Abstract
The tumor suppressor p53 plays a vital role in responding to cell stressors such as DNA damage, hypoxia, and tumor formation by inducing cell-cycle arrest, senescence, or apoptosis. Expression level alterations and mutational frequency implicates p53 in most human cancers. In this review, we show how both computational and experimental methods have been used to provide an integrated view of p53 dynamics, function, and reactivation potential. We argue that p53 serves as an exceptional case study for developing methods in modeling intrinsically disordered proteins. We describe how these methods can be leveraged to improve p53 reactivation molecule design and other novel therapeutic modalities, such as PROteolysis TARgeting Chimeras (PROTACs).
Keywords: p53, molecular dynamics simulations, Markov state models, integrative modeling, NMR, cryptic pockets, flexible receptors
The Opportunities and Challenges of the Tumor Suppressor p53
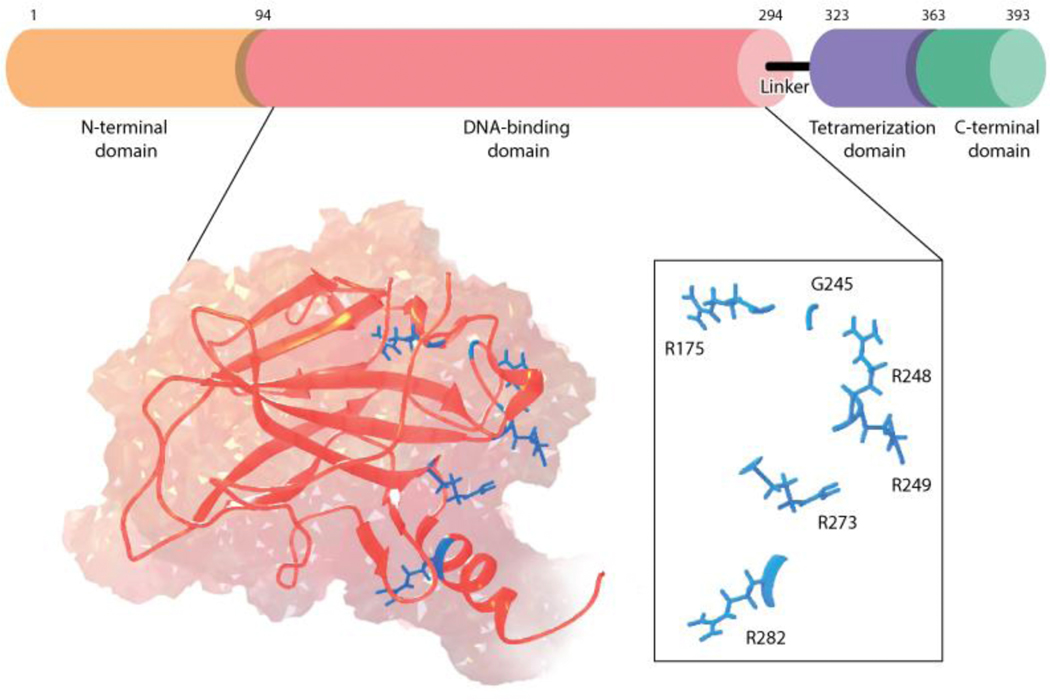
The transcription factor p53 acts as arguably the most important tumor suppressor protein in humans, with more than 50% of all human cancers implicating mutated p53 in their progression. This transcription factor regulates key genes that are involved in cell function and carcinogenesis, such as apoptosis, senescence, and DNA repair. Because of mutant p53’s frequent involvement in cancer progression, it is thought of as a promising and exciting drug target in the field of cancer therapy [1,2]. However, many p53 mutants are known to have significantly altered dynamics and function in comparison to the wildtype protein, challenging traditional structure-based drug discovery methods in the quest for mutant p53 reactivation. Furthermore, p53 consists of multiple domains, adding an additional level of complexity to the characterization of this protein: the intrinsically disordered N-terminal transactivation domain (NTD, residues 1–93) that is responsible for activating target genes, the sequence-specific DNA binding domain (DBD, residues 94–294), the tetramerization domain (TET, residues 323–360) which assists in formation of the functional p53 tetramer, and the intrinsically disordered C-terminal regulatory domain (CTD, residues 363–393) (Figure 1).
Figure 1.
P53 architecture and the hot-spot cancer mutations in p53 DNA-binding domain.
