrosine and lysine residues have been found to be involved in cofactor binding (3). On the basis of similarities with known 4-ketoreductases (DnmV and EryBIV), it is proposed that OleU encodes a 4-ketoreductase involved in l-oleandrose biosynthesis.
(vii) oleNI.
The oleNI gene starts at an ATG codon and ends in a TAG codon. It would code for a polypeptide of 393 amino acids and an Mr of 42,673. The OleNI protein showed similarity with several aminotransferases involved in the biosynthesis of 6-deoxysugars in various antibiotic-biosynthetic pathways: EryCIV from the erythromycin pathway (65% identity) (16, 53), TylB from the tylosin pathway (32.1% identity) (31), DnrJ from the daunorubicin pathway (29.8% identity) (52), and LmbS from the lincomycin pathway (32.6% identity) (38). All of these proteins are similar to the well-characterized AscC dehydratase from the ascarylose pathway in Yersinia pseudotuberculosis (55). Based on the similarity of OleNI with EryCIV, a role is proposed for this protein in the 3,4-dehydration step occurring during d-desosamine biosynthesis. OleN2 (43), homologous to EryCI, would be responsible for the C-3 transamination reaction.
(viii) oleT.
One hundred ten base pairs downstream of oleNI there is a gene, oleT, that would code for a polypeptide of 485 amino acids with an estimated Mr of 53,774. Comparison with databases showed similarity with two proteins involved in macrolide biosynthesis: DesII from the methymycin and pikromycin pathways (57.2% identity) (62) and EryCV from the erythromycin pathway of S. erythraea (56.7% identity) (16, 53). Both enzymes have been proposed to be involved in d-desosamine biosynthesis in these pathways, probably by acting as 3,4-reductases. By analogy, OleT could be responsible for the 3,4-reduction step in d-desosamine biosynthesis in S. antibioticus.
Construction of an S. albus strain expressing the oleandrosyl glycosyltransferase.
In the oleandomycin gene cluster, three genes encoding glycosyltransferases have been described: oleI (43) and oleG1 and oleG2 (35). The first of these codes for a macrolide-inactivating glycosyltransferase responsible for the formation of an inactive glycosylated derivative during oleandomycin biosynthesis (43). The other two glycosyltransferases are involved in the transfer of d-desosamine and l-oleandrose to the oleandolide, as was demonstrated by gene disruption (35). Recently, it has been shown that oleG1 complements an S. erythraea mutant lacking eryCIII and that oleG2, when transformed into a mutant lacking eryBV, produces a new compound that contains a rhamnosyl residue attached to the C-3 position of the erythronolide B (12). Therefore, it has been proposed that OleG1 and OleG2 act as the d-desosaminyl- and l-oleandrosyl-glycosyltransferases, respectively (12). In order to test the capability of the putative oleandrose-biosynthetic genes to make dTDP-l-oleandrose, and the substrate flexibility of the OleG2 glycosyltransferase, we generated an appropriate S. albus strain expressing the OleG2 oleandrosyl glycosyltransferase. The oleG2 gene was subcloned under the control of the ermE* promoter in an integrative bifunctional (Streptomyces-E. coli) plasmid vector, pKCE. In the opposite direction to the oleG2 gene, we also subcloned the erythromycin resistance gene (ermE) and its promoter to provide the new strain with the capability of surviving if new erythromycin-like compounds were synthesized. This strain (NAG2) was now used as a host for transformation with plasmid constructs capable of synthesizing two different dTDP-activated sugars: dTDP-l-olivose (pOLV) and dTDP-l-oleandrose (pOLE).
Construction of plasmids synthesizing dTDP-activated sugars.
To test if the genes assigned were sufficient for dTDP-l-oleandrose biosynthesis, we constructed two different plasmids. The first (pOLE) possesses all the genes that are predicted to be necessary for the biosynthesis of the activated l-oleandrose: genes encoding glucose synthase (oleS), 4,6-dehydratase (oleE), 3,5-epimerase (oleL), 2,3-dehydratase (oleV), 3-reductase (oleW), 3-O-methyltransferase (oleY), and 4-ketoreductase (oleU). All of these genes were under the control of their own promoters, except oleY, which was dependent on ermEp. In the second construct (pOLV), the oleY gene was absent, and therefore this construct would produce dTDP-3-demethyl-l-oleandrose (i.e., dTDP-2-deoxy-l-rhamnose, also designated dTDP-l-olivose). Both plasmids were independently transformed into S. albus strain NAG2, which harbors the oleG2 gene (oleandrosyl glycosyltransferase) integrated into the chromosome (see the preceding section). The choice of S. albus as a host was based on (i) its ability to grow in a well-dispersed way, (ii) the possibility of having good transformation frequencies, and (iii) the fact that it is not known to produce any glycosylated secondary metabolite.
