Abstract
Objectives
This study aimed to explore the role of CD4+ T cells in the mechanisms of COVID-19 related diarrhea.
Methods
We analyzed lymphocyte subsets in patients with COVID-19 and the expression of angiotensin-converting enzyme 2 (ACE2), the transmembrane protease serine 2, and CD4+ T cell-related indicators in the colon were compared between patients with and without diarrhea. Correlation analyses were performed for ACE2 and other indicators to identify the relationship between SARS-CoV-2 infection and CD4+ mediated inflammation. The expression and distribution of CD4+ T cell-associated chemokines and their receptors were detected to determine the possibility of migration of CD4+ T cells to inflammation sites.
Results
The CD4+ T cell counts and percentages and CD4/CD8 ratio showed the most significant differences between the 2 groups. The diarrhea group expressed higher levels of ACE2, T-box expressed in T cells (Tbet), and tumor necrosis factor-alpha (TNFα) at both the mRNA and protein levels, with no difference from the nondiarrhea group for the percentage of ACE2+TNFα+ cells, indicating an indirect association between ACE2 and TNFα. The mRNA expression of CXCL10, CXCL11, and CXCR3 and the number of CD4+CXCR3+T cells were increased in the diarrhea group.
Conclusions
CD4+ T cell-mediated inflammation may contribute to COVID-19 related diarrhea. CXCR3+ mediated migration of CD4+ T cells into the gut may perpetuate inflammation.
Keywords: ACE2, CD4+ T cells, COVID-19, diarrhea, SARS-CoV-2
Introduction
COVID-19, caused by the SARS-CoV-2 strain, has become a global pandemic that is a serious threat to human life and health. The most prominent manifestations of COVID-19 are located in the respiratory system, but an increasing number of extrapulmonary manifestations have also been reported (AlSamman et al., 2020; Gupta et al., 2020). SARS-CoV-2 also affects the gastrointestinal (GI) tract (Ma et al., 2020; Ziegler et al., 2020; Zou et al., 2020), causing digestive symptoms such as anorexia, diarrhea, nausea, vomiting, abdominal pain, and abdominal discomfort (Cheung et al., 2020, Elmunzer et al., 2021). Among these GI presentations, diarrhea is one of the most common symptoms (Cheung et al., 2020; Elmunzer et al., 2021).
In the early phase of the COVID-19 epidemic, diarrhea seemed to be underestimated, with a prevalence of less than 10% (Chen et al., 2020; Guan et al., 2020; Huang et al., 2020; Wang et al., 2020; Xu et al., 2020). As the epidemic progressed, the prevalence of diarrhea during COVID-19 infection was observed to be more than 30% (Elmunzer et al., 2021; Guo et al., 2021), and its importance has since been better described. Further, diarrhea may be associated with the severity of COVID-19, whereby patients with diarrhea have a higher prevalence of required ventilator support, admission to the intensive care unit (Wan et al., 2020), and might be more prone to multiple organ damage than those without diarrhea (Zhang et al., 2021). In addition, based on the potential intestinal diarrheal infection and fecal-oral transmission of SARS-CoV-2, more attention should be paid to diarrhea in the control and prevention of the COVID-19 epidemic, especially when collecting fecal samples or performing endoscopic examinations in these patients.
Although extensive studies have focused on diarrhea in patients with COVID-19, most studies only analyzed the correlations between diarrhea and other clinical data. Possible underlying causes of COVID-19 associated diarrhea have been reported to include the direct cytopathic effects of SARS-CoV-2, robust systemic inflammation or so-called cytokine storm, gut microbiota dysbiosis, and antibiotic-associated diarrhea or adverse effects of antiviral agents such as hydroxy-chloroquine, remdesivir, and ritonavir-lopinavir (de Oliveira et al., 2021, Guo et al., 2021, Jin et al., 2021, Perisetti et al., 2020, Yan et al., 2021). However, to date, no exact mechanism has been proposed to explain the local inflammation in the intestine that leads to diarrhea. During clinical practice, we found that COVID-19 related diarrhea was closely associated with lymphocyte subsets, specifically CD4+ T cells. Therefore, we aimed to explore the role of CD4+ T cells in the mechanism of diarrhea-associated COVID-19.
Method
Study design
We observed a difference in the lymphocyte count and percentage in patients with COVID-19 with diarrhea than those without diarrhea. Retrospective analyses were performed to explore the involvement of lymphocytes in the process of COVID-19 associated diarrhea.
Patients with a positive standard COVID-19 real-time polymerase chain reaction (PCR) and negative influenza virus real-time PCR test were included in the analysis.
Patients with mental illness, pregnancy, infectious disease, systemic disease, food allergy, a history of chronic diarrhea such as inflammatory bowel disease (IBD), celiac disease, and bacillary dysentery before COVID-19 infection were excluded. Further, patients with diarrhea 15 days before COVID-19 infection and those deemed inappropriate by 2 independent doctors for any other conditions were also excluded. Based on these criteria, 145 inpatients were included in this study.
Data collection
Individual characteristics, symptoms on admission, chronic medical illnesses, laboratory results, disease severity, treatment during hospitalization, and hospital stay were collected from patient records in this retrospective study. All data were cross-checked by 2 researchers (WJ and WX) and listed in Table 1 .
Table 1.
