Abstract
Several Artificial Intelligence-based models have been developed for COVID-19 disease diagnosis. In spite of the promise of artificial intelligence, there are very few models which bridge the gap between traditional human-centered diagnosis and the potential future of machine-centered disease diagnosis. Under the concept of human-computer interaction design, this study proposes a new explainable artificial intelligence method that exploits graph analysis for feature visualization and optimization for the purpose of COVID-19 diagnosis from blood test samples. In this developed model, an explainable decision forest classifier is employed to COVID-19 classification based on routinely available patient blood test data. The approach enables the clinician to use the decision tree and feature visualization to guide the explainability and interpretability of the prediction model. By utilizing this novel feature selection phase, the proposed diagnosis model will not only improve diagnosis accuracy but decrease the execution time as well.
Keywords: Explainable artificial intelligence, Human-computer interaction, Disease diagnosis, COVID-19, Decision forest, Feature selection
1. Introduction
In the first four months following the outbreak, the pandemic disease caused by the SARS-CoV-2 virus called COVID-19 has infected between 3 and 5 million people and caused at least 200,000 deaths in more than 200 countries of the world. As a result of the outbreak of COVID-19, governments throughout the world have taken drastic measures like quarantining hundreds of millions of residents [1,2].
Coronavirus is still a worldwide health concern; by 1st March 2022, there had been 438 million positive cases and 5.9 million deaths [3]. Among the essential factors contributing to the increase in deaths caused by COVID-19 infection, one shall mention social disparities in accessing to early diagnosis tests, and shortage of hospital equipment for clinical critically cases.
Currently, more than two years after the COVID-19 pandemic onset, a number of vaccines have been developed, and the vaccination procedure is proceeding at a promising but heterogenous pace between countries. While developed countries are more likely to have access to vaccines, other countries face multiple obstacles to vaccination, such as not having enough vaccine doses to protect vulnerable groups. Additionally, there are no confirmed medications to cure patients infected people. As a result, it remains important to screen patients suspected of being infected with COVID-19.
A primary and trustworthy diagnosis of positive COVID-19 patients is essential to prevent and limit of its prevalence [4]. Reverse transcription polymerase chain reaction, referred RT-PCR, is currently the gold standard for COVID-19 screening and is uniquely recommended by World Health Organization (WHO); but it has main defects as well: delay on turnaround times [5], a deficiency of reagents [6], suffers from a low sensitivity (60–71%) [7], longer waiting time for the results [6,8], a high false-negative rate of 15–20% [6], the need for certified laboratories [6], costly equipment [9], and requiring specialist staff [6]. For these reasons, scientists are looking for alternative faster, more accessible, and affordable diagnosis techniques.
Impressive improvements in machine learning models are rejuvenating the application of Artificial Intelligence (AI) in healthcare that basically started over half a century ago [10]. In the field of COVID-19 disease diagnosis, the utilization of chest X-ray and CT-scan imaging is already populated in many developing countries such as India, Africa, South- America due to insufficient number of RT-PCR test-kits, and the established link between the ground-glass opacity occurrence in the periphery of lungs and SARS-Cov-2 [[11], [12], [13], [14], [15]], although with some limited success [16]. At limited scale, cough sound analysis has been suggested to discriminate COVID-19 patients [17]. Finally, laboratory data including blood test have also been advocated in Ref. [18] because of the identified correlation between parameters such as white blood cells, neutrophils, lymphocytes, basophils, monocytes and others with COVID-19 patients [6]. In essence, the prospect of prognostic biomarkers toward earlier and more targeted treatment has been recognized, especially since some patients with COVID-19 develop intense status, which is associated with a higher risk of hospitalization [19].
Many of the above developments are to a large extent attributed to the development in Artificial Intelligence and availability of relevant large scale clinical dataset related to COVID-19 patients. Indeed, the improvements in computer systems and data storage technologies have substantially increased the accumulation of COVID-19 data, which offers physicians and researchers a unique opportunity to explore simultaneously factors influencing patient diagnosis and comprehend various types of COVID-19, as well as developing new early testing technology for COVID-19 detection. Although, handling such large-scale datasets raises extra challenge of designing effective data processing and analysis, which are computationally appealing, theoretically sound, and easily interpretable [20,21]. Nevertheless, AI is already utilized in COVID-19 decision support systems to aid physicians make diagnosis and prognosis decisions as pointed out in review papers in Refs. [[22], [23], [24]]. Especially, AI can i) improve the quality of physicians' decision-making through augmented visualization tools and expert system like (aid) to decision-making systems; ii) decrease the risk of physicians’ tiredness caused by the overload number of consultations and their criticality, and iii) reduce the problem of simultaneous availability of various clinicians [[25], [26], [27]].
Furthermore, the interpretability and explainability issues relating to the modern AI-based tools should be taken into account, since these could further impede their implementation in healthcare applications. Within machine learning techniques, deep-learning approaches, due to their ability to automatically extract representations from the learning data that are relevant to their predictions, achieved state-of-the-art results in many areas, e.g., computer vision [28,29], speech recognition [30,31], and signal processing [32,33]. This motivated researchers to extend such approach to COVID-19 detection and prediction where it is commonly it is acknowledged that complex machine-learning models such as deep learning and XGBoost perform better than simple models such as logistic regression in the COVID-19 diagnosis [16,34]. Despite their acknowledged performance, the widespread employ of deep learning models is halted by the capacity of such models to provide explanations to their findings, in a way to promote transparency, responsibility and ethical considerations when comprehending the outcomes of such models, especially in the light of the new data protection EU directive on the “right of explanation” [35]. This demand is even more stressed in healthcare sector where any diagnostic error can have fatal consequences on patient life. This raises the need to equip the complex deep-learning black-box models with explanatory modules to accommodate this new need. For example, a physician should understand why a machine learning model provides a given diagnosis and be able to explain it to the patients. This explains why most of deep-learning models implemented in healthcare field rather act as protypes and for aid to decision-support only, offering the possibility to the clinician to bypass the model output or seeking an alternative measurement strategy prior to make clinical decision. This renders several physicians reticent to utilize machine learning and artificial intelligence-based models that are not straightly explainable, interpretable and reliable. Nevertheless, enforcing explainability and transparency in deep-learning models often comes at the expense of increased time complexity and, sometimes, even system accuracy. How to balance accuracy, explainability, and other factors of artificial intelligence in medical applications remains a challenge today. As a result, not only it is necessary to develop complex and efficient model to process such COVID-19 medical data, but also to be able to explain and interpret their decisions. Research in eXplainable AI (XAI) [[36], [37], [38]] aims to provides tools and method for explaining deep-learning models through model approximation, enhanced feature contribution visualization, rule-based generation, among others, enabling either local or global explanation of the model outcome.
In this medical context, XAI framework proposes generating a series of machine learning models that 1) Generate more explainable techniques while preserving a high rate of diagnosis accuracy, and 2) Provide a model for physicians to explain, understand, trust, and effectively manage decisions.
This paper contributes to XAI research in medical context by proposing a new Explainable Random Forest (FSXRF) method that exploits social network graph analysis for feature visualization and optimization for the purpose of COVID-19 detection from blood test samples. The proposed model includes four principal steps. First, the original features of COVID-19 dataset are shown as a graph where each feature is indicated by a node and the links show the similarities between the corresponding features. In the next step, a novel scoring mechanism is proposed for feature importance calculation. The aim of this step is to rank different features based on filtering feature weighting. In the next step, an iterative search mechanism is proposed to choose final features. Therefore, the proposed features selection mechanism while removing redundant features, will also eliminate irrelevant features with the label of COVID-19 dataset. After selecting the final features, in the fourth step, the ensemble Decision Forests classifier is employed to COVID-19 screening in routine blood tests. The proposed strategy has several innovations compared with the previous intelligent COVID-19 prediction approaches:
-
1
In contrast to relatively demanding RT-PCR method, this study uses blood tests, which are faster, more accessible, and less expensive than PCR testing. Therefore, blood tests can potentially provide an alternative tool for the rapid diagnosis of infected cases and compensate for the lack of RT-PCR and CT scan by serving as an early detection tool.
-
2
An explainable artificial intelligence decision system based on Decision Tree (DT) that can support physicians in the COVID-19 diagnosis with a number of simple and explainable rules is developed and put forward.
-
3
Unlike black-box deep learning-based COVID-19 diagnosis models, which are difficult to explain to physicians, the proposed prediction model is based on DT that physicians can trust due to its acknowledged explainability and transparency.
-
4
Many of previous prediction model for COVID-19 diagnosis use a single classifier for final prediction, which reduces their generalization capabilities. In contrast, our model uses a novel Ensemble Learning-based prediction model, which offered increased prediction accuracy.
-
5
Although, previous explainable machine learning models focused on sample-wise (local) explanations, our method focuses on explaining the entire dataset (global explanation) via a single model. In this study, an individual explanation in a graph representation is provided that shows the relative importance of each feature and their interactions.
-
6
The developed approach uses a novel graph mining strategy to find similar features and discard redundant feature, which automatically comprehends the number of relevant feature unlike other clustering methods such as k-means [39] and fuzzy clustering [40] where some prior-knowledge is required.
-
7
Our model uses a novel graph-based technique to measure feature score and feature similarities, while traditional models only measure feature relevance in their feature selection procedure.
-
8
The developed model employs a social network-based technique and the node centrality measure to propose a heuristic search method. In comparison with nature-inspired methods such as [41], the proposed method is enough fast and more accurate and can be applied to medical dataset. The proposed method calculates feature similarities and then applies a scoring mechanism to allocate an importance weight to each feature. Therefore, the developed method satisfies both objectives of feature relevance and feature redundancy in a multi-objective function. Unlike other multi-objective models that choose a set of non-dominated feature in their optimization phase [42,43], the developed search mechanism seeks the optimal feature set in a reasonable amount of time.
The rest of this paper is structured as below: Section 2 reviews the previously artificial intelligence-based model for COVID-19 disease diagnosis as well as discusses the concept of explainable artificial intelligence. The proposed diagnosis model is detailed in Section 3. The experimental results on COVID-19 dataset are described in Section 4 and finally, Section 5 explain the conclusion and future works.