Generation of 3-l-olivosyl-erythronolide B and 3-l-oleandrosyl-erythronolide B.
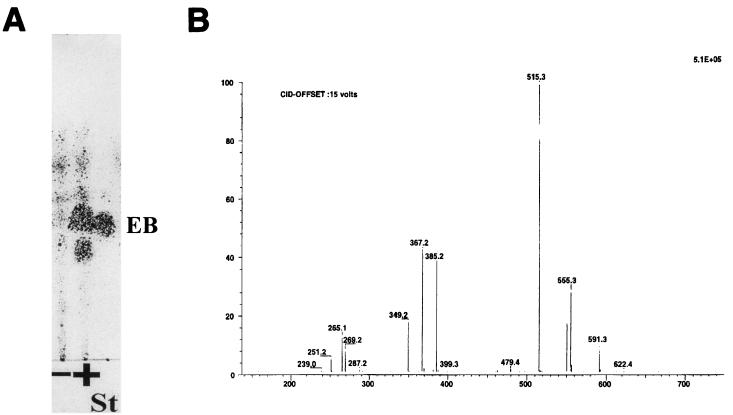
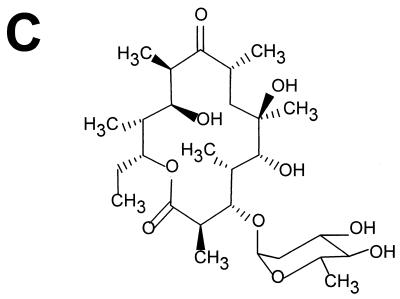
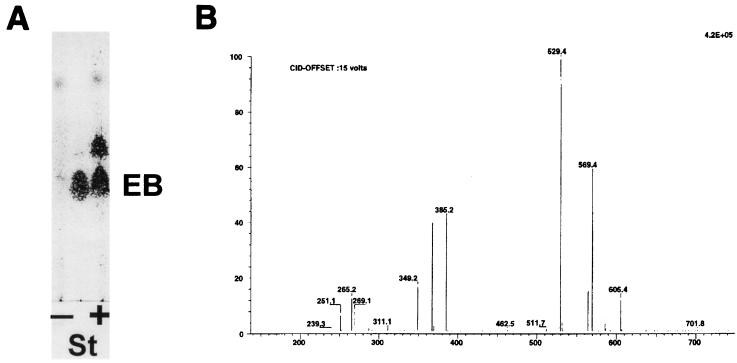
Bioconversion experiments were then carried out to demonstrate that pOLV and pOLE code for the biosynthesis of dTDP-l-olivose and dTDP-l-oleandrose, respectively. S. albus NAG2 containing either pOLV or pOLE (strain IAGS1 or IAGS2, respectively) was incubated in the presence of the erythromycin aglycone (erythronolide B), and after incubation, the presence of glycosylated erythronolide B derivatives was analyzed by thin-layer chromatography and HPLC. In both experiments it was found that approximately 50% of the erythronolide B added was converted into new compounds when biotransformation was carried out on a solid medium. In liquid cultures, the efficiencies obtained were lower. Control experiments were made using as a biotransformation host S. albus NAG2 containing the plasmid vector pIAGO, resulting in no modification of the erythronolide B precursor. In strain IAGS1 (harboring pOLV), the new compound ran slower than erythronolide B on thin-layer chromatography plates, while in strain IAGS2 (harboring pOLE), the new compound ran faster (Fig. 3A and 4A). New peaks were also detected by HPLC analysis (data not shown). The material present in these new HPLC peaks was collected and found to correspond exactly to the new thin-layer chromatography spots. In order to investigate further the structure of the compounds produced by the new recombinant strains, purified extracts made from culture supernatants were analyzed by HPLC coupled to mass spectrometry. In the case of strain IAGS1, one metabolite eluting at 30 min, named M1, gave parent peaks at m/z 555 and 550 for sodium and ammonium ions, respectively, as well as a peak at m/z 515 for the dehydrated H+ ion form (Fig. 3B; Table 2). A fragmentation peak present at m/z 385 can be interpreted as dehydrated erythronolide B (H+ ion). The difference of 130 compared to the 515 parent peak can be traced to the neutral sugar residue, which is consistent with olivose. The M1 compound can therefore be interpreted as 3-l-olivosyl-erythronolide B (Fig. 3C). In the case of strain IAGS2, one metabolite named M2 eluting at 26:36 min gave parent peaks at m/z 569 and 564 for sodium and ammonium ions, respectively, as well as a peak at m/z 529 for the dehydrated H+ ion form (Fig. 4B; Table 2). A fragmentation peak present at m/z 385 which can be interpreted as dehydrated erythronolide B (H+ ion form) indicates a difference of 144 (compared to peak 529), corresponding to the loss of the neutral sugar residue. These results are consistent with the interpretation of M2 as being 3-l-oleandrosyl-erythronolide B (Fig. 4C).