Characteristics of patients with COVID-19 with and without diarrhea
| Variable, N (%) or Median with IQR | OverallN = 145 | With diarrheaN = 21 | Without diarrheaN = 124 | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 66 (45.5) | 8 (38.1) | 58 (46.8) | 0.46 |
| Female | 79 (54.5) | 13 (61.9) | 66 (53.2) | |
| Age (years) | 55.0 (44.0-64.5) | 55.0 (48.0-61.5) | 54.5 (43.5-65.5) | 0.73 |
| Symptoms on admission | ||||
| Fever | 70 (48.3) | 10 (47.6) | 60 (48.4) | 0.95 |
| Cough | 80 (55.2) | 15 (71.4) | 65 (52.4) | 0.11 |
| Expectoration | 25 (17.2) | 4 (19.0) | 21 (16.9) | 0.76 |
| Dyspnea | 24 (16.6) | 5 (23.8) | 19 (15.3) | 0.35 |
| Headache | 10 (6.9) | 1 (4.8) | 9 (7.3) | 1 |
| Fatigue | 44 (30.3) | 5 (23.8) | 39 (31.5) | 0.48 |
| Diarrhea | 21 (14.5) | 21 (100) | 0 | <0.001 |
| Vomiting | 5 (3.4) | 1 (4.8) | 4 (3.2) | 0.55 |
| Chronic medical illness | ||||
| Hypertension | 46 (31.7) | 7 (33.3) | 39 (31.5) | 0.86 |
| Diabetes | 13 (9.0) | 3 (14.3) | 10 (8.1) | 0.40 |
| Cardiovascular diseases | 12 (8.3) | 3 (14.3) | 9 (7.3) | 0.38 |
| Respiratory system disease | 6 (4.1) | 1 (4.8) | 5 (4.0) | 1 |
| Other diseases | 33 (22.8) | 5 (23.8) | 28 (22.6) | 1 |
| Blood routine test | ||||
| RBC (3.8-5.1 × 10^12/L) | 4.04 (3.74-4.43) | 4.08 (3.84-4.54) | 4.02 (3.74-4.43) | 0.35 |
| Hemoglobin (115-150 g/L) | 130.3 (118.3-139.9) | 128.8 (116.5-145.5) | 130.3 (118.7-139.4) | 0.87 |
| Platelet (125-350 × 10^9/L) | 204.0 (175.0-254.0) | 184 (169.5-251.0) | 206 (178-255.5) | 0.28 |
| WBC (3.5-9.5 × 10^9/L) | 5.42 (4.38-6.59) | 5.82 (4.58-6.35) | 5.33 (4.26-6.81) | 0.67 |
| Neutrophils (1.8-6.3 × 10^9/L) | 3.16 (2.28-4.28) | 3.16 (2.42-4.19) | 3.16 (3.16-4.35) | 0.78 |
| Lymphocytes (1.1-3.2 × 10^9/L) | 1.45 (1.20-1.89) | 1.55 (1.27-2.05) | 1.44 (1.16-1.84) | 0.13 |
| Monocytes (0.1-0.6 × 10^9/L) | 0.46 (0.38-0.61) | 0.44 (0.35-0.56) | 0.46 (0.38-0.66) | 0.43 |
| Blood biochemistry | ||||
| ALT (9-50 U/L) | 28.0 (17.0-45.0) | 31.0 (17.5-49.5) | 27.5 (17.0-45.0) | 0.70 |
| AST (15-40 U/L) | 23.0 (19.0-33.0) | 22.0 (17.5-27.5) | 24.0 (19.0-33.8) | 0.34 |
| ALP (30-120 U/L) | 85.0 (68.5-104.5) | 84.0 (65.3-95.5) | 86.0 (69.5-106.8) | 0.34 |
| GGT (8-57 U/L) | 30.0 (17.0-52.0) | 24.0 (18.0-49.0) | 32.0 (16.8-60.0) | 0.51 |
| Albumin (40-55 g/L) | 40.0 (36.6-43.4) | 40.1 (38.2-43.1) | 40.0 (36.4-43.4) | 0.56 |
| Globulin (20-30 g/L) | 29.0 (26.3-31.4) | 27.3 (23.4-29.3) | 29.6 (26.4-31.9) | <0.01 |
| TBIL (5-21μmol/L) | 11.4 (9.4-14.7) | 11.5 (11.0-16.4) | 11.4 (9.3-14.7) | 0.39 |
| Creatinine (49-90 μmol/L) | 60.3 (50.3-71.5) | 60.0 (45.1-70.7) | 60.4 (50.5-72.0) | 0.24 |
| Glucose (3.9-6.1 mmol/L) | 5.12 (4.64-5.7) | 5.21 (4.78-6.01) | 5.10 (4.57-5.74) | 0.51 |
| Creatinine kinase (<145 U/L) | 74.0 (53.0-100.0) | 69.0 (56.8-96.5) | 75.0 (51.5-101.0) | 0.89 |
| CKMB (0-25 U/L) | 9.0 (7.0-13.0) | 10.0 (7.5-14.0) | 9.0 (6.5-12.5) | 0.29 |
| LDH (110-245 U/L) | 172.0 (144.0-197.0) | 170 (139.5-188.0) | 174 (146.0-197.0) | 0.50 |
| Potassium ion (3.5-5.3 mmol/L) | 4.29 (4.03-4.58) | 4.20 (4.04-4.35) | 4.30 (4.01-4.59) | 0.27 |
| Sodium ion (137-147 mmol/L) | 139.9 (138.4-141.3) | 140.7 (139.1-141.9) | 139.8 (138.3-141.2) | 0.18 |
| Fibrinogen (238-498 mg/dL) | 379.0 (323.0-425.0) | 349.0 (292.0-394.0) | 380.0 (324.0-435.0) | 0.09 |
| D-dimer (0-500 ng/mL) | 180.0 (100.0-297.0) | 139.0 (98.0-218.0) | 184.0 (100.0-305.0) | 0.43 |
| Blood inflammatory indicators | ||||
| CRP (0-10 mg/L) | 2.3 (1.20-7.60) | 2.3 (1.0-5.0) | 2.2 (1.2-8.9) | 0.38 |
| IL-6 (0.1-2.9 pg/mL) | 2.85 (0.81-5.61) | 1.85 (1.21-4.14) | 3.20 (0.71-6.88) | 0.39 |
| Lymphocyte subsets | ||||
| CD3+ T cells (805-4459/μL) | 1052.0 (789.0-1427.0) | 1261.0 (1001.0-1516.0) | 1040.0 (727.0-1369.0) | 0.02 |
| Percentage of CD3+ T cells (38.56-70.06, %) | 69.21 (61.24-74.39) | 74.14 (69.83-76.38) | 67.99 (60.55-73.15) | <0.01 |
| CD3+CD4+ T cell (345-2350/μL) | 598.0 (426.0-844.0) | 818.0 (652.0-964.0) | 547.5 (390.5-772.5) | <0.001 |
| Percentage of CD4+ T cells (14.21-36.99, %) | 37.83 (32.36-41.78) | 48.40 (41.22-52.13) | 36.79 (31.56-40.65) | <0.001 |
| CD3+CD8+ T cell (345-2350/μL) | 411.0 (306.0-572.0) | 376.0 (296.5-553.0) | 428.0 (302.0-572.0) | 0.55 |
| Percentage of CD8+ T cells (13.24-38.53, %) | 27.56 (22.08-32.51) | 24.14 (16.40-27.96) | 28.17 (22.37-33.45) | 0.02 |
| CD19+ B cell (240-1317/μL) | 197.0 (104.0-279.0) | 239.0 (162.5-302.5) | 169.0 (102.3-277.3) | 0.02 |
| Percentage of CD19+ B cells (10.86-28.03, %) | 12.67 (7.59-16.57) | 14.07 (10.99-16.87) | 12.45 (7.30-16.44) | 0.14 |
| CD16+CD56+ NK cell (210-1514/μL) | 240.0 (162.0-390.0) | 196.0 (153.0-296.5) | 244.0 (160.5-408.5) | 0.18 |
| Percentage ofCD16+CD56+ NK cells (7.92-33.99, %) | 16.25 (10.93-23.64) | 10.93 (8.35-15.65) | 16.99 (11.58-24.70) | <0.01 |
| CD4/CD8 ratio (0.96-2.05) | 1.36 (1.07-1.80) | 2.00 (1.62-3.02) | 1.28 (1.03-1.65) | <0.001 |
| Disease severity | ||||
| Common illness | 130 (89.7) | 19 (90.5) | 111 (89.5) | 1 |
| Serious illness | 14 (9.7) | 2 (9.5) | 12 (9.7) | |
| Critical illness | 1 (0.7) | 0 | 1 (0.8) | |
| Treatment during hospitalization | ||||
| Corticosteroid | 26 (17.9) | 2 (9.5) | 24 (19.4) | 0.37 |
| Antiviral drugs | 111 (76.6) | 16 (76.2) | 95 (76.6) | 1 |
| Antibacterial drugs | 67 (46.2) | 13 (61.9) | 54 (43.5) | 0.12 |
| Chinese medicine | 115 (79.3) | 18 (85.7) | 97 (78.2) | 0.57 |
| Oxygen inhalation | 25 (17.2) | 3 (14.3) | 22 (17.7) | 1 |
| Mechanical ventilation | 1 (0.7) | 0 | 1 (0.8) | 1 |
| Hospital stay (days) | 11.0 (9.0-16.0) | 12.0 (10.5-17.0) | 11.0 (9.0-15.0) | 0.51 |
*ALP = alkaline phosphatase; ALT = alanine amiotransferase; AST = aspartate aminotransferase; CKMB = creatine kinase-MB; CRP = C reactive protein; GGT = glutamyl transpeptadase; IL-6 = interleukin-6; IQR = interquartile range; LDH = lactate dehydrogenase; N = number; RBC = red blood cell; TBIL = total bilirubin; WBC = white blood cell.