2. Background
2.1. Machine learning for Covid-19 detection
Artificial intelligence-based models are the promising approaches employed to aid physicians in the early screening of COVID-19 positive cases. Moreover, these models decrease the workload of the physicians, increase the accuracy prediction, gives a timely response and precise treatment for the COVID-19 positive cases. Artificial intelligence-based models are used to prevent and mitigate COVID-19 pandemics by screening, identifying viruses, and disease diagnosis, repurposing or repositioning drugs, and predicting and forecasting their future spread. In the area research of medical prediction of COVID-19, intelligent and machine learning-based models grounded on biomarkers can help optimize the screening of patients with severe disease, minimizing mortality and hospitalization, and decreasing care delays [44].
Deep learning and machine learning are the two major branches of artificial intelligence. The following subsections discuss the applications of machine learning and deep learning models to combat and mitigate the COVID-19 outbreak. Fig. 1 demonstrates the schematic diagram of artificial intelligence approaches for related COVID-19 outbreak tasks.
Fig. 1.
Schematic diagram of artificial intelligence approaches for related COVID-19 outbreak tasks.
These approaches were promising areas of research and development for the decision-making process related to COVID-19 and many studies are performed to review them as extensively reviewed in several review papers, e.g., Refs. [16,45]. We therefore focus herein on works that tackle the issue of feature selection and optimization in machine learning for COVID-19 detection and prediction, which seems to be overlooked in previous reviews.
For the early prediction and diagnosis of COVID-19 positive, the authors of [46] proposed a method based on SVM classifier utilizing X-ray patient data. There are 40 lungs X-ray images in this dataset, 15 of which are normal lung images and the remaining 25 are COVID-19 infected chest X-ray images. The developed method has high efficiency (sensitivity = 95.76%, specificity = 99.7%, and accuracy = 97.48%), indicating the SVM-based method can be utilized efficiently for the diagnosis of COVID-19 cases.
During the past few years, Decision Tree's reputation has increased in the medical research and health sector. For example in Ref. [47], a model-based decision tree is proposed for the severity identification of COVID-19 in children. They obtained reports on 105 children who were infected between February 1 and March 3 of 2020 from the Chinese hospital. There were 105 positive children among the 105, including 41 girls and 64 boys. The developed method has high performance.
Too et al. [48] presented a new feature selection method using Hyper Learning Binary Dragonfly Algorithm search strategy for predicting the condition of COVID-19 patient with a decreasing number of selected features with high performance accuracy.
Using time-dependent parameters, the authors of [49] proposed a novel approach for forecasting the dynamic spread of COVID-19. Their approach advocates an epidemiologic model in time domain to develop the nonlinear model for dynamic approximation of COVID-19 prevalence.
Using an improved fuzzy clustering algorithm, a novel time series forecasting method is developed in Ref. [50] for the upcoming COVID-19 patients and deaths in India. Essentially, this technique consists of two steps. In the first step, an improved fuzzy clustering algorithm is used to create initial intervals, and then these initial intervals are updated in the second step in order to create new sub-intervals. This developed technique was evaluated using available COVID-19 and the results demonstrated that this method was superior to previous methods in terms of mean square error, root mean square error, and average forecasting error rate.
In [51], different artificial intelligence-based techniques for prediction of COVID-19 positivity and severity where K nearest neighbor classifier, Neural Networks, Decision Tree and Partial Least Squares Discriminant Analysis techniques are compared. Their experimental results demonstrated that COVID-19 severity can be diagnosed and predicted using all these classifiers with acceptable accuracy.
In [52], a machine learning-based model is proposed for future intubation prediction among positive COVID-19 cases where the model forecasts future probability of intubation based on prior vitals, laboratory, and demographic patient information. The model uses a supervised prediction technique that employs a sliding-window technique to predict the possibility of intubation 72 h after the end of the 24-h sampling period.
Pahar et al. [53] uses AI model to classify COVID-19 cough using smartphone audio recordings where several machine learning models are compared. The authors showed that the residual neural networks classification model can differentiate between positive coughs and healthy coughs with an area under the ROC curve of 0.98.
In [54], three machine learning models are proposed to forecast the likelihood of prolonged length of stay utilizing electronic health record data from COVID-19 patients and to help hospital systems prepare for bed capacity needs.
Zhang et al. [55] analyzed the clinical features and outcome of different positive COVID-19 where nine mortality factors are identified utilizing a least absolute shrinkage and selection operator regression technique that are then tested by an artificial neural network algorithm.
In [56], classic machine learning-based classification model for Sentiment Analysis on 72,000 COVID-19 related tweets is evaluated where several models are compared. The authors show that SVC, Perceptron, Passive Aggressive Classifier, and Logistic Regression can achieve higher than 98% prediction rate in Sentiment Analysis.
Singh et al. [57] examined the performance of transfer learning technique for intelligent prediction of COVID-19. In this presented model, a deep learning-based approach is developed for COVID-19 CT image screening. This approach utilized VGG16 and PCA for feature extraction and feature selection from CT scan data, respectively. Additionally, four classification models are evaluated in the prediction phase, including Convolutional neural networks, Extreme Learning, online sequential Extreme Learning Machine, and Bagging Ensemble with SVM. Finding of this paper indicated that the bagging ensemble and SVM had the highest prediction accuracy in the experiments.
In [58], a novel Joint Classification and Segmentation (JCS) model was developed for real-time and explainable COVID- 19 diagnosis using chest CT images.
Yang et al. [59] proposed a new model for analyzing clinical characteristics and predicting death outcomes in severe COVID-19 patients. The authors developed a clinically useful and easily interpretable DT-based model to help clinicians rapidly identify COVID-19 patients with high mortality risks.
Using human respiratory sounds such as voice, dry cough, and breath, Lella and Pja [60] introduced a deep learning-based method to diagnose COVID-19 disease. The method employs multi feature channels to extract deep features from the patient data, which are fed to a Deep Convolutional Neural Network for final disease diagnosis after an initial preprocessing.
In [61], a depth-wise deep learning method was proposed to reorganize of COVID-19 affected lungs regions.
Roy et al. [62] investigated the application of deep learning-based model in the lung ultrasonography (LUS) images analysis of COVID-19 patients where a new deep learning technique, extracted from Spatial Transformer Networks, that diagnoses the patient status intensity, was put forward.
In [63], Convolutional Neural Networks (CNN)-based techniques were employed for deep feature extraction using chest X-ray and CT images. Then, these features are sent to transfer learning-based approach to diagnosis positive COVID-19 cases.
In recent years, many researchers have suggested Long Short-Term Memory (LSTM) networks for COVID-19 detection, diagnosis, classification, prediction, and forecasting. In Ref. [64], a deep learning-based LSTM method was developed for COVID-19 prediction utilizing X-ray image data. In this method, Convolutional Neural Networks was trained to select the deep features and based on these selected features, the deep model was trained for the final prediction of COVID-19.
In [65], another CNN-based method was proposed to detect the COVID-19 positive cases based on X-ray images. This dataset includes X-ray images of 135 COVID-19 patient and 320 from viral and bacterial pneumonia cases. The reported result indicated that this developed method achieved an accuracy of 89.2%.
Moreover Ahmadian et al. [66] developed a novel two-phase improved deep neuroevolution model to COVID-19 diagnosis from chest X-ray data. The deep neuroevolution algorithm developed in this paper is tested on a real-world dataset, and its performance was indicated by comparing different evaluation metrics.
In Table 1 , a selection of the main previous machine learning models employed for COVID-19 pandemic related tasks and their techniques, tasks, data types, accuracy and explainability are detailed. For Explainability categorization, we distinguished High Explainability models (e.g., Decision Tree, Random Forest), Medium Explainability models (e.g., KNN, Joint Classification) and Low Explainability models, which include Deep Learning, Neural network and other similar black box models.
Table 1.
Outlining the reviewed machine learning-based models in COVID-19 pandemic related tasks.
| Paper | Technique | Task | Data type | Accuracy | Explainability |
|---|---|---|---|---|---|
| Mahdy et al. [46] | SVM | Covid-19 lung image classification | X-ray image | High | Low |
| Yu et al. [47], | Decision Tree | Severity detection of COVID-19 paediatric cases | Chest radiography and CT images | Medium | High |
| Too and Mirjalili [48]. | KNN | Prediction of the death and recovery conditions | The patients' information (Gender, Age, Country, etc.) and their symptoms | Medium | Medium |
| Song et al. [49] | Time-dependent model parameters. | forecasting the dynamic spread of COVID-19 | Daily reported cases in China and the United States | High | Low |
| Kumar and Kumar [50] | Fuzzy clustering and time series model | Prediction of COVID-19 infected cases and deaths | Daily reported cases in India | Medium | Low |
| Cobre et al. [51] | KNN, Neural Networks, Partial Least Squares Discriminant Analysis, etc. | Diagnosis and prediction of COVID-19 severity | Biochemical, hematological, and urinary biomarkers | Medium | Low |
| Arvind et al. [52] | Sliding-window approach | Prediction of intubation among hospitalized patients | laboratory and vitals data COVID-19+ patients | Medium | Low |
| Pahar et al. [53] | Residual neural networks | Classification of COVID-19 cough | Coughing sounds recorded during or after the acute phase of COVID-19 | Medium | Low |
| Ebinger et al. [54] | Logistic regression, SVM, KNN, etc. | Prediction of duration of hospitalization in COVID-19 patients | Electronic health record data from COVID-19 patients | Medium | Low |
| Zhang et al. [55] | Least absolute shrinkage and selection operator regression and least absolute shrinkage and selection operator neural network models. | Identification and validation of prognostic factors in COVID-19 patients | Demographic data including, clinical data including and outcome (28-day mortality) | Medium | Low |
| Gulati et al. [56] | Linear SVC, Perceptron, Passive Aggressive, Logistic Regression, etc. | Sentiment classification of discussion related to COVID-19 pandemic | Tweets related to COVID-19 pandemic | Medium | Low |
| Singh et al. [57] | Ensemble Support Vector Machine | COVID-19 detection | Lung tomography scan data | High | Low |
| Wu et al. [58] | Joint Classification and Segmentation | COVID-19 diagnosis | Chest CT images | Medium | Medium |
| Yang et al. [59] | Decision Tree | Death outcome prediction | Medical records (demographics, clinical characteristics, and laboratory test results) | Medium | High |
| Lella and Pja [60] | Deep Convolutional Neural Network | Diagnosis of COVID-19 disease | Human respiratory sounds such as voice, dry cough, and breath, | High | Low |
| Qayyum et al. [61] | Depth-wise deep learning | Detection and diagnosis of COVID-19 infection | Lungs X-rays images | High | Low |
| Roy et al. [62] | Spatial Transformer Networks-based Deep learning | Classification and Localization of COVID-19 Markers | Lung ultrasonography (LUS) images. | High | Low |
| Shamsi et al. [63] | Deep transfer learning | Diagnosis of COVID-19 | Chest X-ray and CT images | High | Low |
| Islam et al. [64] | Deep Convolutional Neural Network and LSTM | Detection of COVID-19 | X-ray images | High | Low |
| Hall et al. [65] | Deep Convolutional Neural Network | Detection of COVID-19 | Chest x-rays | High | Low |
| Ahmadian et al. [66] | Deep Neuroevolution | Diagnosis of COVID-19 | Chest x-rays | High | Low |
In overall, we noticed that in the diagnosis of COVID-19, complex machine-learning models such as deep learning perform better than simple models such as linear regression and decision trees. Nevertheless, the deep learning-based approaches proposed in previous works were indeed black boxes that did not explain their prediction in a manner a human could understand [[67], [68], [69], [70]]. It is therefore important to endow the highly performing deep-learning models with explainability and interpretability ability to accommodate the new EU data protection directive and ensure their widespread adoption by healthcare authorities. Moreover, in the next subsection previous XAI-based model are reviewed.