FIG. 3.
Analysis of the products of the biotransformation of erythronolide B by strain IAGS1. (A) Thin-layer chromatography analysis of the reaction. EB, erythronolide B; St, standard; + and −, incubation in the presence and absence of EB, respectively. (B) Mass spectrometry analysis of compound M1. (C) Structure of 3-l-olivosyl-erythronolide B.
FIG. 4.
Analysis of the products of the biotransformation of erythronolide B by strain IAGS2. (A) Thin-layer chromatography analysis of the reaction. EB, erythronolide B; St, standard; + and −, incubation in the presence and absence of EB, respectively. (B) Mass spectrometry analysis of compound M2. (C) Structure of 3-l-oleandrosyl-erythronolide B.
TABLE 2.
Mass spectrometry data for metabolites produced by recombinant S. albus strains IAGS1 and IAGS2
| Iona | m/z | Result
for strain:
|
|
|---|---|---|---|
| IAGS1 | IAGS2 | ||
| [M1]Na+ | 555 | ++ | |
| [M1]NH4+ | 550 | ++ | |
| [M1–H2O]H+ | 515 | ++ | |
| [EB–H2O]H+ | 385 | ++ | ++ |
| [EB–2H2O]H+ | 367 | ++ | ++ |
| [EB–3H2O]H+ | 349 | ++ | ++ |
| [M2]Na+ | 569 | ++ | |
| [M2]NH4+ | 564 | ++ | |
| [M2–H2O]H+ | 529 | ++ | |
EB, erythronolide B.
DISCUSSION
A number of antibiotics and antitumor agents possess deoxysugars attached to their aglycones; in many cases, these deoxysugars are essential for their biological activity. They belong to the family of 6-deoxyhexoses and are commonly synthesized through dTDP-activated intermediates (27, 39, 56). However, the biochemical pathways involved in 6-deoxyhexose biosynthesis have not been characterized in most cases. Several authors have reported the cloning of different genes involved in the biosynthesis of 6-deoxyhexoses in antibiotic and antitumor pathways: streptomycin (40), erythromycin (10, 16, 46, 53), tylosin (15, 19, 31), daunosamine (36, 37), lincomycin (38), mithramycin (28), methymycin-pikromycin (62), and granaticin (4, 22). Macrolide antibiotics contain mono- or disaccharides attached to the macrolactone rings which are important for activity. Oleandomycin shares some structural features with erythromycin. They both belong to the 14-unit macrolides and contain d-desosamine attached to the aglycone via the hydroxyl group at C-5. However, they differ in the other sugar at C-3, l-mycarose in erythromycin and l-oleandrose in oleandomycin. In previous papers we have reported the isolation of several genes of the oleandomycin gene cluster in S. antibioticus (34, 35, 43, 45, 54). The eight additional genes we report here (oleW, oleV, oleL, oleS, oleE, oleU, oleNI, and oleT), together with other six genes previously reported (oleN2, oleP1, oleG1, oleG2, oleM1, and oleY) (35, 43), constitute the set of genes encoding the enzymatic functions required for the biosynthesis and transfer of the two 6-deoxyhexoses in the oleandomycin molecule. The involvement of the isolated gene cluster in oleandomycin biosynthesis came from the following evidence: (i) the high similarities of the gene products identified with homologous proteins in the erythromycin cluster, (ii) the presence in the cloned cluster of resistance genes specifically conferring resistance to oleandomycin and not to other macrolides (34, 43, 44), and (iii) the complete abolishment of oleandomycin biosynthesis by the inactivation of one of the genes of the cluster, oleG1 (35).
Of the eight genes described in this paper, four would code for l-oleandrose-specific biosynthetic enzymes (oleW, oleV, oleL, and oleU) and two would code for d-desosamine-specific biosynthetic enzymes (oleNI and oleT). The other two genes (oleS and oleE) would code for two enzymes catalyzing early steps common to the biosynthesis of both deoxysugars: a glucose-1-phosphate: TTP thymidylyl transferase (OleS) catalyzing the activation of glucose-1-phosphate into dTDP-glucose and a dTDP-glucose 4,6-dehydratase (OleE) responsible for the conversion of dTDP-glucose into dTDP-4-keto-6-deoxyglucose. These two enzymes are well conserved in most of the sugar-biosynthetic pathways so far characterized, with the exception of the erythromycin pathway, where no gene encoding a synthase or a dehydratase has been identified within the cluster. The dTDP-4-keto-6-deoxyglucose intermediate generated by the action of these two enzymes is a common intermediate in the biosynthesis of both dTDP-d-desosamine and dTDP-l-oleandrose.