Other diseases included immune system diseases, urinary system diseases, neurological diseases, mental illness, liver disease, thyroid diseases, hysteromyoma, lower extremity deep vein thrombosis, thromboangiitis obliterans, lumbar compression fracture, cervical spondylosis, gastritis, and psoriasis.
The collection of human colon tissues
The retrospective analysis indicated that CD4+ T cells might be associated with diarrhea in patients with COVID-19. Therefore, the intestinal mucosa was collected for analysis from patients with COVID-19 with and without diarrhea, patients with IBD, and healthy control patients.
All patients were recruited at the Digestive Endoscopy Center of Zhongnan Hospital between January 2020 and December 2020, and their clinical characteristics are listed in Supplementary Table S1. The diagnosis of COVID-19 was based on the World Health Organization's interim guidance.
Patients who had been diagnosed with COVID-19, with no previous history of diarrhea (>15 days), that presented with diarrhea (including that which persisted past viral infection) considered to be associated with COVID-19 (as opposed to other causes) were included in this part of the study.
The exclusion criteria were as follows:
Patients with incomplete clinical data, endoscopy, imaging and histological results, follow-up information, taking drugs, radiotherapy or chemotherapy before biopsy, with a history of chronic diarrhea such as IBD or celiac disease before COVID-19 infection, with mental illness, pregnancy, infectious disease, and food allergy were excluded from the study.
A total of 41 patients met these criteria and were included in the analysis, and underwent a biopsy. Biopsies were collected using electronic colonoscopies and the 41 biopsy samples were divided into 5 groups, namely ulcerative colitis (n = 8), Crohn's disease (n = 8), COVID-19 diarrhea (n = 6), COVID-19 nondiarrhea (n = 8), and healthy controls (n = 11).
Real-time PCR
According to the guidelines, the human colon tissues were ground, extracted by Trizol reagent (Invitrogen), and centrifuged to obtain total RNA. Reverse transcription of first-strand cDNA was performed according to the manufacturer's instructions (Thermo Fisher Scientific). Gel electrophoresis confirmed that the primers produced only 1 specific band. SYBR Green dye (CoWin BioSciences) was used, and PCR was performed using a LightCycler® 96 instrument (Roche Holding AG). Primers were obtained from TSINGKE Biosciences (Beijing, China), and glyceraldehyde 3-phosphate dehydrogenase was used as the control. Primer sequences used are listed in Supplementary Table S2. Data were quantified using a LightCycler analyzer (Roche Diagnostics) and normalized using the 2−ΔΔCT method.
Western blotting
The human colon tissues were lysed with an ultrasonic crusher, and the cell lysate (which contained protease and phosphatase inhibitors; Servicebio Technology Co, Ltd) was used to split them. After centrifugation, the supernatant was collected and used as the protein extract. The protein was denatured by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis protein loading buffer. An electrophoresis apparatus (Bio-Rad Laboratories) was used to separate the proteins. The protein was transferred to a cellulose nitrate (PVDF) membrane (Millipore Corporation) using a membrane transfer instrument (Bio-Rad). Membranes were blocked with 5% bovine serum albumin at 20°C for 2 hours and incubated with primary antibodies overnight at 4°C. The membranes were incubated with secondary antibodies at 20°C for 1 hour. Finally, the membranes were treated with chemiluminescent detection reagents (Advansta Inc.) and imaged. The optical density of the protein was analyzed using the ImageJ software. All western blotting experiments were conducted in triplicate.
Immunofluorescence
Fresh human biopsy samples were fixed with 4% paraformaldehyde and embedded in paraffin. Tissues were dewaxed and rehydrated with xylene, alcohol, and distilled water. After blocking the tissue with 5% bovine serum albumin for 30 minutes, the sections were incubated overnight at 4°C with the primary antibodies. After incubation and washing, the tissue was incubated with secondary antibodies at 20°C for 1 hour and kept away from light. After washing with phosphate-buffered saline, Vectashield with 4′,6-diamidino-2-phenylindole (DAPI) (Baiqiandu Biotechnology Co.) was used to cover the slides, and the staining results were observed using a fluorescence microscope. A double-blinded method was used in this experiment. The details of all the antibodies are provided in supplementary table S3 and supplementary table S4.
Statistical analysis
All data were analyzed using GraphPad Prism Version 6.0 (GraphPad Software, Inc.). Categorical variables were described as counts and percentages and compared with the chi-square test. Continuous variables with normal distribution were described as the mean ± SD and compared using 1-way ANOVA with a post hoc test (Bonferroni or Tukey). If not, continuous variables were presented as interquartile ranges and compared using the Kruskal-Wallis test with the Nemenyi post hoc test. Spearman's correlation coefficients were used to assess the correlation between the expression of angiotensin-converting enzyme-2 (ACE2) and inflammatory cytokines in patients with COVID-19. A P value <0.05 was considered statistically significant.
Ethical review
All study steps were in line with ethical requirements, and the Ethics Committee of the Zhongnan Hospital of Wuhan University (No. 2021052) approved the study. Written informed consent was waived, and oral consent was obtained from all patients. The clinical courses were followed until July 11, 2021.
Results
Comparisons of clinical characteristics and biomarkers between patients with COVID-19 with and without diarrhea
A total of 145 cases were included in the retrospective study, of which 21 were patients with COVID-19 with diarrhea and 124 were patients with COVID-19 without diarrhea. As listed in Table 1, there was no significant difference between the 2 groups in terms of demographic features, concomitant symptoms, co-morbidities, routine blood test results, blood inflammatory indicators, disease severity, treatment, and hospital stay.
Among the blood biochemistry indices, the diarrhea group showed lower globulin levels than the nondiarrhea group, whereas the other indices were comparable in both groups.
The lymphocyte subsets in the 2 groups showed significant differences. Compared with the nondiarrhea group, the diarrhea group had increased CD3+ and higher CD4+ T cell counts and percentages. In addition, CD8+ T cell counts were lower, and the CD4/CD8 ratio was higher in the diarrhea group than in the nondiarrhea group. Furthermore, the diarrhea group showed higher numbers of CD19+ B cells and lower numbers of CD16+CD56+ natural killer (NK) cells than the nondiarrhea group.
Among these laboratory indicators, CD4+ T cell counts, CD4+ T cell percentage, and the CD4/CD8 ratio showed the most significant differences between the 2 groups. Furthermore, the results for patients with common and serious illnesses were provided separately. The results showed no significant difference in diarrhea between patients with common illnesses and all patients (Supplementary Table S5 and Supplementary Table S6). As there were only 2 patients with severe diarrhea, the number of cases was too small to represent the whole of severe diarrhea, which was considered of no statistical significance.