2.2. Explainable artificial intelligence
The lack of explainability and transparency of AI-based methods in medical environments is one of their major limitations. In many healthcare applications, it is necessary to know how the prediction model made a specific decision, allowing the healthcare stakeholders (e.g., physicians, specialists, patients, researchers and public) to trust the model. In healthcare domain, questions such as “What makes this prediction trustworthy?” or “How did this intelligent model achieve this result?” need to be responded for specialists and physicians to entirely embrace the application of artificial intelligence-based model in assisting them with early diagnosis. It is crucial that every model should also be able to provide a rationale for the diagnosis or recommendation it made. Although some prediction techniques like decision trees are transparent, the vast majority of artificial intelligence applications in medicine using deep learning techniques are black box in essence and have therefore no explanation for their prediction. This has led to the creation of several explainable AI methods in the past few years [[70], [71], [72]]. Accordingly, a new research area called Explainable AI aims to increase the explainability of black box models.
Explainable AI refers to AI and machine learning approaches that can provide human-understandable explanation for their models' behavior. XAI is a rapidly growing research area that is aimed at providing a justifiable, transparent, interpretable, trustable, and traceable intelligent model [73].
Explainable AI model can focus on several types of explanations. These types of explanations can be classified according to their scope, origin, and application. Depending on the scope of the explanations, they can be either global or local. While global explanations attempt to explain the whole model at once, local explanations focus on a small area around a specific sample. Explanations may have intrinsic origins or be post-hoc. An explanation is inherent when the ML model is transparent and can be understood due to its simple structure (for example, Linear Regression or Decision Tree). Conversely, post-hoc explanation techniques attempt to obtain explanations from trained models. Moreover, an explanation artificial intelligence can be model-agnostic if it applies to different learning algorithms meeting several requirements, or it can be model-specific if it is crafted for a particular artificial intelligence model.
As opposed to previous explainable artificial intelligence models that analyzed local and post-hoc explanations, our method explains the entire dataset (global explanation) through a single model. We propose a global XAI model for generating predication methods that increase the users' trust in the diagnosis. Moreover, visual representations of the entire model provide a global explanation for the developed model. In the next section the detail of developed explainable artificial intelligence-based model for COVID-19 disease diagnosis are described.
3. Proposed XAI-based model for COVID-19 diagnosis
In this section, our explainable AI-model for COVID-19 diagnosis is developed by combining Feature Selection with Explainable Random Forest (FSXRF). The developed FSXRF method is grouped as a model explanation of the RF outcomes that i) calculates the nature of the dependency between different features of COVID-19 dataset, ii) ranks the importance of each feature, iii) discards redundant features to optimize the feature-space and reduce burden complexity and, iv) visualizes the various dependency in a way to ease explanation with clinicians by providing a decision tree like analysis.
The conceptual framework of the developed model is donated in Fig. 2 . The developed XAI COVID-19 diagnosis model focuses on explaining the entire procedure of generating the prediction model and the result of the developed model is provided by a combination of the rules, numerical and visual information.
Fig. 2.
Explainable artificial intelligence approach for COVID-19 diagnosis.
The developed FSXRF achieves both explainability and feature optimization, which are known to enhance attractiveness in medical diagnosis. Indeed, irrelevant and redundant features in medical dataset have presented serious challenges to the existing artificial intelligence-based prediction model, impacting accuracy and prediction [[74], [75], [76], [77]]. Irrelevant and redundant features also increase the probability of overfitting and increase the computational complexity [[78], [79], [80], [81]]. As a result, our model adds a feature selection phase to the main phase of prediction for eliminating the redundant and irrelevant features.
Moreover, Fig. 3 shows the overall flow diagram of the proposed FSXRF model. In overall, FSXRF consist of four main steps: (1) Graph representation of COVID-19 features, (2) Ranking COVID-19 features, (3) Identifying the final feature set and (4) Final COVID-19 diagnosis using Explainable Random Forest and considering the selected features. The aim of the first step is to represent the features of COVID-19 diagnosis problem as a network graph where each node corresponds to a given feature of COVID-19 dataset, and the edges demonstrates the feature similarities. In the next step of the FSXRF, all the original features of COVID-19 dataset are ranked by utilizing a filter-based feature weighting measure. The aim of the next step is to score different factors of COVID-19 diagnosis by employing a feature ranking technique. In the third step, to select non-redundant and relevant features, those of high scores and dissimilar feature are chosen using a novel feature selection strategy, while the remaining features are removed. Finally, in fourth step, an Explainable Random Forest-based classifier is used to diagnosis COVID-19 cases considering the selected features on the previous steps.
Fig. 3.
Flowchart of the proposed model.
This developed explainable prediction model has two main phases: the feature section (i.e., 1–3 steps) and the developed explainable random forest predication model (i.e., 4 step). In the first phase, the features of COVID-19 data are illustrated by a graph and a set of relevant and non-redundant of initial features is selected for final diagnosis phase, and then in the second phase, a novel approach to increase the interpretability of the random forest-based predictions is used and an effective artificial intelligence-based predictor for COVID-19 disease diagnosis is developed using routine blood tests. In the reminder of this section the details of these phases are described. Moreover, the nomenclature and parameters of the developed prediction model are provided in Table 2 .
Table 2.
Nomenclature and parameters of the developed COVID-19 diagnosis model.
| Symbol | Description |
|---|---|
| Feature node | |
| Feature vector | |
| Feature graph | |
| Link between original features | |
| Number of initial features | |
| Similarity between features and | |
| Feature Vector | |
| Average of feature vector | |
| Set of dataset samples | |
| Average of all the calculated similarities | |
| Variance of all the calculated similarities | |
| Importance Value of feature | |
| Fisher Score of features | |
| Node Centrality of feature | |
| Set of all classes in a dataset (i.e. Positive and Negative) | |
| Number of patterns on the class | |
| Average of feature vector on class | |
| Variance of feature vector on class | |
| Laplacian Energy | |
| Average value of all the samples related to the feature | |
| Set of all classes in a dataset (i.e. Positive and Negative) | |
| Number of samples on the class | |
| Variance and average of feature on class | |
| Average of feature on class | |
| Decision tree | |
| Set of rules in Decision tree | |
| Number of trees in decision forest |
3.1. Graph presentation
To apply the proposed feature selection method, the feature of the COVID-19 data should be shown using a weighted graph. For this aim, the initial features are illustrated with a graph , where is a set of initial features in which each feature corresponds to a node in the graph, shows the set of edges of the graph, and denotes the similarity between two features and that are connected by the edge . In this paper, Pearson similarity criteria [82] is used for the feature similarities calculation. The similarity between the two features and is computed as follows:
| (1) |
where and denote the vectors of features and for all samples, respectively. Variables and indicate the average of vectors and , over all of the COVID-19 samples (i.e. set), respectively. If these two features are very similar, the Pearson criterion will be close to one, while if these two features are very dissimilar, the Pearson criterion will be close to zero. After similarity calculation, SoftMax normalization [83] is used to scale these values into a unit interval as below:
| (2) |
where, is the similarity value between features and , and are the average and variance for all calculated similarities, respectively, and shows the normalized similarity between features and .
This similarity measure maps the feature space of a COVID-19 data into a fully weighted and connected graph. To make the graph sparser, the edges with associated weights lower than some threshold value are removed. is an adjustable parameter that takes values in the unit interval [0 1]. When value is small (resp. large), more (resp. fewer) edges will be considered in the next steps.
3.2. Feature scoring
The main goal of this step is to score the initial features of COVID-19 dataset using a filter-based feature selection measure. In the proposed model, weights are assigned to features in each cluster according to a scoring mechanism. Therefore, removing both irrelevant and redundant features is achieved by the proposed method. In fact, the number of the high important and dissimilar features are selected, while the reminder features are removed. During this step, the Fisher Score (FS) and the Node Centrality (NC) are integrated to determine the score of each feature. The Feature Importance (FI) of -the feature, i.e., is measured as below:
| (3) |
where and denote the Fisher Score and Node Centrality of feature , respectively. Also, shows the number of initial features. In this study, Fisher Score (FS) feature weighting measure is utilized for feature ranking. Fisher Score is used to identify the features that are most relevant to the target class. The Fisher Score scores feature according to their predictive and discriminatory power. Accordingly, this criterion assigns a higher value for features with higher separation characteristics. The Fisher Score of feature is calculated as below:
| (4) |
where is the mean value of all the samples regarding the feature , is a set of all classes in the COVID-19 dataset (i.e. positive and negative), is the number of samples on the class , and and indicates the variance and average of feature on class , respectively.