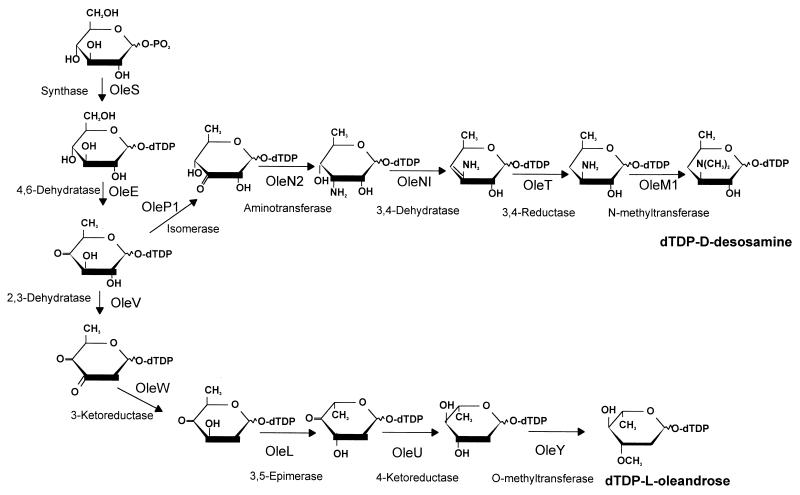
d-Desosamine is an aminosugar which is present in many macrolide antibiotics (erythromycin, oleandomycin, methymycin-pikromycin, etc.). Desosamine-biosynthetic genes from the erythromycin producer (eryC genes) have been independently characterized by different laboratories (16, 46, 53). We have found five genes (oleNI, oleT, oleN2, oleP1, and oleM1) in the oleandomycin cluster showing high similarities to eryC genes; this indicates that probably this aminosugar is synthesized in both producer strains through similar biosynthetic pathways (Fig. 5).
FIG. 5.
Proposed pathways for the biosynthesis of d-desosamine and l-oleandrose by S. antibioticus.
l-Oleandrose is a neutral sugar not frequently found in bioactive compounds. It is present as a monosaccharide in oleandomycin and as a disaccharide in the antihelmintic avermectins. Several enzymatic steps are required for the conversion of dTDP-4-keto-6-deoxyglucose into dTDP-l-oleandrose: C-2 deoxygenation, epimerization at C-3 and C-5, C-4 ketoreduction, and C-3 O-methylation. l-Oleandrose and l-mycarose are 2,6-deoxyhexoses, and therefore their biosynthesis will require enzymes catalyzing C-2 deoxygenation. In the erythromycin pathway in S. erythraea, it has been proposed that two enzyme activities are involved in C-2 deoxygenation during l-mycarose biosynthesis: the EryBVI (2,3-dehydratase) and EryBII (2,3-reductase) proteins (16, 53). The oleandomycin cluster also contains a gene encoding an EryBVI-like protein (oleV). However, we did not find an eryBII-like gene either within the oleandomycin cluster or in the chromosome. Our attempts to identify such a gene in the chromosome of S. antibioticus were unsuccessful, both by use of the eryBII gene from S. erythraea as a probe and by PCR amplification using oligoprimers derived from the eryBII sequence. Interestingly, an eryBII-like gene for l-oleandrose biosynthesis has been described in the avermectin cluster of Streptomyces avermitilis (cited in reference 29). However, it has recently been shown by in vitro studies that 2-deoxygenation during 6-deoxyhexose biosynthesis requires the participation of two enzymes (Orf10 and Orf11) in S. antibioticus Tü99 and two enzymes (Gra-Orf27 and Gra-Orf26) in S. violaceoruber Tü22 (13). The OleV and OleW proteins show high similarity to Gra-Orf27 (50.7% identical amino acids) and Gra-Orf26 (49.1% identical amino acids). Consequently, 2-deoxygenation during l-oleandrose biosynthesis in S. antibioticus ATCC 11891 could be mediated by OleV and OleW without the involvement of an EryBII-like protein.