The mRNA expressions of inflammatory indicators in colonic mucosa
Using real-time PCR, we compared the relative mRNA expression of ACE2, transmembrane serine protease 2 (TMPRSS2), and CD4+ T cell-related cytokines and transcription factors in the colonic mucosa of patients with COVID-19 with and without diarrhea, patients with IBD, and healthy controls.
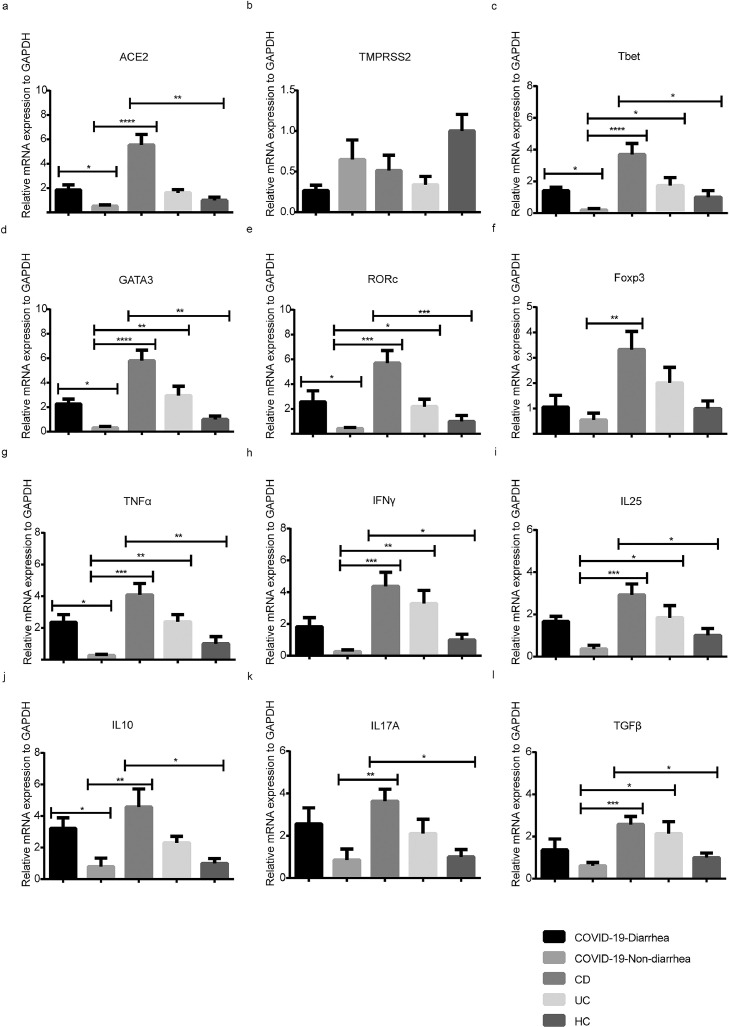
As shown in Figure 1 a, the relative level of ACE2 was significantly different between the diarrhea and nondiarrhea groups (P = 0.049). Compared with ACE2, there were no remarkable changes in TMPRSS2 expression between the 2 groups (Figure 1b).
Figure 1.
Quantitation of mRNA expression of ACE2 and inflammatory cytokines in patients with COVID-19 with and without diarrhea. (a, b) The mRNA expression of ACE2 and TMPRSS2; (c-f) The mRNA expression of CD4+ T cells related transcription factor (Tbet, GATA3, RORc, and Foxp3); (g-l) The mRNA expression of CD4+ T cells related cytokines (TNFα, IFNγ, IL25, IL10, IL17A, and TGFβ). “HC” represented Healthy control. (Kruskal-Wallis test with Nemenyi post hoc test; n = 5-10 per group). Error bars denote the mean ± SEM. ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. ACE2 = angiotensin-converting enzyme 2; Foxp3 = forkhead box protein 3; GATA3 = GATA binding protein 3; IL = interleukin; mRNA = messenger RNA; RORc = retinoic acid receptor-related orphan receptor C; SEM = standard error of the mean; Tbet = T-box expressed in T cells; TGF = transforming growth factor; TMPRSS2 = transmembrane protease, Serine 2; TNFα = tumor necrosis factor-alpha.
Among different CD4+ T cell subsets, T-box expressed in T cells (Tbet) of T helper type (Th) 1 (P = 0.041; Figure 1c), GATA binding protein 3 (GATA3) of Th2 (P = 0.025; Figure 1d), and retinoic acid receptor-related orphan receptor C (RORc) of Th17 (P = 0.047; Figure 1e) in the diarrhea group were observed to have significantly increased expression at the mRNA level, whereas the expression of forkhead box protein 3 (Foxp3) in regulatory T cells (Tregs) (Figure 1f) was not upregulated.
Moreover, the expression of tumor necrosis factor-alpha (TNFα) (P = 0.031; Figure 1g) and interleukin-10 (IL10) (P = 0.018; Figure 1j) in the diarrhea group was higher than that in the nondiarrhea group. These results suggest that diarrhea in patients with COVID-19 is related to ACE2 and the cytokines secreted by Th1, Th2, and Th17 cells at the mRNA level.
The protein expressions of inflammatory indicators in colonic mucosa
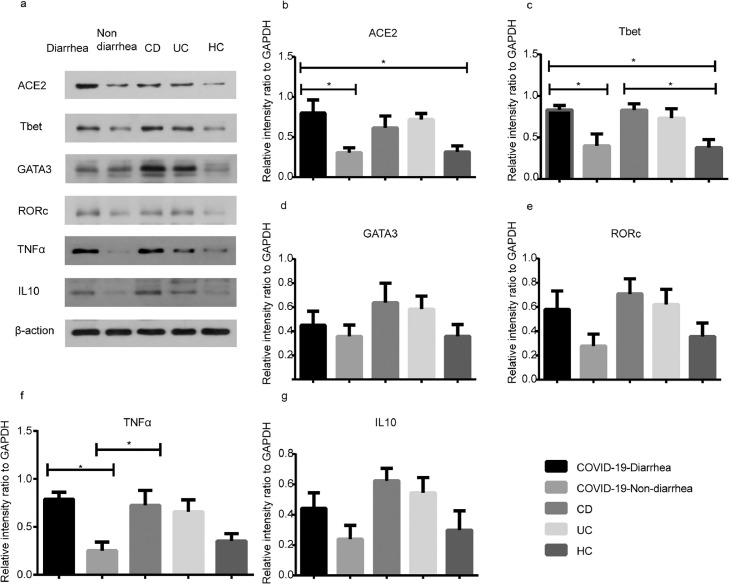
Inflammatory factors with significant differences at the mRNA level were selected for Western Blot testing. The results showed that ACE2 protein levels in the diarrhea group were significantly higher than those in both the nondiarrhea (P = 0.041) and healthy control (HC) groups (P = 0.047; Figure 2 b). When compared with both the nondiarrhea (P = 0.049) and the HC group (P = 0.036), the protein expression of Tbet (Figure 2c) and TNFα (P = 0.018; Figure 2f) was remarkably upregulated in the diarrhea group. The expression of cytokines in other pathways was not significantly increased (Figure 2d; Figure 2e; Figure 2g). These results indicate that the pathogenesis of diarrhea in patients with COVID-19 was related to the ACE2 and Th1 pathways at the protein level.
Figure 2.