Furthermore, in our developed feature selection method, as opposed to previous models [[84], [85], [86]] where only feature relevance is employed to select final features, a subset of features with high importance and relevancy will be chosen. In this developed feature selection method, the centrality of nodes is employed to calculate the influential features of the dataset. In the analysis of social networks, identifying the more influential or “central” nodes has been an important challenge [87,88]. In many areas of social network analysis, detecting influential and more central nodes has been used to characterize network properties. Our model employs Laplacian Centrality (LC) [88] for node centrality calculation.
3.3. Identifying the final feature set
In most of the previous proposals for eliminating redundant features, only feature relevance has been used, but in this paper, we evaluate the correlation between features by integrating average similarity and node centrality. Specifically, all features are sorted according to their Feature Importance (FI) scores. First, feature with the highest FI score is added to the selected feature set as the first representative of original features. Then, the next feature with the highest FI is considered as the candidate feature, and the average similarity of this feature with the previously selected features is calculated using the Pearson similarity criterion. If the similarity of the candidate feature with one of the previously selected features was greater than the value of threshold , this feature is removed from the original feature graph , and the next feature with high importance value from the initial features is considered as the next candidate feature. This process continues until all the features have been checked and the reminding features in the graph are sent to Random Forest-based COVID-19 diagnosis.
3.4. Final COVID-19 diagnosis
In this subsection the details of the fourth step of developed method are described. A decision tree is a prediction algorithm that performs a set of test conjunctions where each test evaluates a feature score with a threshold value or a set of feasible values to decide whether to maintain or discard the underlined feature. In order to divide the dataset into disjoint subsets, test nodes are created starting from the root node. The recursive process repeats itself until no further division is necessary. Since each leaf corresponds to a combination of features, it is easy to interpret local decisions. These capabilities make DT widely employed for different applications that require a comprehension of both the model construction and its prediction. Although decision tree-based prediction models are highly interpretable, these intelligent decision-making models have limited prediction performance due to the nearsightedness characteristic of their induction models [89,90]. When complex interactions exist among input features, DT models usually fail to capture these, leading to essential biases. To deal with this issue, a decision forest, or ensemble of decision trees, is adopted in our approach.
Decision Forest (DF) is a powerful ensemble learning algorithm for integrating the results of several machine learning algorithms into a single decision.
Several factors motivate this choice. First, the risk of trapping local minima is reduced when several predictors are integrated. Further, when only a small amount of data is available, a single algorithm can choose an incorrect hypothesis, which provides additional ability to handle small-size data scenarios. Finally, the combination of different classifiers may also result in a wider search space, specifically in problems where the optimal hypothesis lies away from individual models.
In this work, decision forests that combine multiple decision trees towards providing a single decision is employed for final COVID-19 diagnose.
A new technique to convert a DF into a single DT is developed in this study. Based on the original decision forest, the resulting decision tree approximates its prediction accuracy and provides more explainable and faster classification. A tree decision model was chosen to be the outputted model since it can be explained both in terms of its graphic prediction structure as well as its separability. As compared to previous prediction models, the developed model can be applied to all sizes of forests and does not need complicated hyperparameter setting.
Suppose each datum , where is the total number of samples, is represented in a feature space of dimension . Let be the output indicating the class label of the datum . Then, given the decision set , a DT classifier is defined as , where denotes the used features in tree, contains a set of rules and is an input sample. Moreover, a DF classifier is defined as , where corresponds to the number of trees in the DF.
In our proposed method, the are limited to neighboring nodes in graph . Through this regularization, features that are functionally related are placed on the same Decision Tree. Therefore, the generated classifier using this tree is more trustable and explainable for physicians, and simultaneously is more generalizable. Therefore, the developed DF is defined as , where is a set of features specified using random walk on graph . In our proposed method, a greedy-based scenario is developed to transform a DF into an explainable DT. For each greedy step the performance for all , DT is evaluated (see Line 22 of Algorithm 1). Here, the accuracy measure is utilized for evaluation and if the accuracy of -th DT in iteration is lower than the accuracy of the DT in the prior iteration, the suggested DT and its corresponding nodes are eliminated. On the other hand, if DT has a higher accuracy, a random walk on a subgraph, determined using the features in in the iteration , is initialized. It should be noted that the depth of this walk has now reduced by one. Finally, after updating the DTs, a new set of trees are sorted considering their accuracy in order to initialize a selection procedure. These repetitions continue until a specified number of iterations is reached.
Fig. 4 indicates the pseudo-code of the developed COVID-19 diagnosis model.
Fig. 4.
Flow pseudo-code of the developed model.
4. Experimental results
In this section, our experimental setup for COVID-19 disease diagnosis is highlighted and the results are reported. The efficiency of the presented model is compared with some well-known prediction model including XGBoost [91], SVM, MLP together with the state-of-the-art COVID-19 prediction model that aims to propose understandable approach based on eXplainable Decision Trees (XDT) [92] for COVID-19 diagnosis. The results are evaluated using a set of criteria: Accuracy, F1–score, Sensitivity, Specificity and AUROC.
To ensure a more accurate and trustworthy validation, a 10-fold validation test is conducted. For this purpose, at each iteration, one set is considered as a test data while other sets were considered as train data. Then, we ran the experiment 30 times. Moreover, in all experiments both average and standard deviation values are recorded.
For fair experiments, different models should be evaluated on the same training, validation and testing dataset. These experiments report the standard deviation of the accuracy in ten independent runs together with the average accuracy since train and test samples are randomly separated.
In the reminder of this section, the detail of the used COVID-19 data, experimental results, sensitivity analysis, statistical analysis, and discussion are explained.
4.1. Dataset
In this work, we used public COVID-19 dataset [93] to demonstrate the effectiveness and robustness of the developed COVID-19 diagnosis model.1 This dataset includes unknown data from patients who present COVID-19 symptoms and requested to accomplish the SARS-CoV-2 RT-PCR and supplementary tests during their stay at the hospital. This COVID-19 dataset included 5644 patients and 111 features (includes 69 Decimal features, 37 String features and 5 Universally unique identifier features) associated with blood tests (e.g. Red blood Cells, Red blood cell distribution width, venous blood gas analysis, lymphocytes, Mean corpuscular hemoglobin concentration, Urea, Proteina C reativa, Creatinine, Potassium, Sodium, etc.), urine (e.g. Esterase, Aspect, Hemoglobin, Ketone Bodies, Density, Protein, Leukocytes, Red blood cells, Granular cylinders, etc.), and tests for the presence of other viruses (e.g. Influenza A, Influenza B, Parainfluenza 1, etc.). During the hospital visit, RT-PCR and DNA sequencing are used to diagnose Covid-19 positive cases. The dataset demonstrates the complexity of decision making during real healthcare problems, compared to what happens in more theoretical experiments. As a result data sparsity is to be expected. The dataset demonstrates the complexity of decision making during real healthcare problems, compared to what happens in more theoretical experiments. As a result data sparsity is to be expected.
Since this dataset contains features with missing values, to handle these missing data in our experiments, we replaced each missing datum with the mean of the available data on the feature set [94].
4.2. Experimental results
In first experiments, the performance of the developed prediction model is evaluated over COVID-19 dataset. Table 3 summarizes the average Accuracy, F1–score, Sensitivity, Specificity and AUROC over ten separate and autonomous runs of the different prediction model (i.e. XGBoost, SVM, MLP and XDT). In this table, the best average values are marked in boldface. The reported results of Table 3 show that in all cases the developed prediction model performs better than the other COVID-19 disease diagnosis models. For example, the reported results of this Table reveals that the average classification accuracy of the developed approach data was 89.97%, which is 1.56% higher than the average classification accuracy for the second-ranked method (i.e., XDT). Moreover, Table 3 shows that the developed model superior to the second-best model (i.e., XDT model) with a difference of 1.96%, 4.44%, 1.97% and 2.19% for F1-Score, Sensitivity, Specificity and AUROC measures, respectively. Moreover, the boxplot of 10-fold validation of these independent runs is shown in Fig. 5 .
Table 3.
Average performance, standard deviation (shown in parenthesis) and p-value of different predications model based on 10-fold validation in 30 independent runs.
| Method | Accuracy | F1-Score | Sensitivity | Specificity | AUROC |
|---|---|---|---|---|---|
| XGBoost | 87.71 (1.34) | 71.45 (1.42) | 67.52 (1.38) | 90.82 (1.26) | 89.36 (1.24) |
| SVM | 84.79 (1.31) | 72.48(1.71) | 67.01 (1.36) | 88.96 (0.67) | 87.69 (1.31) |
| MLP | 85.25 (1.29) | 69.92 (1.37) | 62.21 (1.03) | 88.17 (1.43) | 88.38 (1.36) |
| XDT | 88.41 (0.59) | 76.17 (0.67) | 67.21 (0.82) | 91.02 (0.69) | 90.62 (1.03) |
| Proposed Model | 89.97 (1.08) | 78.13 (1.21) | 71.65 (0.76) | 92.99 (0.82) | 92.81 (1.06) |
| P-value | 0.0034218 | 0.0037548 | 0.003295 | 0.004606 | 0.004438 |
Fig. 5.
Boxplot of 10-fold validation in 30 independent runs.
Moreover, based on 30 independent executions, Table 4 shows the number of times the best performance was achieved by different prediction models. The values of this table show that in most cases the developed COVID-19 diagnosis model achieved the highest performance compared to the other diagnosis models on the different measures.
Table 4.
Number of times the best results are achieved by different prediction models in 30 independent runs.
| Method | Accuracy | F1-Score | Sensitivity | Specificity | AUROC |
|---|---|---|---|---|---|
| XGBoost | 2 | 1 | 1 | 2 | 2 |
| SVM | 0 | 1 | 0 | 0 | 1 |
| MLP | 1 | 0 | 1 | 1 | 0 |
| XDT | 2 | 1 | 1 | 1 | 2 |
| Proposed Model | 25 | 27 | 27 | 26 | 25 |
Moreover, in Table 5 , the normalized confusion matrices per class are investigated. In this Table different COVID-19 diagnosis model are compared in terms of True-Negative (The actual class is Negative and predicted class is Negative), True-Positive, False-Negative, False-Positive. As it can be seen from this Table, the proposed model had the highest performance. The results of this Table show that the differences between the obtained performance of the proposed prediction model in term of True-Negative and the second-best ones (XDT) and third-best ones (XGBoost) are calculated 1.61 (i.e., 92.99–91.38) and 2.28 (i.e., 92.99–90.71), respectively. Furthermore, based on reported result of Table 5 and for the True-Positive (The actual class is Positive and predicted class is Positive) criterion, the developed model gained the first rank with an average True-Positive of 71.98, and the XDT and XGBoost prediction model were ranked second and third with an average True-Positive of 68.81 and 65.57, respectively. Moreover, in terms of False-Negative and False-Positive, respectively. Given that these two criteria are calculated based on false predictions, the lowness of this criterion indicates the superiority of that method. The reported results show that in both criteria, the developed prediction model was more accurate than the other COVID-19 diagnosis models.