Erythronolide B was biotransformed by S. albus strains IAGS2 (containing oleG2 and pOLE) into 3-l-oleandrosyl erythronolide B. This demonstrates that the set of genes formed by oleS, oleE, oleV, oleW, oleL, oleU, and oleY is able to synthesize l-oleandrose. Interestingly, another strain (IAGS1) containing the same set of genes, but lacking the 3-O-methyltransferase (oleY gene), transferred l-olivose to erythronolide B, generating 3-l-olivosyl-erythronolide B. This indicates that the oleandrose genes are also able to synthesize dTDP-l-olivose, an unmethylated form of dTDP-l-oleandrose, suggesting that dTDP-l-olivose could be an intermediate in dTDP-l-oleandrose biosynthesis (Fig. 5). Several eryB and eryC mutants have been found to incorporate some intermediates in d-desosamine and l-mycarose biosynthesis into the erythromycin aglycone, but as minor compounds with low efficiency (17, 46). Interestingly, the formation of the two glycosylated derivatives of erythronolide B reported in this work was quite efficient, with a bioconversion rate of approximately 40% of the originally added precursor and no significant difference between the transfer of l-oleandrose and that of l-olivose. These macrolide derivatives are the first two hybrid glycosylated macrolides generated by biotransformation of a macrolide aglycone in a non-macrolide-producing host in which a 6-deoxyhexose-biosynthetic gene cluster has been expressed. Generation of novel glycosylated derivatives of medically important bioactive compounds by genetic engineering requires both the construction of a plasmid synthesizing activated sugars and the use of glycosyltransferases that are flexible in some way. The two constructs shown in this work (pOLV and pOLE) are the first two plasmids reported capable of synthesizing dTDP-activated sugars, and so they have the potential to be used for modifying different aglycones. This will require the use of “flexible” glycosyltransferases, and in this context, a few examples of glycosyltransferase substrate flexibility have been reported (8, 46, 61). The S. antibioticus OleG2 glycosyltransferase has been shown to transfer l-oleandrose and l-olivose (this work) and also l-rhamnose (12). This “substrate flexibility” of the OleG2 glycosyltransferase opens up the possibility of using this sugar transfer enzyme as a genetic tool for further studies leading to the generation of novel glycosylated macrolides.
ACKNOWLEDGMENTS
This work was supported by grants from the European Community (Biotech BIO4-CT96-0080) and the Plan FEDER (1FD97-0040) to J.A.S.
We thank M. C. Raynal and R. Legrand for help with the mass spectrometry analyses.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.August P R, Tang L, Yoon Y J, Ning S, Muller R, Yu T W, Taylor M, Hoffmann D, Kim G G, Zhang X, Hutchinson C R, Floss H G. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterraneiS699. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 3.Bauer A J, Rayment I, Frey P A, Holden H M. The molecular structure of UDP-galactose-4-epimerase from Escherichia colidetermined at 2.5 Å resolution. Proteins. 1992;12:372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- 4.Bechthold A, Sohng J K, Smith T M, Chu X, Floss H G. Identification of Streptomyces violaceoruber Tü22 genes involved in the biosynthesis of granaticin. Mol Gen Genet. 1995;248:610–620. doi: 10.1007/BF02423457. [DOI] [PubMed] [Google Scholar]
- 5.Bullock W O, Fernández J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia colistrain with β-galactosidase selection. BioTechniques. 1987;5:376. [Google Scholar]
- 6.Chater K F, Wilde L C. Streptomyces albus G mutants defective in the SalGI restriction-modification system. J Gen Microbiol. 1980;116:323–334. doi: 10.1099/00221287-116-2-323. [DOI] [PubMed] [Google Scholar]
- 7.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Squares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosisfrom the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 8.Decker H, Haag S, Udvarnoki G, Rohr J. Novel genetically engineered tetracenomycins. Angew Chem Int Ed Engl. 1995;34:1107–1110. [Google Scholar]
- 9.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhillon N, Hale R S, Cortés J, Leadlay P F. Molecular characterization of a gene from Saccharopolyspora erythraea (Streptomyces erythreus) which is involved in erythromycin biosynthesis. Mol Microbiol. 1989;3:1405–1414. doi: 10.1111/j.1365-2958.1989.tb00123.x. [DOI] [PubMed] [Google Scholar]