The protein expression of ACE2 and CD4 + T cell-related factors in patients with COVID-19 with and without diarrhea. The diarrhea group represented the COVID-19-diarrhea group, whereas the nondiarrhea group represented the COVID-19-nondiarrhea group. (a) The expression of ACE2 and CD4+ T cells related factors; (b-e) Statistics of ACE2 and related transcription factors (Tbet, GATA3, and RORc) expression; (f, g) Statistics of CD4+ T cells’ cytokines expression (TNFα, IL10). “HC” represented Healthy control. (one-way ANOVA test with Tukey post hoc test; n = 5 per group). Error bars denote the mean ± SEM. ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. ACE2 = angiotensin-converting enzyme 2; ANOVA = analysis of variance; GATA3 = GATA binding protein 3; IL = interleukin; RORc = retinoic acid receptor-related orphan receptor C; SEM = standard error of the mean; Tbet = T-box expressed in T cells; TNFα = tumor necrosis factor-alpha.
The results of the correlation analysis performed between ACE2 and CD4+ T cell-related cytokines suggested that ACE2 was moderately correlated with Th1-associated indicators, including Tbet and TNFα, at both mRNA and protein expression levels (r values ≥0.5; P < 0.05; Supplementary Table S7; Supplementary Table S8; Supplementary Table S9; Supplementary Table S10). These results suggested a close correlation between ACE2 and Th1 cells. Therefore, to explore whether ACE2 was directly associated with CD4+ T cell-mediated inflammation, we performed double immunofluorescence of ACE2 and TNFα in the colonic mucosa.
Double immunostaining of ACE2 and TNFα in colonic mucosa
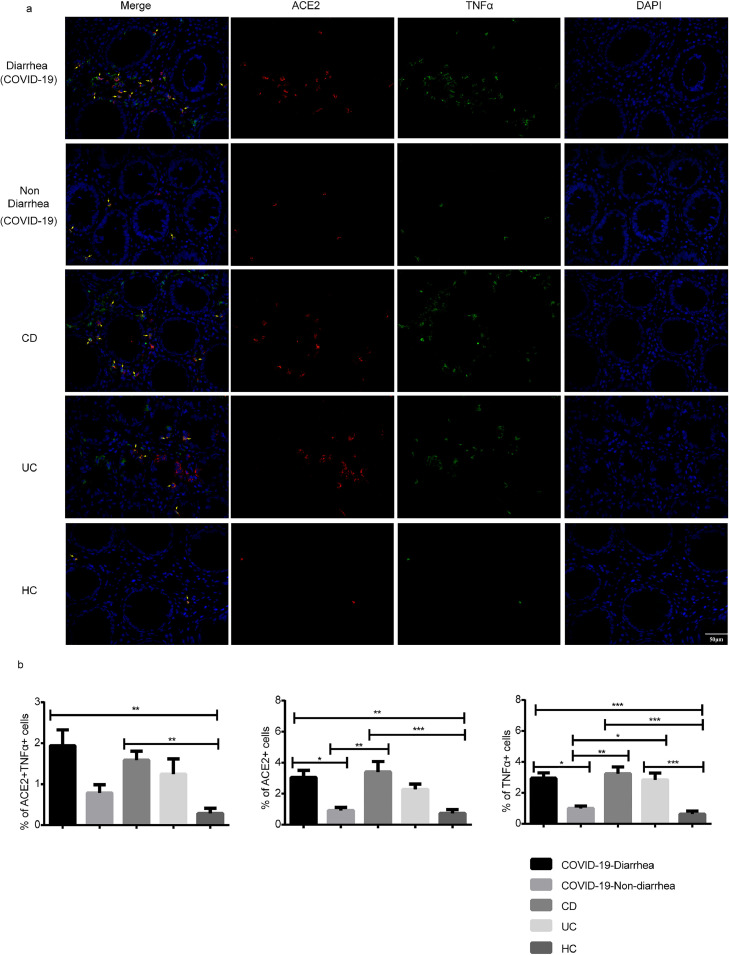
It was observed that ACE2 and TNFα were prominently expressed by lamina propria cells of colonic mucosa from patients with COVID-19 with diarrhea and IBD (Figure 3 ). There were a significantly higher amount of ACE2 positive cells in the diarrhea group than in both the nondiarrhea (P = 0.034) and HC groups (P = 0.004). The number of TNFα-positive cells in the diarrhea group was also higher than that in both the nondiarrhea (P = 0.012) and HC groups (P < 0.001). The number of ACE2+TNFα+ cells in the diarrhea group was significantly higher than that in the HC group (P = 0.001). However, there were no significant differences in ACE2+TNFα cells observed between the diarrhea and nondiarrhea groups (P = 0.090).
Figure 3.
Distribution of ACE2 and TNFα in colons of patients with COVID-19 with and without diarrhea. Representative fluorescence images (a) and quantification of optical density of ACE2 and TNFα (b). Panels from left to right: COVID-19-diarrhea (n = 6), COVID-19-nondiarrhea (n = 6), Crohn's disease (n = 8), ulcerative colitis (n = 8), and HC (n = 11). “HC” represented Healthy control. Scale bar corresponds to 50 μm. ACE2 (red), TNFα (green), and nucleus (DAPI; blue). (1-way ANOVA test with Bonferroni post hoc test; n = 6-11 per group) P < 0.05 was considered statistically significant. Error bars denote the mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. ACE2 = angiotensin-converting enzyme 2; ANOVA = analysis of variance; DAPI = 4′,6-diamidino-2-phenylindole; SEM = standard error of the mean; TNFα = tumor necrosis factor-alpha.
Gene expression of chemokine and corresponding receptors in colonic mucosa
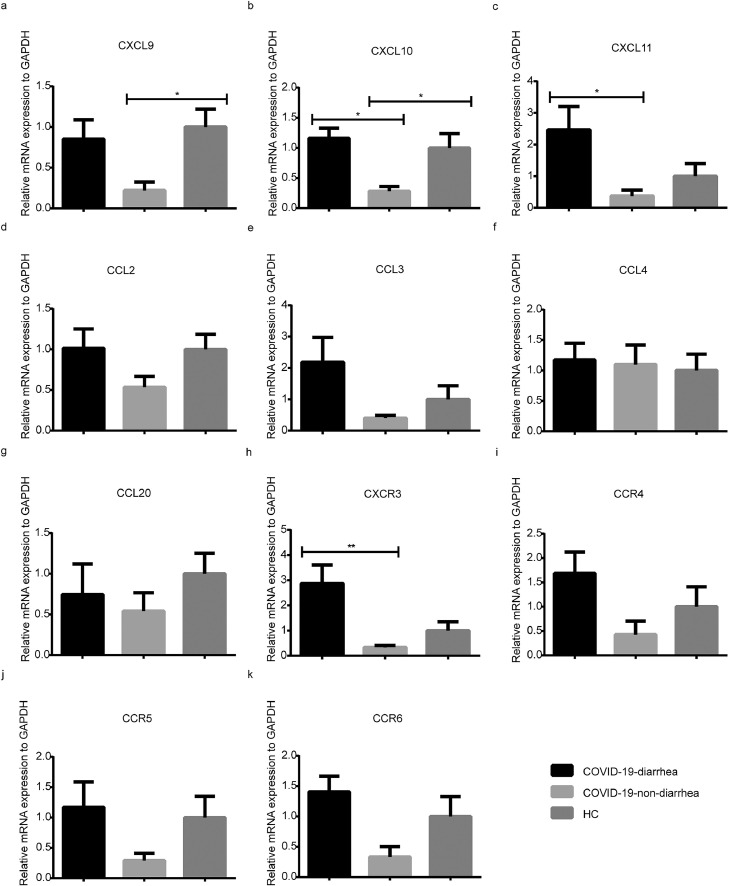
The mRNA expression of chemokine receptors CXCR3, CCR5, CCR4, CCR6, and their ligands (CXCL9, CXCL10, CXCL11, CCL2, CCL3, CCL4, and CCL20) were determined with real-time PCR to assess the chemotactic activity of the CD4+ T cell axis. Total mRNA was extracted from the colon tissues of the patients, and 21 cases were included in the analyses. Compared with the patients with COVID-19 without diarrhea, the mRNA expression of CXCL10 (P = 0.028; Figure 4 b), CXCL11 (P = 0.020; Figure 4c), and CXCR3 (P = 0.004; Figure 4h) in patients with COVID-19 with diarrhea was significantly higher. No significant changes were observed in the expression of other chemokines and their receptors.