Table 5.
Normalized confusion matrices for different COVID-19 diagnosis model.
| Method | True-Negative | True-Positive | False-Negative | False-Positive |
|---|---|---|---|---|
| XGBoost | 92.18 | 66.87 | 8.71 | 34.71 |
| SVM | 89.79 | 67.36 | 11.27 | 37.21 |
| MLP | 90.71 | 65.57 | 10.01 | 38.37 |
| XDT | 91.38 | 69.81 | 9.75 | 32.56 |
| Proposed Model | 92.99 | 71.98 | 8.12 | 30.72 |
Moreover, Fig. 6 displays the average ROC curve acquired by the developed model. Based on changing the decision threshold, the curve is calculated for both 1-Specificity (False Positive Rate) and sensitivity (True Positive Rate). With a model that is close to 1, its discrimination capability is greater in the prediction test. These reported results indicated that, with a sensitivity of 0.826 and specificity of 0.802, the ROC curve had a maximum average sensitivity and specificity.
Fig. 6.
The ROC Curve for the developed model.
As explained earlier, one of the main parts of the proposed method is feature selection phase, which prevents the selection of redundant and irrelevant features. Typically, a large portion of this COVID-19 data is irrelevant or redundant, decreasing the predictability of the model. Therefore, the performance of the prediction model is significantly influenced by feature selection. In this study, an efficient feature selection method has been proposed utilizing the feature similarity and node centrality techniques. This mechanism identifies a subset of dissimilar features that have the highest correlation with the target class of diseases. In this developed feature selection model, in feature selection phase, 21 features are selected, and the reminder features are eliminated. These features are listed in Table 6 .
Table 6.
The selected features sorted based on their importance.
| Number | Feature |
|---|---|
| 1 | PLT |
| 2 | EOS |
| 3 | MPV |
| 4 | CRP |
| 5 | AST |
| 6 | CREAT |
| 7 | WBC |
| 8 | MONO |
| 9 | LYM |
| 10 | RBC |
| 11 | NEU |
| 12 | NA |
| 13 | ALT |
| 14 | HCT |
| 15 | HGB |
| 16 | RWD |
| 17 | UREA |
| 18 | K+ |
| 19 | MCV |
| 20 | MCH |
| 21 | MCHC |
It should be noted that the features of this listed are sorted based on their importance values (FIs). Moreover, in these selected features the features of PLT, EOS, MPV, CRP, AST, CREAT, WBC, MONO, LYM and AST obtained the highest importance value compared to others features.
Moreover in Fig. 7 part of the final extracted tree for explainable COVID-19 diagnosis is shown. Considering this route in the derived decision tree, the features of PLT, MPV, EOS, WBC, LYM, ALT and HGB can be used for final COVID-19 disease diagnosis. Based on these extracted features and generated rules, Decision Tree explanation is as follows:
Fig. 7.
Part of explainable tree for COVID-19 diagnosis.
if (PLT ≤ 0.10) and (MPV > −1.02) and (EOS ≤ −0.66) and (WBC ≤-0.52) and (LYM > −1.11) and (ALT > −0.51) and (HGB > 0.96) then the COVID-19 diagnosis is positive.
4.3. Comparison with other feature selection methods
In this subsection, the performance of the proposed feature selection method is evaluated. The performance of the developed feature selection method is compared with four well-known feature selection methods including Fisher Score (FS) [86], Laplacian Score [95], Relevance-Redundancy Feature Selection (RRFS) [96] and Minimal-Redundancy–Maximal-Relevance (MRMR) as well as four state-of-the-art feature selection methods including Five-way Joint Mutual Information (FJMI) [97], Adaptive Hypergraph Embedded Dictionary Learning (AHEDL) [98], Artificial Bee Colony Algorithm based on Dominance (ABCD) [99] and Multi-objective PSO (MPSO) [79] methods. In this experiment, in the feature selection phase, the method proposed in each paper is used and for a fair comparison, in classification phase, for all of them, a common classifier is used.
Fig. 8 shows the average classification accuracy of different feature selection methods on various classifiers in 30 independent runs. The reported results of this figure indicated that the performance of the developed model is higher than those from all other feature selection models. As an example, on SVM classifier, the classification accuracy of FS, LS, RRFS, MRMR, FJMI, AHEDL, ABCD and MPSO feature selection methods are 75.17%, 73.19%, 83.81%, 84.93%, 86.17%, 85.19%, 86.71% and 88.09%, respectively. However, the classification accuracy of proposed feature selection method yields 89.96% accuracy.
Fig. 8.
Average classification accuracy of different feature selection methods on various classifiers.
In the next experiment, different feature selection methods are compared in term of execution times. In these experiments, corresponding execution times (in ms) for each feature selection method are shown in Fig. 9 . Due to the fact that the feature selection phase and the final prediction phase are separate, only the execution time for feature selection phase is calculated in this figure. It can be seen from the recorded data that generally the single variate feature selection approaches (FS and LS) are much faster than the multivariate feature selection approaches (i.e., RRFS, MRMR, FJMI, AHEDL, ABCD, MPSO and proposed method). This is because univariate methods do not consider the possible dependency between features in feature selection, therefore, they are computationally less costly than multivariate approaches. It should be noted that compared to multivariate feature selection approaches, univariate ones are less accurate since they ignore feature dependencies, as demonstrated in Fig. 8. Moreover, the reported results revealed that the between the state-of-the-art feature selection approaches, the proposed feature selection has the lowest average execution time.
Fig. 9.
Average execution time (in ms) of different feature selection approaches over 30 independent runs.
4.4. Comparison with other explainable RF-based models
RFs integrate multiple DTs towards providing a single output in supervised prediction duties. RFs have gained popularity among data scientists due to their ability to combine different hypotheses into a single model and their effectiveness in dealing with any type of relational dataset. Every prediction model that is made by a RF must go over a wide variety of trees. Consequently, the end-user does not have a clear understanding of the model's predictions. Additionally, the model structure is practically made up of numerous single models, which makes it difficult for the end user to comprehend. Several researchers, developed models to transform a RF into a single DT. In this subsection, the performance of the proposed method for transforming a RF to a single DT is compared with four state-of-the-art methods. The details of these methods are explained in Table 7 . In this experiment, in the feature selection phase, for a fair comparison a common feature selection method (the proposed method in this paper) is used. and in transformation phase (transform a RF to a single DT), the method proposed in each paper is employed. Table 8 reports the average performance of different transforming techniques on in 30 independent runs. The reported results of this table indicated that the performance of proposed transforming technique is higher than those from all other transforming techniques. As an example, the classification accuracy of Counterfactual Sets [100], Rule Conjunctions [90], Construction and Filtering of Conjunction Sets [89] and Explainable Matrix–Visualization [101] methods are 87.81%, 84.79%, 85.12%, and 88.19%, respectively. However, the classification accuracy of proposed transforming technique yields 89.53%.
Table 7.
Characteristics of comparative transforming techniques.
Table 8.
Average performance and standard deviation (shown in parenthesis) of transforming techniques.
| Method | Accuracy | F1-Score | Sensitivity | Specificity |
|---|---|---|---|---|
| Counterfactual Sets | 87.81 (1.24) | 72.32 (2.31) | 68.51 (2.41) | 91.86 (1.93) |
| Rule Conjunctions | 84.79 (2.31) | 75.31(1.72) | 69.51 (3.31) | 88.82 (3.12) |
| Filtering of Conjunction Sets | 85.12 (3.27) | 69.17 (3.12) | 51.15 (1.28) | 89.13 (2.84) |
| Matrix–Visualization | 88.19 (2.51) | 76.13 (2.13) | 68.21 (2.81) | 91.54 (1.71) |
| Proposed Method | 89.53 (1.12) | 78.21 (1.32) | 71.38 (2.69) | 92.79 (1.76) |
| p-value | 0.0034784 | 0.0037691 | 0.0039877 | 0.004897 |
4.5. Sensitivity analysis of the parameters
The developed COVID-19 prediction model has two parameters and , where their corresponding optimum values must be justified by the user.
The value is a threshold for edge removing of initial generated graph of feature space value. This threshold is employed to make the graph sparser. If parameter is set to a low value, fewer edges will be removed, and a denser graph emerges for subsequent steps. Similarly, when is set to a high value, more edges will be removed, and a sparser graph is resulted or the next steps. The value is a threshold for average similarity value that governs the final feature selection process. This parameter can be set to any value in the range . If this parameter is set to a high value, the number of features and the portability risk of selection of similar features will be increased. Moreover, when is set to a low value, the number of features will be reduced. It is therefore important to identify appropriate value of these parameters in a way to maximize the prediction accuracy.
To investigate the optimal value for these parameter, different experiments were performed to examine how the performance impacts the parameter selection.
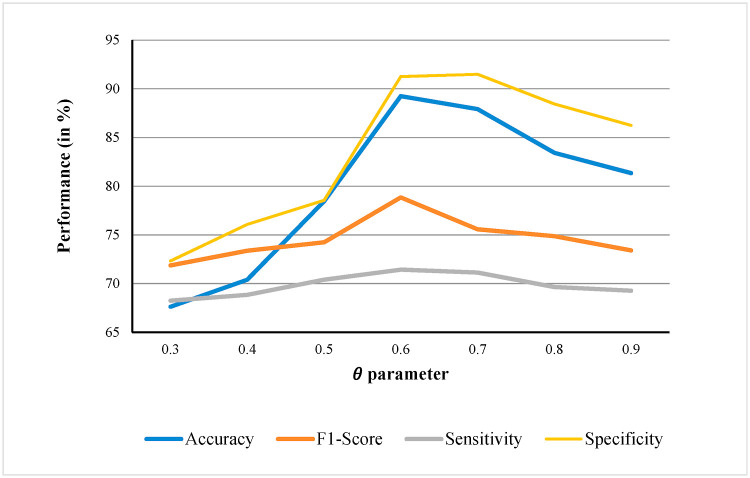
Fig. 10 exhibits the parameter sensitivity analysis for Accuracy, F1-Score, Sensitivity and Specificity measures. The results showed that in most cases when the is adjusted to 0.6, the developed COVID-19 diagnosis model yields the best performance.