- 11.Dickens M L, Ye J, Strohl W R. Cloning, sequencing and analysis of aklaviketone reductase from Streptomycessp. strain C5. J Bacteriol. 1996;178:3384–3388. doi: 10.1128/jb.178.11.3384-3388.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doumith M, Legrand R, Lang C, Salas J A, Raynal M C. Interspecies complementation in Saccharopolyspora erythraea: elucidation of the function of oleP1, oleG1 and oleG2 from the oleandomycin biosynthetic gene cluster of Streptomyces antibioticusand generation of new erythromycin derivatives. Mol Microbiol. 1999;34:1039–1048. doi: 10.1046/j.1365-2958.1999.01666.x. [DOI] [PubMed] [Google Scholar]
- 13.Draeger G, Park S-H, Floss H G. Mechanism of the 2-deoxygenation step in the biosynthesis of the deoxyhexose moieties of the antibiotics granaticin and oleandomycin. J Am Chem Soc. 1999;121:2611–2612. [Google Scholar]
- 14.Fernández E, Weibbach U, Sánchez Reillo C, Braña A F, Méndez C, Rohr J, Salas J A. Identification of two genes from Streptomyces argillaceusencoding two glycosyltransferases involved in the transfer of a disaccharide during the biosynthesis of the antitumor drug mithramycin. J Bacteriol. 1998;180:4929–4937. doi: 10.1128/jb.180.18.4929-4937.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouces R, Mellado E, Diez B, Barredo J L. The tylosin biosynthetic cluster from Streptomyces fradiae: genetic organization of the left region. Microbiology. 1999;145:855–868. doi: 10.1099/13500872-145-4-855. [DOI] [PubMed] [Google Scholar]
- 16.Gaisser S, Böhm G, Cortés J, Leadlay P F. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol Gen Genet. 1997;256:239–251. doi: 10.1007/s004380050566. [DOI] [PubMed] [Google Scholar]
- 17.Gaisser S, Böhm G, Doumith M, Raynal M C, Dhillon N, Cortés J, Leadlay P F. Analysis of eryBI, eryBIII, and eryBVII from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol Gen Genet. 1998;258:78–88. doi: 10.1007/s004380050709. [DOI] [PubMed] [Google Scholar]
- 18.Gallo M A, Ward J, Hutchinson C R. The dnrM gene in Streptomyces peucetiuscontains a naturally occurring frameshift mutation that is suppressed by another locus outside of the daunorubicin-production gene cluster. Microbiology. 1996;142:269–275. doi: 10.1099/13500872-142-2-269. [DOI] [PubMed] [Google Scholar]
- 19.Gandecha A R, Large S L, Cundliffe E. Analysis of four tylosin biosynthetic genes from the tylLM region of the Streptomyces fradiaegenome. Gene. 1997;184:197–203. doi: 10.1016/s0378-1119(96)00595-1. [DOI] [PubMed] [Google Scholar]
- 20.Hernández C, Olano C, Méndez C, Salas J A. Characterization of a Streptomyces antibioticusgene cluster encoding a glycosyltransferase involved in oleandomycin inactivation. Gene. 1993;134:139–140. doi: 10.1016/0378-1119(93)90189-a. [DOI] [PubMed] [Google Scholar]
- 21.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces. A laboratory manual. Norwich. England: The John Innes Foundation; 1985. [Google Scholar]
- 22.Ichinose K, Bedford D J, Tornus D, Bechthold A, Bibb M J, Revill W P, Floss H G, Hopwood D A. The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tü22: sequence analysis and expression in a heterologous host. Chem Biol. 1998;5:647–659. doi: 10.1016/s1074-5521(98)90292-7. [DOI] [PubMed] [Google Scholar]
- 23.Janssen G, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia colicolonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 24.Kushtoss S, Richardson M A, Rao R N. Plasmid cloning vectors that integrate site-specifically in Streptomycesspp. Gene. 1991;97:143–146. doi: 10.1016/0378-1119(91)90022-4. [DOI] [PubMed] [Google Scholar]
- 25.Leskiw B K, Lawlor E J, Fernández-Abalos J M, Chater K F. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative Streptomycesmutants. Proc Natl Acad Sci USA. 1991;88:184–189. doi: 10.1073/pnas.88.6.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linton K J, Jarvis B W, Hutchinson C R. Cloning of the genes encoding thymidine diphosphoglucose 4,6-dehydratase and thymidine diphospho-4-keto-6-deoxyglucose 3,5-epimerase from the erythromycin-producing Saccharopolyspora erythraea. Gene. 1995;153:33–40. doi: 10.1016/0378-1119(94)00809-7. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Thorson S. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu Rev Microbiol. 1994;48:223–256. doi: 10.1146/annurev.mi.48.100194.001255. [DOI] [PubMed] [Google Scholar]