Figure 4.
Quantitation of mRNA expression of CD4+ T cells chemokines and receptors in patients with COVID-19 with and without diarrhea. The mRNA expression of chemokines CXCL9(a),CXCL10(b),CXCL11(c), CCL2(d), CCL3(e), CCL4(f), CCL20(g); The mRNA expression of chemokines receptors CXCR3(h), CCR4(i), CCR5(j), CCR6(k). “HC” represented Healthy control. (Kruskal-Wallis test with Nemenyi post hoc test; n = 5-9 per group). Error bars denote the mean ± SEM. ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. mRNA = Messenger RNA; SEM = standard error of the mean.
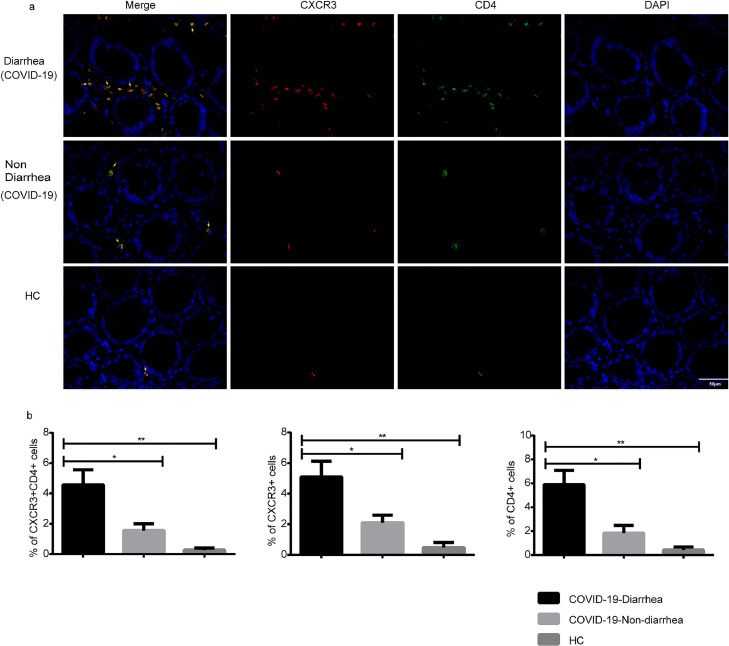
Double immunostaining of CXCR3 and CD4 in colonic mucosa
Finally, we examined the expression and distribution of CXCR3 and CD4 by double IF analysis. Both CXCR3 and CD4 were distributed in the lamina propria of the colon (Figure 5 a). CXCR3+ cells in the diarrhea group showed a significant increase compared with those in both the nondiarrhea (P = 0.027) and HC groups (P = 0.002). The diarrhea group also showed a higher amount of CD4+ T cells than that in the nondiarrhea (P = 0.010) and HC groups (P = 0.001). Moreover, the number of CXCR3+CD4+ cells in the diarrhea group was significantly higher than those in the nondiarrhea (P = 0.017) and HC groups (P = 0.002) (Figure 5b).
Figure 5.
Distribution of CXCR3 and CD4 in colons of patients with COVID-19 with and without diarrhea. Representative fluorescence images (a) and quantification of optical density of CXCR3 and CD4 (b). Panels from left to right: COVID-19-diarrhea (n = 6), COVID-19-nondiarrhea (n = 6), HC (n = 5). “HC” represented Healthy control. Scale bar corresponds to 50 μm. CXCR3 (red), CD4 (green), and nucleus (DAPI; blue). (1-way ANOVA test with Bonferroni post hoc test; n = 5-6 per group) P < 0.05 was considered statistically significant. Error bars denote the mean ± SEM. ns = not significant, * P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. ANOVA = analysis of variance; SEM = standard error of the mean.
Discussion
Although diarrhea may be common in patients with COVID-19, the exact pathogenesis is still unclear. Some studies have detected SARS-CoV-2 RNA in the feces of patients with COVID-19 using rapid real-time PCR (Holshue et al., 2020, van Doorn et al., 2020), and some positive fecal samples remained positive even after respiratory samples tested negative (van Doorn et al., 2020), which indicates the possibility of delayed clearance of SARS-CoV-2 in the GI tract. Another study detected 4 SARS-CoV-2 RNA-positive fecal samples using electron microscopy, and a live virus was observed in 2 specimens from patients who did not have diarrhea (Wang W. et al., 2020). In addition, SARS-CoV-2 RNA can also be detected in biopsies of the esophagus, stomach, duodenum, and rectum through endoscopy (Lin et al., 2020). Further, positive viral nucleocapsid protein expression was identified by histological and immunofluorescent staining in the cytoplasm of gastric, duodenal, and rectal glandular epithelial cells (Xiao et al., 2020), which enhances the evidence for GI infection of SARS-CoV-2. Furthermore, ACE2, which controls intestinal inflammation and diarrhea (Liang et al., 2020), is a viral receptor for the entry of SARS-CoV-2 into human cells (Lan et al., 2020). Different studies have found that ACE2 is highly expressed in digestive organs, except the stomach (Liang et al., 2020, Zhang et al., 2020, Zou et al., 2020), suggesting an increased susceptibility of digestive systems to SARS-CoV-2 infection and potential risk of GI symptoms such as diarrhea after infection. Based on this evidence, we speculated that the direct cytopathic effects of SARS-CoV-2 might play an important role in the pathogenesis of COVID-19 associated diarrhea.
Both systemic and local inflammation in the GI tract have been observed in patients with COVID-19, and many studies have found that hyperinflammation in peripheral blood is associated with disease severity and mortality (Carter et al., 2020, Gustine and Jones, 2021). For local inflammation, numerous infiltrating lymphocytes and plasma cells with interstitial edema were observed in the lamina propria of the stomach, duodenum, and rectum biopsies obtained from patients with COVID-19 through endoscopy (Xiao et al., 2020). To seek clues related to diarrhea in patients with COVID-19, we performed a case-control analysis based on the symptom of ‘diarrhea’ in patients with COVID-19. Our study revealed significant differences in lymphocyte subsets and globulin levels between patients with COVID-19 with and without diarrhea, with no significance for all other indicators, suggesting that immune factors might play a role in the occurrence of diarrhea in COVID-19. In lymphocyte subset detection, patients with diarrhea had different patterns of CD3+, CD4+, and CD8+ T cell percentages, CD19+ B cell counts, CD16+CD56+ NK cell percentages, and CD4/CD8 ratios. In view of the most significant differences observed in CD4+ count, percentage, and CD4/CD8 ratio between groups, and the change patterns of other subsets, we speculated that CD4+ T cells are a key factor associated with diarrhea in patients with COVID-19.