Fig. 10.
Average performance (in %) over 30 independent runs, with different values for Accuracy, F1-Score, Sensitivity and Specificity measures.
Likewise, Fig. 11 shows the parameter sensitivity analysis for Accuracy, F1-Score, Sensitivity and Specificity measures. The results indicated that in most cases when the is adjusted to 0.7, the proposed COVID-19 diagnosis model achieves the best performance.
Fig. 11.
Average performance (in %) over 30 independent runs, with different values for Accuracy, F1-Score, Sensitivity and Specificity measures.
4.6. Statistical analysis of the proposed method
In this subsection, the Friedman test [102] is employed to perform the statistical analysis of the experimental results. The Friedman test is a non-parametric statistical test introduced by Milton Friedman to detect differences between multiple treatments, as is the case with parametric measures. By ranking each row together and then evaluating the values of the ranks based on columns, the ranking procedure is completed. For this purpose, each COVID-19 prediction model is ranked for each measure. We used SPSS statistics to run this statistical test. Based on reported results of Table 3, the average ranking for different prediction models (i.e., XGBoost, SVM, MLP, XDT and Proposed Model) on each measure is indicated in Table 9 . These results indicate that the proposed model has the best performance. Moreover, Table 10 shows the results of Friedman test for these compared COVID-19 diagnosis models. The reported results demonstrated that the p-value of 0.0034218, 0.0037548, 0.003295, 0.004606 and 0.004438 on Accuracy, F1-Score, Sensitivity and Specificity and AUROC measures, correspondingly. Considering these values are lower than 0.05, a conclusion can be drawn of the statistical significance of the outperformance of our model with respect to alternative models shown in the table. Moreover, based on the result of Table 8, the p-value for Accuracy, F1-Score, Sensitivity and Specificity measures were 0.0034784, 0.0037691, 0.0039877 and 0.004897. These reported values demonstrated that, in terms of statistical significance, our model outperforms other transforming techniques (i.e., Counterfactual Sets, Rule Conjunctions, Filtering of Conjunction Sets and Matrix–Visualization).
Table 9.
Average ranks of the different COVID-19 prediction models on different measures.
| Measure | Compared COVID-19 diagnosis models |
||||
|---|---|---|---|---|---|
| XGBoost | SVM | MLP | XDT | Proposed Model | |
| Accuracy | 4.79 | 4.06 | 2.68 | 2.10 | 1.34 |
| F1-Score | 4.79 | 3.93 | 2.79 | 2.17 | 1.31 |
| Sensitivity | 4.68 | 3.89 | 2.82 | 2.31 | 1.27 |
| Specificity | 4.62 | 4.06 | 2.72 | 2.20 | 1.37 |
| AUROC | 4.58 | 4.03 | 2.79 | 2.27 | 1.32 |
Table 10.
The results of the Friedman statistics test.
| Measure |
|||||
|---|---|---|---|---|---|
| Accuracy | F1-Score | Sensitivity | Specificity | AUROC | |
| Chi-Square | 10.3858 | 13.9218 | 10.2319 | 15.0436 | 15.0321 |
| df | 4 | 4 | 4 | 4 | 4 |
| Asymp.Sig (p-value) | 0.0034218 | 0.0037548 | 0.003295 | 0.004606 | 0.004438 |
5. Discussion
In this paper, an available dataset of routine blood tests is used, which includes positive COVID-19 cases as well as negative COVID-19 cases. Our machine learning-based method is trained and evaluated for COVID-19 disease diagnosis. The average Accuracy, F1-Score, Sensitivity, Specificity and AUROC of our model were 89.98%, 78.12%, 71.69%, 92.96% and 92.88%, respectively, which is higher than other classical and state-of-the-art COVID-19 disease diagnosis (Table 3, Table 4).
Moreover, in terms of the average normalized confusion matrix, the developed method was compared with other methods, and the reported results showed that in all cases, the developed COVID-19 diagnosis model had better performance (Table 5).
Furthermore, the performance of the proposed feature selection method in terms of accuracy and execution time is compared with four well-known feature selection methods as well as four state-of-the-art feature selection methods. The reported results indicated that the accuracy of the developed method is higher than other feature selection methods. Moreover, in terms of execution time the reported results revealed that, the univariate feature selection methods (i.e. FS and LS) have less execution time than multivariate methods due to the fewer calculations they perform in their selection process. That is because in univariate feature selection approaches, the possible similarity between features is not considered in the feature selection process. Therefore, these methods have low accuracy in real applications and high-dimensional datasets. On the other hand compared to state-of-the-art feature selection approaches, the reported results revealed that, the proposed feature selection method has the lowest average execution time (Figs. 8 and 9).
The reported results of feature selection phase indicated that, PLT, EOS, MPV, CRP, AST, CREAT, WBC, MONO, LYM and RBC were the ten most important features, respectively (Table 6). This finding demonstrates the importance of AST, WBC, CRP, RBC and MONO features in COVID-19 diagnosis, that is in line with the results of other previous studies [7,92].
Based on these reported results, it can be said that the developed prediction method in this study is one of the most accurate and fastest models presented to date. Our proposed model is an intelligent model that can help physicians to diagnose COVID-19 positive cases. Through our understandable prediction and feature selection phase, it is possible to determine which features of datasets were more important in prediction. This explainable COVID-19 disease diagnosis model has higher transparency and explainability than previous black box methods [[67], [68], [69], [70],[103], [104], [105]] that can improve the acceptance rate and trustworthy of intelligent model for physicians.
In the reminder of this section the reasons for the enhanced performances of the developed COVID-19 prediction model compared to other prediction methods are discussed. These are grounded on a key innovation that are incorporated into the developed, which made the model perform better than many state-of-the-art methods:
-
1
Unlike many previous methods of COVID-19 diagnosis, which use PCR and imaging approaches (i.e., Chest X-ray, and Chest CT) and have drawbacks such as costly equipment, need to specialist staff, and certified labs, our developed model make use of routinely available blood test results, which is much faster, accessible, cheaper, and affordable than previous methods.
-
2
This paper proposes an artificial intelligence decision system to provide physicians with a simple and human-interpretable set of rules for diagnosing COVID-19 positive cases in the same spirit as Decision Tree model together with social graph visualization to ensure transparency and explainability for any clinician.
-
3
In many previous COVID-19 prediction models, the final prediction is made by a single classifier, which means that their generalization ability is limited. Contrary to these previous models, a model based on Ensemble Learning is developed in this study. As a result of the ensemble decision forest model, the prediction accuracy is improved, and also the probability of overfitting is reduced.
-
4
Irrelevant features, as well as redundant features, strongly affect the performance of learning model and the result of COVID-19 diagnosis. Therefore, an intelligent prediction model should recognize and remove irrelevant and redundant features as far as possible. All of the initial features have been used by many previous methods of diagnosing COVID-19. Therefore, the accuracy and generalizability of these methods will be reduced, as well as their computational complexity will be increased. In our proposed model, by incorporating an additional feature selection phase, a new COVID-19 diagnosis model is developed which will enhance the final model's performance.
-
5
Though decision forest has a high level of prediction performance, its inherent limitations cannot be hidden as well. This can be summarized in two factors. First, because the decision forest generates many trees instead of one, the classification of decision forest is usually inefficient and in real-time prediction systems, this property creates a significant vulnerability. Second, a DF must explore a variety of trees when it makes a classification. Therefore, the end-user cannot clearly justify the model's predictions and they cannot understand the model structure as it is composed of numerous single models. As a result, standard decision forests are usually prone to limitation when a straightforward and real time explanations are required in the health-system application. However, when real time is not an important aspect, such a limitation can be ignored.
6. Conclusion and future works
A severe respiratory disease called COVID-19 has been reported by the WHO. Since the beginning of the pandemic until 1st March 2022, more than 5.9 million people have died as a result of the COVID-19. Recently, artificial intelligence has emerged as a breakthrough of current strategy, and it can be utilized to diagnose COVID-19 positive cases, detect, and predict their mortality. A complex machine-learning model like deep learning performs better than simple algorithms for COVID-19 diagnosis. Although deep learning models perform well, their decisions cannot be justified by explanations, which may limit their effectiveness in medical applications.
To overcome this limitation, explainable AI proposes developing a suite of machine learning models that have a high level of accuracy and are easily explained by physicians. The proposed method includes two main phases. In the first phase, a set of relevant and non-redundant feature are selected for final prediction. In this developed feature selection mechanism, the feature relevance is calculated using the node centrality and Fisher Score, whereas the redundancy of features is calculated using feature similarities. Then, in the second phase, after selecting the final features, in the fourth step, the Decision Forests-based classifier is employed to COVID-19 prediction by employing routine blood tests. In contrast to previous deep learning-based COVID-19 diagnosis models, which are difficult to explain for physicians due to their black box nature, the developed prediction model is transparent through its explainable decision trees. Moreover, by employing an ensemble learning-based prediction model, a new COVID-19 diagnosis model is developed that will improve the final prediction accuracy. The proposed method has been compared to the well-known and newest COVID-19 diagnosis models including XGBoost, SVM, MLP and eXplainable Decision Trees model with respect to four different performance metrics: Classification Accuracy, F1–Score, Sensitivity and Specificity. The experimental results indicate that the developed prediction model achieved outperformed the state-of-the-art methods and baseline algorithms.
The developed model opens the door for the use of explainable AI in healthcare applications. Inherent limitations have also been examined and thoroughly discussed. In future works, we will attempt to extend out proposal model beyond the inherent limitations of DF algorithm by endowing a set of rule-based approach that each the integration of various DT in DF mechanism to ensure further transparency.
Declaration of competing interest
The authors declare that they have no known competing financial interestsor personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This research is connected to the DigiHealth-project, a strategic profiling project at the University of Oulu. The project is supported by the Academy of Finland (project number 326291) and the University of Oulu Academy of Finland Profi5 on Digihealth.