- 28.Lombó F, Siems K, Braña A F, Méndez C, Bindseil K, Salas J A. Cloning and insertional inactivation of Streptomyces argillaceusgenes involved in earliest steps of sugar biosynthesis of the antitumor polyketide mithramycin. J Bacteriol. 1997;179:3354–3357. doi: 10.1128/jb.179.10.3354-3357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madduri K, Kennedy J, Rivola G, Inventi-Solari A, Filippini S, Zanuso G, Colombo A L, Gewain K M, Occi J L, MacNeil D J, Hutchinson C R. Production of the antitumor drug epirubicin (4′-epidoxorubicin) and its precursor by a genetically engineered strain of Streptomyces peucetius. Nat Biotechnol. 1998;16:69–74. doi: 10.1038/nbt0198-69. [DOI] [PubMed] [Google Scholar]
- 30.Macpherson D F, Manning P A, Morona R. Characterization of the dTDP-rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol. 1994;11:281–292. doi: 10.1111/j.1365-2958.1994.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 31.Merson-Davies L A, Cundliffe E. Analysis of five tylosin biosynthetic genes from the tylIBA region of the Streptomyces fradiaegenome. Mol Microbiol. 1994;13:349–355. doi: 10.1111/j.1365-2958.1994.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 32.Niemi J, Mantsala P. Nucleotide sequences and expression of genes from Streptomyces purpurascens that cause the production of new anthracyclines in Streptomyces galilaeus. J Bacteriol. 1995;177:2942–2945. doi: 10.1128/jb.177.10.2942-2945.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Normark S, Bergström S, Edlund T, Grundström T, Jaurin B, Lindberg F P, Olsson O. Overlapping genes. Annu Rev Genet. 1983;58:229–241. doi: 10.1146/annurev.ge.17.120183.002435. [DOI] [PubMed] [Google Scholar]
- 34.Olano C, Rodriguez A M, Méndez C, Salas J A. A second ABC transporter is involved in oleandomycin resistance and secretion by the producer organism Streptomyces antibioticus. Mol Microbiol. 1995;16:333–343. doi: 10.1111/j.1365-2958.1995.tb02305.x. [DOI] [PubMed] [Google Scholar]
- 35.Olano C, Rodriguez A M, Michel J M, Méndez C, Raynal M C, Salas J A. Analysis of a Streptomyces antibioticuschromosomal region involved in oleandomycin biosynthesis, which encodes two glycosyltransferases responsible for glycosylation of the macrolactone ring. Mol Gen Genet. 1998;259:299–308. doi: 10.1007/s004380050816. [DOI] [PubMed] [Google Scholar]
- 36.Otten S L, Liu X, Ferguson J, Hutchinson C R. Cloning and characterization of the Streptomyces peucetius dnrQSgenes encoding a daunosamine biosynthesis enzyme and a glycosyl transferase involved in daunorubicin biosynthesis. J Bacteriol. 1995;177:6688–6692. doi: 10.1128/jb.177.22.6688-6692.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otten S L, Gallo M A, Madduri K, Liu X, Hutchinson C R. Cloning and characterization of the Streptomyces peucetius dnmZUV genes encoding three enzymes required for biosynthesis of the daunorubicin precursor thymidine diphospho-l-daunosamine. J Bacteriol. 1997;179:4446–4450. doi: 10.1128/jb.179.13.4446-4450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peschke U, Schmidt H, Zhang H Z, Piepersberg W. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis78-11. Mol Microbiol. 1995;16:1137–1156. doi: 10.1111/j.1365-2958.1995.tb02338.x. [DOI] [PubMed] [Google Scholar]
- 39.Piepersberg W. Pathway engineering in secondary metabolite-producing actinomycetes. Crit Rev Biotechnol. 1994;14:251–285. doi: 10.3109/07388554409079835. [DOI] [PubMed] [Google Scholar]
- 40.Pissowotzki K, Mansouri K, Piepersberg W. Genetics of streptomycin production in Streptomyces griseus: molecular structure and putative functions of strELMB2N. Mol Gen Genet. 1991;231:113–123. doi: 10.1007/BF00293829. [DOI] [PubMed] [Google Scholar]
- 41.Quirós L M, Hernández C, Salas J A. Purification and characterization of an extracellular enzyme from Streptomyces antibioticusthat converts inactive glycosylated oleandomycin into active antibiotic. Eur J Biochem. 1993;222:129–135. doi: 10.1111/j.1432-1033.1994.tb18850.x. [DOI] [PubMed] [Google Scholar]
- 42.Quirós L M, Salas J A. Biosynthesis of the macrolide oleandomycin by Streptomyces antibioticus. Purification and kinetic characterization of an oleandomycin glucosyltransferase. J Biol Chem. 1995;270:18234–18239. doi: 10.1074/jbc.270.31.18234. [DOI] [PubMed] [Google Scholar]
- 43.Quirós L M, Aguirrezabalaga I, Olano C, Méndez C, Salas J A. Two glycosyltransferases and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol Microbiol. 1998;28:1177–1186. doi: 10.1046/j.1365-2958.1998.00880.x. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez A M, Olano C, Vilches C, Méndez C, Salas J A. Streptomyces antibioticuscontains at least three oleandomycin resistance determinants one of which shows similarity with proteins of the ABC-transporter superfamily. Mol Microbiol. 1993;8:571–582. doi: 10.1111/j.1365-2958.1993.tb01601.x. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez A M, Olano C, Méndez C, Hutchinson C R, Salas J A. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol Lett. 1995;127:117–120. doi: 10.1111/j.1574-6968.1995.tb07459.x. [DOI] [PubMed] [Google Scholar]