To clarify the role of CD4+ T cell-mediated inflammation in COVID-19 associated diarrhea, we collected colonic mucosa from patients with COVID-19 with and without diarrhea, taking IBD patients as positive controls and healthy subjects as negative controls. CD4+ T cells are believed to play an important role in both innate and adaptive immune responses (Ruterbusch et al., 2020), and have been identified as the main drivers of IBD and players perpetuating intestinal inflammation (Imam et al., 2018).
ACE2 is a viral receptor for the entry of SARS-CoV-2 that is highly expressed in the intestine. Thus, the detection of ACE2 expression in the intestine reflects the invasion of SARS-CoV-2 into the intestine. TMPRSS2 is highly expressed in lung, heart, prostate, kidney, and intestinal tissues (Comperat et al., 2020; Hamming et al., 2004; Thunders and Delahunt, 2020). SARS-CoV-2 uses ACE2 for entry and TMPRSS2 for S protein priming (Hoffmann et al., 2020), and as a host cell factor, TMPRSS2 is essential for SARS-CoV-2 transmission (Hoffmann et al., 2020). Therefore, TMPRSS2 is usually detected together with ACE2 as evidence of SARS-CoV-2 infection. CD4+ T cell-related factors to be detected include specific transcription factors and inflammatory effector cytokines that interact with ACE2 to facilitate viral entry and activation (Baughn et al., 2020). The expression of inflammatory factors in the intestinal tract reflects the occurrence of a CD4+ T cell-mediated immune response. Chemokines play an important role in the priming, differentiation, and regulation of specific effector T cell subsets after acute pathogen challenge (Eberlein et al., 2020). Thus, specific transcription factors of different CD4+ T cell subsets, such as Tbet of Th1, GATA3 of Th2, RORc of Th17, and Foxp3 of Tregs (Jin et al., 2018, Ruterbusch et al., 2020), and inflammatory effector cytokines such as TNFα, interferon γ, IL10, IL17, IL25, and transforming growth factor β were detected at both the mRNA and protein levels, with detection of ACE2 and TMPRSS2.
Our results revealed that patients with COVID-19 with diarrhea had a higher expression of ACE2, Tbet, and TNFα at both mRNA and protein levels than those patients without diarrhea and HCs (Figure 1 and Figure 2), and a higher expression of GATA3, RORc, and IL10 at the mRNA level. High expression of these transcription factors and cytokines indicated that CD4+ T cells, especially Th 1 cells, might participate in the modulation of intestinal inflammation in patients with COVID-19 with diarrhea. In addition, the high expression of ACE2 mRNA and protein indicates the possibility of direct infection of SARS-CoV-2 in the intestinal mucosa. Since there was a close association between ACE2 and SARS-CoV-2 infection, we performed correlation analyses for ACE2 and other indicators to further investigate the potential relevance of SARS-CoV-2 infection in the intestine and CD4+ mediated local inflammation. The results indicated that the expression levels of ACE2 were positively correlated with those of CD4+ T cell-associated indicators, especially with moderate correlations between ACE2 and Th1-associated indicators, including Tbet and TNFα, at both mRNA and protein expression levels (r values ≥0.5; P < 0.05; Supplementary Table S7; Supplementary Table S8; Supplementary Table S9; Supplementary Table S10).
On the basis of the previously mentioned findings, we speculated that the close association between ACE2 and CD4+ T cells might be explained by 2 possibilities: 1) SARS-CoV-2 directly infects CD4+ T cells in the intestine, resulting in the occurrence of CD4+ T cell-mediated immune responses; or 2) SARS-CoV-2 infection occurs in the intestinal mucosa, which led to local intestinal inflammation and involvement of CD4+ T cells to perpetuate intestinal inflammation. Although ACE2 is rarely expressed in immune cells (Berthelot et al., 2020), a study has shown that immune cells, including CD4+ and CD8+ T cells, monocytes, and B cells, are susceptible to SARS-CoV-2 infection (Borsa and Mazet, 2020). An immunofluorescence assay of dual-labeling for ACE2 and TNFα was conducted to exclude the direct association of ACE2 with CD4+ T cell-mediated inflammation. The results revealed that the percentages of ACE2+ cells and TNFα+ cells in the diarrhea group were higher than those in the nondiarrhea group, although no statistical difference in the percentage of ACE2+TNFα+ cells between the 2 groups was observed (Figure 3), indicating an indirect association between ACE2 and TNFα secretion that usually originates from CD4+ T cells.
Chemokines play an important role in the priming, differentiation, and regulation of specific effector T cell subsets after acute pathogen challenge (Eberlein et al., 2020). Thus, the expression of chemokines and their corresponding receptors was measured at the mRNA level, and it was found that patients with COVID-19 with diarrhea had higher expression levels of CXCL10, CXCL11, and CXCR3 than those without diarrhea (Figure 4). Interestingly, CXCL10 and CXCL11 are both ligands for CXCR3, which was found to be expressed on differentiated Th1 cells to modulate their migration into the gut in a murine colitis model (Wadwa et al., 2016). Furthermore, to determine the potential role of CXCR3-expressing CD4+ T cells COVID-19 associated diarrhea in more detail, we analyzed the presence of CD4+CXCR3+ T cells in biopsy samples from the gut of patients with COVID-19 with and without diarrhea and healthy subjects. The level of CD4+CXCR3+ T cells was increased in the diarrhea group (Figure 5), suggesting that the migration of CD4+CXCR3+ effector T cells to the site of inflammation might promote intestinal inflammation and lead to diarrhea during COVID-19 infection.
In summary, our findings showed that both enhanced systemic and local inflammation in the GI tract mediated by CD4 + T cells were identified in patients with COVID-19 with diarrhea and that the occurrence and persistence of inflammation in the gut might be closely associated with SARS-CoV-2 infection. CXCR3+-mediated migration of CD4+ T cells into the gut may contribute to perpetuating intestinal inflammation. Future investigations may be performed to elucidate more detailed mechanisms of COVID-19 associated diarrhea in animal models and in vitro to clarify the potential therapeutic benefits of targeting CXCR3.
Acknowledgments
Competing interests
There are no conflicts of interest to be declared.
Funding
This work was supported by the National Natural Science Fund of China (grant number: 81870390). The funding sources had no sponsors.
Acknowledgments
We thank all the patients included in this study.
Contributors
Qiu Zhao and Xiaobing Wang designed this study. Zhu and RuiZhou collected the epidemiological and clinical data. Xiaobing Wang and Jun Xiao collected the mucosa samples of the included patients. Feng Ding, Liuqing Ge, Ruiping Zhu, and Liping Chen participated in the data collection. Jia Wei performed the detections as described in the manuscript. Xiaobing Wang and Jia Wei drafted the manuscript, and Qiu Zhao revised the final manuscript.
Xiaobing Wang and Jia Wei contributed equally to this paper.