Footnotes
The datasets and sourcecode are available from the corresponding author (Mehrdad Rostami, E-mail: Mehrdad.Rostami@oulu.fi) on reasonable request.
References
- 1.Barnawi A., et al. Artificial intelligence-enabled Internet of Things-based system for COVID-19 screening using aerial thermal imaging. Future Generat Comput Syst. 2021;124:119–132. doi: 10.1016/j.future.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Viviani M., et al. Assessing vulnerability to psychological distress during the COVID-19 pandemic through the analysis of microblogging content. Future Generat Comput Syst. 2021;125:446–459. doi: 10.1016/j.future.2021.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puxeddu M.G., Petti M., Astolfi L. A comprehensive analysis of multilayer community detection algorithms for application to EEG-based brain networks. Front Syst Neurosci. 2021;15 doi: 10.3389/fnsys.2021.624183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saberi-Movahed F., et al. 2021. Decoding clinical biomarker space of covid-19: exploring matrix factorization-based feature selection methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Z., et al. Development and clinical application of a rapid IgM‐IgG combined antibody test for SARS‐CoV‐2 infection diagnosis. J Med Virol. 2020;92(9):1518–1524. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinati D., et al. Detection of COVID-19 infection from routine blood exams with machine learning: a feasibility study. J Med Syst. 2020;44(8):1–12. doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babaei Rikan S., et al. Biomedical Signal Processing and Control; 2021. COVID-19 diagnosis from routine blood tests using artificial intelligence techniques. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsharif W., Qurashi A. Effectiveness of COVID-19 diagnosis and management tools: a review. Radiography. 2021;27(2):682–687. doi: 10.1016/j.radi.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalane P., et al. Automatic detection of COVID-19 disease using U-Net architecture based fully convolutional network. Biomed Signal Process Control. 2021;67:102518. doi: 10.1016/j.bspc.2021.102518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nasiri E., et al. A novel link prediction algorithm for protein-protein interaction networks by attributed graph embedding. Comput Biol Med. 2021;137 doi: 10.1016/j.compbiomed.2021.104772. [DOI] [PubMed] [Google Scholar]
- 11.Vantaggiato E., et al. Covid-19 recognition using ensemble-cnns in two new chest x-ray databases. Sensors. 2021;21(5):1742. doi: 10.3390/s21051742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turkoglu M. COVIDetectioNet: COVID-19 diagnosis system based on X-ray images using features selected from pre-learned deep features ensemble. Appl Intell. 2021;51(3):1213–1226. doi: 10.1007/s10489-020-01888-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berrimi M., et al. 2021 international conference of women in data science at taif university (WiDSTaif) IEEE; 2021. COVID-19 detection from Xray and CT scans using transfer learning. [Google Scholar]
- 14.Sun L., et al. Adaptive feature selection guided deep forest for covid-19 classification with chest ct. IEEE J Biochem Health Informat. 2020;24(10):2798–2805. doi: 10.1109/JBHI.2020.3019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaban W.M., et al. A new COVID-19 Patients Detection Strategy (CPDS) based on hybrid feature selection and enhanced KNN classifier. Knowl Base Syst. 2020;205:106270. doi: 10.1016/j.knosys.2020.106270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ji T., et al. Detection of COVID-19: a review of the current literature and future perspectives. Biosens Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian S., et al. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J Thorac Oncol. 2020;15(5):700–704. doi: 10.1016/j.jtho.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari D., et al. Routine blood tests as a potential diagnostic tool for COVID-19. Clin Chem Lab Med. 2020;58(7):1095–1099. doi: 10.1515/cclm-2020-0398. [DOI] [PubMed] [Google Scholar]
- 19.Sharma M.K., et al. Mediative fuzzy logic mathematical model: a contradictory management prediction in COVID-19 pandemic. Appl Soft Comput. 2021;105:107285. doi: 10.1016/j.asoc.2021.107285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alharthi A.M., Lee M.H., Algamal Z.Y. Gene selection and classification of microarray gene expression data based on a new adaptive L1-norm elastic net penalty. Inform Med Unlocked. 2021;24 [Google Scholar]
- 21.Wang W., et al. Gene selection for cancer detection using graph signal processing. Inform Med Unlocked. 2021;25 [Google Scholar]
- 22.Vaishya R., et al. Artificial Intelligence (AI) applications for COVID-19 pandemic. Diabetes Metabol Syndr: Clin Res Rev. 2020;14(4):337–339. doi: 10.1016/j.dsx.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swapnarekha H., et al. Role of intelligent computing in COVID-19 prognosis: a state-of-the-art review. Chaos, Solit Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oussalah M.A.I. International conference on pattern recognition. Springer; 2021. Explainability. A bridge between machine vision and natural language processing. [Google Scholar]
- 25.Huerta E.B., Duval B., Hao J.-K. Fuzzy logic for elimination of redundant information of microarray data. Dev Reprod Biol. 2008;6(2):61–73. doi: 10.1016/S1672-0229(08)60021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao C., et al. Deep learning and its applications in biomedicine. Dev Reprod Biol. 2018;16(1):17–32. doi: 10.1016/j.gpb.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma A., Rani R. C-HMOSHSSA: gene selection for cancer classification using multi-objective meta-heuristic and machine learning methods. Comput Methods Progr Biomed. 2019;178:219–235. doi: 10.1016/j.cmpb.2019.06.029. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Sun M., Liu L. Future Generation Computer Systems; 2021. Basketball shooting angle calculation and analysis by deeply-learned vision model. [Google Scholar]
- 29.Deng L., Li J., Han Z. Online defect detection and automatic grading of carrots using computer vision combined with deep learning methods. Lebensm Wiss Technol. 2021;149 [Google Scholar]
- 30.Dokuz Y., Tufekci Z. Mini-batch sample selection strategies for deep learning based speech recognition. Appl Acoust. 2021;171 [Google Scholar]
- 31.Ma J., et al. Deep dual-side learning ensemble model for Parkinson speech recognition. Biomed Signal Process Control. 2021;69 [Google Scholar]
- 32.Idowu O.P., et al. An integrated deep learning model for motor intention recognition of multi-class EEG Signals in upper limb amputees. Comput Methods Progr Biomed. 2021;206 doi: 10.1016/j.cmpb.2021.106121. [DOI] [PubMed] [Google Scholar]
- 33.Gabeff V., et al. Interpreting deep learning models for epileptic seizure detection on EEG signals. Artif Intell Med. 2021;117 doi: 10.1016/j.artmed.2021.102084. [DOI] [PubMed] [Google Scholar]
- 34.Shamshirband S., et al. A review on deep learning approaches in healthcare systems: taxonomies, challenges, and open issues. J Biomed Inf. 2020 doi: 10.1016/j.jbi.2020.103627. [DOI] [PubMed] [Google Scholar]
- 35.Goodman B., Flaxman S. European Union regulations on algorithmic decision-making and a “right to explanation”. AI Mag. 2017;38(3):50–57. [Google Scholar]
- 36.Barredo Arrieta A., et al. Explainable Artificial Intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Inf Fusion. 2020;58:82–115. [Google Scholar]
- 37.Adadi A., Berrada M. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI) IEEE Access. 2018;6:52138–52160. [Google Scholar]
- 38.Molnar C. Lulu. com; 2020. Interpretable machine learning. [Google Scholar]
- 39.Anusha M., Sathiaseelan J.G.R. Feature selection using K-means genetic algorithm for multi-objective optimization. Procedia Comput Sci. 2015;57:1074–1080. [Google Scholar]
- 40.Marcelloni F. Feature selection based on a modified fuzzy C-means algorithm with supervision. Inf Sci. 2003;151:201–226. [Google Scholar]
- 41.Moradi P., Rostami M. Integration of graph clustering with ant colony optimization for feature selection. Knowl Base Syst. 2015;84:144–161. [Google Scholar]
- 42.González J., et al. A new multi-objective wrapper method for feature selection – accuracy and stability analysis for BCI. Neurocomputing. 2019;333:407–418. [Google Scholar]
- 43.Xue Bing, Zhang Mengjie, Browne W.N. Particle swarm optimization for feature selection in classification: a multi-objective approach. IEEE Trans Cybern. 2013;43(6):1656–1671. doi: 10.1109/TSMCB.2012.2227469. [DOI] [PubMed] [Google Scholar]
- 44.Jalali S.M.J., et al. An oppositional-Cauchy based GSK evolutionary algorithm with a novel deep ensemble reinforcement learning strategy for COVID-19 diagnosis. Appl Soft Comput. 2021;111:107675. doi: 10.1016/j.asoc.2021.107675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan M., et al. Applications of artificial intelligence in COVID-19 pandemic: a comprehensive review. Expert Syst Appl. 2021;185:115695. doi: 10.1016/j.eswa.2021.115695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahdy L.N., et al. MedRxiv; 2020. Automatic x-ray covid-19 lung image classification system based on multi-level thresholding and support vector machine; p. 2020. 03. 30.20047787. [Google Scholar]
- 47.Yu H., et al. Data-driven discovery of a clinical route for severity detection of COVID-19 paediatric cases. IET Cybersyst Robot. 2020;2(4):205–206. [Google Scholar]
- 48.Too J., Mirjalili S. A hyper learning binary dragonfly algorithm for feature selection: a COVID-19 case study. Knowl Base Syst. 2021;212 [Google Scholar]