- 46.Salah-Bey K, Doumith M, Michel J M, Haydock S, Cortes J, Leadlay P F, Raynal M C. Targeted gene inactivation for the elucidation of deoxysugar biosynthesis in the erythromycin producer Saccharopolyspora erythraea. Mol Gen Genet. 1998;257:542–553. doi: 10.1007/s004380050680. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 48.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scotti C, Hutchinson C R. Enhanced antibiotic production by manipulation of the Streptomyces peucetius dnrH and dnmTgenes involved in doxorubicin (adriamycin) biosynthesis. J Bacteriol. 1996;178:7316–7321. doi: 10.1128/jb.178.24.7316-7321.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scrutton N S, Berry A, Perham R N. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990;343:38–43. doi: 10.1038/343038a0. [DOI] [PubMed] [Google Scholar]
- 51.Stockmann M, Piepersberg W. Gene probes for the detection of 6-deoxyhexose metabolism in secondary metabolism-producing streptomycetes. FEMS Microbiol Lett. 1992;90:185–190. doi: 10.1016/0378-1097(92)90626-y. [DOI] [PubMed] [Google Scholar]
- 52.Stutzman-Engwall K J, Otten S L, Hutchinson C R. Regulation of secondary metabolism in Streptomyces spp. and overproduction of daunorubicin in Streptomyces peucetius. J Bacteriol. 1992;174:144–154. doi: 10.1128/jb.174.1.144-154.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Summers R G, Donadio S, Staver M J, Wendt-Pienkowski E, Hutchinson C R, Katz L. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in l-mycarose and d-desosamine production. Microbiology. 1997;143:3251–3262. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- 54.Swan D G, Rodriguez A M, Vilches C, Méndez C, Salas J A. Characterisation of a Streptomyces antibioticusgene encoding a type I polyketide synthase which has an unusual coding sequence. Mol Gen Genet. 1993;242:358–362. doi: 10.1007/BF00280426. [DOI] [PubMed] [Google Scholar]
- 55.Thorson J S, Lo S F, Ploux O, Liu H W. Studies of the biosynthesis of 3,6-dideoxyhexoses: molecular cloning and characterization of the asc (ascarylose) region from Yersinia pseudotuberculosisserogroup VA. J Bacteriol. 1994;176:5483–5493. doi: 10.1128/jb.176.17.5483-5493.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trefzer A, Salas J A, Bechthold A. Genes and enzymes of deoxysugar biosyntheses. Nat Prod Rep. 1999;16:283–299. doi: 10.1039/a804431g. [DOI] [PubMed] [Google Scholar]
- 57.Vara J, Lewandowska-Skarbek M, Wang Y G, Donadio S, Hutchinson C R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus) J Bacteriol. 1989;171:5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vilches C, Hernández C, Méndez C, Salas J A. Role of glycosylation and deglycosylation in biosynthesis of and resistance to oleandomycin in the producer organism, Streptomyces antibioticus. J Bacteriol. 1992;174:161–165. doi: 10.1128/jb.174.1.161-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Westrich L, Domann S, Faust B, Bedford D, Hopwood D A, Bechthold A. Cloning and characterization of a gene cluster from Streptomyces cyanogenusS136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 60.Wierenga R K, Terpstra P, Hol W G J. Interaction of pyrophosphate moieties with α-helices in dinucleotide binding proteins. J Mol Biol. 1985;24:1346–1357. [Google Scholar]
- 61.Wohlert S-E, Blanco G, Lombó F, Fernández E, Braña A F, Reich S, Udvarnoki G, Méndez C, Decker H, Salas J A, Rohr J. Novel hybrid tetracenomycins through combinatorial biosynthesis using a glycosyltransferase encoded by the elm-genes in cosmid 16F4 which shows a broad sugar substrate specificity. J Am Chem Soc. 1998;41:10596–10601. [Google Scholar]
- 62.Xue Y, Zhao L, Liu H W, Sherman D H. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc Natl Acad Sci USA. 1998;95:12111–12116. doi: 10.1073/pnas.95.21.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 64.Ylihonko K, Tuikkanen J, Jussila S, Cong L, Mäntsälä P. A gene cluster involved in nogalamycin biosynthesis from Streptomyces nogalater: sequence analysis and complementation of early-block mutations in the anthracycline pathway. Mol Gen Genet. 1996;251:113–120. doi: 10.1007/BF02172908. [DOI] [PubMed] [Google Scholar]