All authors listed have approved the manuscript that is enclosed.
Ethics statement
This study was reviewed and approved by the ethics committee of Zhongnan Hospital of Wuhan University (No. 2021052). This study was an observational study. Informed verbal consent was obtained from all patients in our study as the ethics committee waived written consent.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.04.006.
Appendix. Supplementary materials
References
- AlSamman M, Caggiula A, Ganguli S, Misak M, Pourmand A. Non-respiratory presentations of COVID-19, a clinical review. The American journal of emergency medicine. 2020;38(11):2444–2454. doi: 10.1016/j.ajem.2020.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clinic proceedings. 2020;95(9):1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthelot JM, Liote F, Maugars Y, Sibilia J. Lymphocyte Changes in Severe COVID-19: Delayed Over-Activation of STING? Frontiers in immunology. 2020;11 doi: 10.3389/fimmu.2020.607069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsa M, Mazet JM. Attacking the defence: SARS-CoV-2 can infect immune cells. Nature reviews Immunology. 2020;20(10):592. doi: 10.1038/s41577-020-00439-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MJ, Fish M, Jennings A, Doores KJ, Wellman P, Seow J, et al. Peripheral immunophenotypes in children with multisystem inflammatory syndrome associated with SARS-CoV-2 infection. Nature medicine. 2020;26(11):1701–1707. doi: 10.1038/s41591-020-1054-6. [DOI] [PubMed] [Google Scholar]
- Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung KS, Hung IFN, Chan PPY, Lung KC, Tso E, Liu R, et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology. 2020;159(1):81–95. doi: 10.1053/j.gastro.2020.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comperat E, Wasinger G, Oszwald A, Kain R, Cancel-Tassin G, Cussenot O. The Genetic Complexity of Prostate Cancer. Genes. 2020;11(12) doi: 10.3390/genes11121396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira GLV, Oliveira CNS, Pinzan CF, de Salis LVV, Cardoso CRB. Microbiota Modulation of the Gut-Lung Axis in COVID-19. Frontiers in immunology. 2021;12 doi: 10.3389/fimmu.2021.635471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberlein J, Davenport B, Nguyen TT, Victorino F, Jhun K, van der Heide V, et al. Chemokine Signatures of Pathogen-Specific T Cells I: Effector T Cells. Journal of immunology. 2020;205(8):2169–2187. doi: 10.4049/jimmunol.2000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmunzer BJ, Spitzer RL, Foster LD, Merchant AA, Howard EF, Patel VA, et al. Digestive Manifestations in Patients Hospitalized With Coronavirus Disease 2019. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2021;19(7):1355–1365. doi: 10.1016/j.cgh.2020.09.041. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. The New England journal of medicine. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Tao W, Flavell RA, Zhu S. Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nature reviews Gastroenterology & hepatology. 2021;18(4):269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, et al. Extrapulmonary manifestations of COVID-19. Nature medicine. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PubMed] [Google Scholar]
- Gustine JN, Jones D. Immunopathology of Hyperinflammation in COVID-19. The American journal of pathology. 2021;191(1):4–17. doi: 10.1016/j.ajpath.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of pathology. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, et al. First Case of 2019 Novel Coronavirus in the United States. The New England journal of medicine. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam T, Park S, Kaplan MH, Olson MR. Effector T Helper Cell Subsets in Inflammatory Bowel Diseases. Frontiers in immunology. 2018;9:1212. doi: 10.3389/fimmu.2018.01212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Singh R, Ha SE, Zogg H, Park PJ, Ro S. Pathophysiological mechanisms underlying gastrointestinal symptoms in patients with COVID-19. World journal of gastroenterology. 2021;27(19):2341–2352. doi: 10.3748/wjg.v27.i19.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Chen H, Li Y, Zhong H, Sun W, Wang J, et al. Maresin 1 improves the Treg/Th17 imbalance in rheumatoid arthritis through miR-21. Annals of the rheumatic diseases. 2018;77(11):1644–1652. doi: 10.1136/annrheumdis-2018-213511. [DOI] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin Z, et al. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut. 2020;69(6):1141–1143. doi: 10.1136/gutjnl-2020-320832. [DOI] [PubMed] [Google Scholar]
- Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69(6):997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- Ma C, Cong Y, Zhang H. COVID-19 and the Digestive System. The American journal of gastroenterology. 2020;115(7):1003–1006. doi: 10.14309/ajg.0000000000000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisetti A, Gajendran M, Goyal H. Putative Mechanisms of Diarrhea in COVID-19. Clinical gastroenterology and hepatology: the official clinical practice journal of the American Gastroenterological Association. 2020;18(13):3054–3055. doi: 10.1016/j.cgh.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruterbusch M, Pruner KB, Shehata L, In Pepper M. Vivo CD4(+) T Cell Differentiation and Function: Revisiting the Th1/Th2 Paradigm. Annual review of immunology. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2) Journal of clinical pathology. 2020;73(12):773–776. doi: 10.1136/jclinpath-2020-206987. [DOI] [PubMed] [Google Scholar]
- van Doorn AS, Meijer B, Frampton CMA, Barclay ML, de Boer NKH. Systematic review with meta-analysis: SARS-CoV-2 stool testing and the potential for faecal-oral transmission. Alimentary pharmacology & therapeutics. 2020;52(8):1276–1288. doi: 10.1111/apt.16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadwa M, Klopfleisch R, Adamczyk A, Frede A, Pastille E, Mahnke K, et al. IL-10 downregulates CXCR3 expression on Th1 cells and interferes with their migration to intestinal inflammatory sites. Mucosal immunology. 2016;9(5):1263–1277. doi: 10.1038/mi.2015.132. [DOI] [PubMed] [Google Scholar]
- Wan Y, Li J, Shen L, Zou Y, Hou L, Zhu L, et al. Enteric involvement in hospitalised patients with COVID-19 outside Wuhan. The lancet Gastroenterology & hepatology. 2020;5(6):534–535. doi: 10.1016/S2468-1253(20)30118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. Jama. 2020;323(18):1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831. doi: 10.1053/j.gastro.2020.02.055. -3 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XW, Wu XX, Jiang XG, Xu KJ, Ying LJ, Ma CL, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS-Cov-2) outside of Wuhan, China: retrospective case series. Bmj. 2020;368:m606. doi: 10.1136/bmj.m606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Jiang X, Wang X, Liu B, Ding H, Jiang M, et al. CCL28 mucosal expression in SARS-CoV-2-infected patients with diarrhea in relation to disease severity. The Journal of infection. 2021;82(1):e19–e21. doi: 10.1016/j.jinf.2020.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li HB, Lyu JR, Lei XM, Li W, Wu G, et al. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. International journal of infectious diseases: IJID: official publication of the International Society for Infectious Diseases. 2020;96:19–24. doi: 10.1016/j.ijid.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Han C, Zhang S, Duan C, Shang H, Bai T, et al. Diarrhea and altered inflammatory cytokine pattern in severe coronavirus disease 2019: Impact on disease course and in-hospital mortality. Journal of gastroenterology and hepatology. 2021;36(2):421–429. doi: 10.1111/jgh.15166. [DOI] [PubMed] [Google Scholar]
- Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181(5):1016. doi: 10.1016/j.cell.2020.04.035. -35 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Frontiers of medicine. 2020;14(2):185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.