- 49.Song J., et al. Maximum likelihood-based extended Kalman filter for COVID-19 prediction. Chaos, Solit Fractals. 2021;146 doi: 10.1016/j.chaos.2021.110922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar N., Kumar H. A novel hybrid fuzzy time series model for prediction of COVID-19 infected cases and deaths in India. ISA (Instrum Soc Am) Trans. 2021;In Press, Corrected Proof doi: 10.1016/j.isatra.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cobre A.d.F., et al. Diagnosis and prediction of COVID-19 severity: can biochemical tests and machine learning be used as prognostic indicators? Comput Biol Med. 2021;134 doi: 10.1016/j.compbiomed.2021.104531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arvind V., et al. Development of a machine learning algorithm to predict intubation among hospitalized patients with COVID-19. J Crit Care. 2021;62:25–30. doi: 10.1016/j.jcrc.2020.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pahar M., et al. COVID-19 cough classification using machine learning and global smartphone recordings. Comput Biol Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebinger J., et al. A machine learning algorithm predicts duration of hospitalization in COVID-19 patients. Intel Based Med. 2021;5 doi: 10.1016/j.ibmed.2021.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang S., et al. Identification and validation of prognostic factors in patients with COVID-19: a retrospective study based on artificial intelligence algorithms. J Intensive Med. 2021;1:103–109. doi: 10.1016/j.jointm.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gulati K., et al. Comparative analysis of machine learning-based classification models using sentiment classification of tweets related to COVID-19 pandemic. Mater Today Proc. 2021;51:38–41. [Google Scholar]
- 57.Singh M., et al. Transfer learning–based ensemble support vector machine model for automated COVID-19 detection using lung computerized tomography scan data. Med Biol Eng Comput. 2021;59(4):825–839. doi: 10.1007/s11517-020-02299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu Y.H., et al. JCS: an explainable COVID-19 diagnosis system by Joint classification and segmentation. IEEE Trans Image Process. 2021;30:3113–3126. doi: 10.1109/TIP.2021.3058783. [DOI] [PubMed] [Google Scholar]
- 59.Yang Q., et al. Clinical characteristics and a decision tree model to predict death outcome in severe COVID-19 patients. BMC Infect Dis. 2021;21(1):783. doi: 10.1186/s12879-021-06478-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lella K.K., Pja A. Automatic diagnosis of COVID-19 disease using deep convolutional neural network with multi-feature channel from respiratory sound data: cough, voice, and breath. Alex Eng J. 2021;61:1319–1334. [Google Scholar]
- 61.Qayyum A., et al. Depth-wise dense neural network for automatic COVID19 infection detection and diagnosis. Ann Oper Res. 2021;302:1–21. doi: 10.1007/s10479-021-04154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roy S., et al. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE Trans Med Imag. 2020;39(8):2676–2687. doi: 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 63.Shamsi A., et al. An uncertainty-aware transfer learning-based framework for COVID-19 diagnosis. IEEE Transact Neural Networks Learn Syst. 2021;32(4):1408–1417. doi: 10.1109/TNNLS.2021.3054306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Islam M.Z., Islam M.M., Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hall L.O., et al. 2020. Finding covid-19 from chest x-rays using deep learning on a small dataset. arXiv preprint arXiv:2004.02060. [Google Scholar]
- 66.Ahmadian S., et al. A novel deep neuroevolution-based image classification method to diagnose coronavirus disease (COVID-19) Comput Biol Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jo Y.-Y., et al. Detection and classification of arrhythmia using an explainable deep learning model. J Electrocardiol. 2021;67:124–132. doi: 10.1016/j.jelectrocard.2021.06.006. [DOI] [PubMed] [Google Scholar]
- 68.Peng J., et al. An explainable artificial intelligence framework for the deterioration risk prediction of hepatitis patients. J Med Syst. 2021;45(5):61. doi: 10.1007/s10916-021-01736-5. [DOI] [PubMed] [Google Scholar]
- 69.Valente F., et al. A new approach for interpretability and reliability in clinical risk prediction: acute coronary syndrome scenario. Artif Intell Med. 2021;117:102113. doi: 10.1016/j.artmed.2021.102113. [DOI] [PubMed] [Google Scholar]
- 70.Neves I., et al. Interpretable heartbeat classification using local model-agnostic explanations on ECGs. Comput Biol Med. 2021;133 doi: 10.1016/j.compbiomed.2021.104393. [DOI] [PubMed] [Google Scholar]
- 71.Hu Y., et al. Understanding risk factors for postoperative mortality in neonates based on explainable machine learning technology. J Pediatr Surg. 2021;56(12):2165–2171. doi: 10.1016/j.jpedsurg.2021.03.057. [DOI] [PubMed] [Google Scholar]
- 72.Jiang H., et al. Learning efficient, explainable and discriminative representations for pulmonary nodules classification. Pattern Recogn. 2021;113 [Google Scholar]
- 73.Shin D. The effects of explainability and causability on perception, trust, and acceptance: implications for explainable AI. Int J Hum Comput Stud. 2021;146 [Google Scholar]
- 74.Moradi P., Rostami M. A graph theoretic approach for unsupervised feature selection. Eng Appl Artif Intell. 2015;44:33–45. [Google Scholar]
- 75.Rostami M., Berahmand K., Forouzandeh S. A novel method of constrained feature selection by the measurement of pairwise constraints uncertainty. J Big Data. 2020;7(1):83. [Google Scholar]
- 76.Rostami M., Berahmand K., Forouzandeh S. A novel community detection based genetic algorithm for feature selection. J Big Data. 2021;8(1):2. [Google Scholar]
- 77.Mokhtia M., Eftekhari M., Saberi-Movahed F. Feature selection based on regularization of sparsity based regression models by hesitant fuzzy correlation. Appl Soft Comput. 2020;91 [Google Scholar]
- 78.Rostami M., et al. Review of swarm intelligence-based feature selection methods. Eng Appl Artif Intell. 2021;100 [Google Scholar]
- 79.Rostami M., et al. Integration of multi-objective PSO based feature selection and node centrality for medical datasets. Genomics. 2020;112(6):4370–4384. doi: 10.1016/j.ygeno.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 80.Rostami M., M P. Information and Knowledge Technology (IKT); 2014. A clustering based genetic algorithm for feature selection; pp. 112–116. [Google Scholar]
- 81.Mokhtia M., Eftekhari M., Saberi-Movahed F. Dual-manifold regularized regression models for feature selection based on hesitant fuzzy correlation. Knowl Base Syst. 2021;229 [Google Scholar]
- 82.MonirulKabir Md, Shahjahan Md, Murase K. A new local search based hybrid genetic algorithm for feature selection. Neurocomputing. 2011;74(17):2914–2928. [Google Scholar]
- 83.Theodoridis S., Koutroumbas K. Academic Press; Oxford.: 2008. Pattern recognition. [Google Scholar]
- 84.Yeo G.F.A., Aksakalli V. A stochastic approximation approach to simultaneous feature weighting and selection for nearest neighbour learners. Expert Syst Appl. 2021;185 [Google Scholar]
- 85.Zhu R., Dornaika F., Ruichek Y. Joint graph based embedding and feature weighting for image classification. Pattern Recogn. 2019;93:458–469. [Google Scholar]
- 86.Gu Quanquan, Li Zhenhui, Han J. Proceedings of the international conference on uncertainty in artificial intelligence. 2011. Generalized Fisher score for feature selection. [Google Scholar]
- 87.Yan Xiangbin, Li Zhaia, Fan W. C-index: a weighted network node centrality measure for collaboration competence. J Informat. 2013;7(1):223–239. [Google Scholar]
- 88.Qi Xingqin, et al. vol. 194. Information Sciences; 2012. pp. 240–253. (Laplacian centrality: a new centrality measure for weighted networks). [Google Scholar]
- 89.Sagi O., Rokach L. Approximating XGBoost with an interpretable decision tree. Inf Sci. 2021;572:522–542. [Google Scholar]
- 90.Sagi O., Rokach L. Explainable decision forest: transforming a decision forest into an interpretable tree. Inf Fusion. 2020;61:124–138. [Google Scholar]
- 91.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. Association for Computing Machinery; San Francisco, California, USA: 2016. XGBoost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 92.Alves M.A., et al. Explaining machine learning based diagnosis of COVID-19 from routine blood tests with decision trees and criteria graphs. Comput Biol Med. 2021;132 doi: 10.1016/j.compbiomed.2021.104335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hospital Israelita Albert Einstein Diagnosis of covid-19 and its clinical spectrum - ai and data science supporting clinical decisions. https://www.kaggle.com/einsteindata4u/covid19 (From 28th Mar to 3st Apr)
- 94.Rostami M., et al. Gene selection for microarray data classification via multi-objective graph theoretic-based method. Artif Intell Med. 2022;123:102228. doi: 10.1016/j.artmed.2021.102228. [DOI] [PubMed] [Google Scholar]
- 95.He Xiaofei, Deng Cai, Niyogi1 P. Laplacian score for feature selection. Adv Neural Inf Process Syst. 2005;18:507–514. [Google Scholar]
- 96.Ferreira Artur J., Figueiredo M.A.T. An unsupervised approach to feature discretization and selection. Pattern Recogn. 2012;45(9):3048–3060. [Google Scholar]
- 97.Tang X., Dai Y., Xiang Y. Feature selection based on feature interactions with application to text categorization. Expert Syst Appl. 2019;120:207–216. [Google Scholar]
- 98.Zheng X., et al. Gene selection for microarray data classification via adaptive hypergraph embedded dictionary learning. Gene. 2019;706:188–200. doi: 10.1016/j.gene.2019.04.060. [DOI] [PubMed] [Google Scholar]
- 99.Coleto-Alcudia V., Vega-Rodríguez M.A. Artificial Bee Colony algorithm based on Dominance (ABCD) for a hybrid gene selection method. Knowl Base Syst. 2020;205 [Google Scholar]
- 100.Fernández R.R., et al. Random forest explainability using counterfactual sets. Inf Fusion. 2020;63:196–207. [Google Scholar]
- 101.Neto M.P., Paulovich F.V. Explainable matrix - visualization for global and local interpretability of random forest classification ensembles. IEEE Trans Visual Comput Graph. 2021;27(2):1427–1437. doi: 10.1109/TVCG.2020.3030354. [DOI] [PubMed] [Google Scholar]
- 102.Friedman M. A comparison of alternative tests of significance for the problem of m rankings. Ann Math Stat. 1940;11(1):86–92. [Google Scholar]
- 103.Kukar M., et al. COVID-19 diagnosis by routine blood tests using machine learning. Sci Rep. 2021;11(1):1–9. doi: 10.1038/s41598-021-90265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Plante T.B., et al. Development and external validation of a machine learning tool to rule out COVID-19 among adults in the emergency department using routine blood tests: a large, multicenter, real-world study. J Med Internet Res. 2020;22(12) doi: 10.2196/24048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alakus T.B., Turkoglu I. Comparison of deep learning approaches to predict COVID-19 infection. Chaos, Solit Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110120. [DOI] [PMC free article] [PubMed] [Google Scholar]