In this review, we describe how modern computational methodologies, tightly integrated with experiment, are emerging as capable of providing avenues for reactivation of p53 cancer mutants. We discuss recent computational and experimental advances in characterizing each of these domains separately as well as in the context of the larger oligomeric, quaternary complex. Lastly, we discuss the therapeutic implications in terms of mutant p53 reactivation and protein-protein interactions, and provide a perspective on how the approaches developed to study this important system can be useful to guide work on similar challenging systems, including intrinsically disordered proteins (IDPs) more generally and in the context of emerging therapeutic modalities, such as PROteolysis TARgeting Chimeras (PROTACs). This mini-review covers the most recent developments in p53 reactivation research; for more comprehensive reviews, we refer readers to Refs. [1–5].
Insight into individual p53 domains
Despite lacking a fixed three-dimensional structure, intrinsically disordered proteins (IDPs) can adopt one or more secondary structures upon target binding (protein, small molecule, RNA, DNA or ions), post-translational modifications or chemical environment changes. Given the inherent instability of p53, its multidomain architecture with long intrinsically disordered stretches and the challenges it poses to experimental studies, molecular dynamics (MD) simulations have provided a useful window into the function and dynamics of the wildtype p53 protein and its oncogenic mutants. Computational costs associated with all-atom simulations, however, limit the size of the systems under investigation, and thus, the great majority of studies have focused on individual protein domains.
DBD
The p53 DBD is intrinsically unstable, often aggregating with small perturbations in temperature, ionic environment, or mutations. This domain is a hotspot region for cancer mutations (Figure 1) and has been the focus of several recent computational studies highlighting the diverse effects of the single-point mutations. MD simulations of several structural mutants evidenced distinct conformational sampling in the S6/S7 loop, located at the vicinity of an aggregation-prone region, compared to wildtype [6]. Changes in the loop’s interaction network due to the mutations are suggested to drive the destabilization of the DBD, leading to unfolding and protein aggregation. The presence of allostery within the DBD that explains the inactivating effect of mutations located distant from the DNA binding surface has also been proposed for the R249S mutant using MD simulations [7] and the Y220C mutant following extensive sampling and Markov state models (MSMs) [8].
The flexibility of the DBD was highlighted in MD simulations of 20 mutants, which identified exacerbation of wildtype’s inherent structural vulnerabilities by the mutations and suggested a general mechanism for p53 rescue by targeting the so-called “structural-disruption motifs” [9]. Using NMR in combination with equilibrium denaturation experiments, Bej et al [10] probed the backbone dynamics of the DBD, showing a linear correlation between the equilibrium denaturation parameters with the extent of changes in conformational entropy. Upon studying aggregation of p53 mutants using MD and coarse-grained simulations of a DBD peptide segment containing the aggregation nucleating region in conjunction with biophysical experiments, Lima et al found polymorphisms in the β-sheet peptide aggregates [11]. Distinct structural destabilization and aggregation properties were also evident in experimental and computational studies of the DNA-contact R273 mutants, indicating that conformational instability is widespread among p53 mutants and the loss of contact with DNA may not be the single molecular basis of altered p53 function [12].
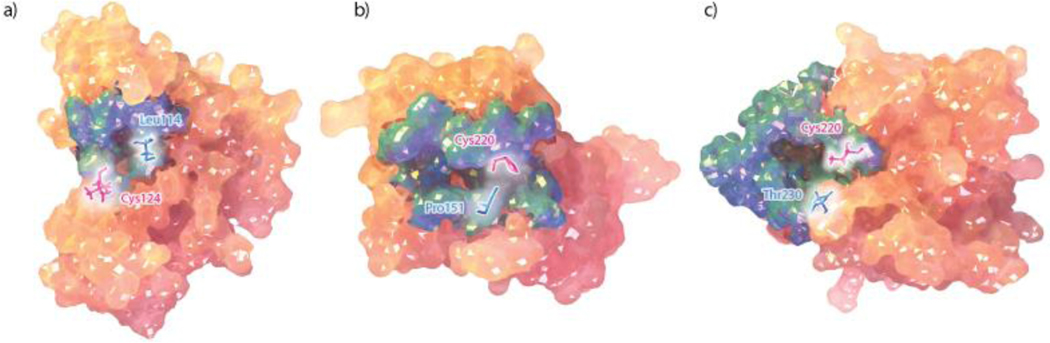
Significant research aims to develop wildtype p53 reactivating small molecules through the stabilization of the native, folded state over the unfolded, aggregated state [13]. Two small molecules, PRIMA-1 and its analogue APR-246, were identified in a cell-based screen from the National Cancer Institute library [14,15]. Both molecules have been shown to target mutant p53 proteins, restoring wild-type p53 transcriptional activity, and are currently in clinical trials [16]. Another class of stabilizers, termed cavity binders, have been developed to target the elongated surface crevice created by the Y220C mutation [5,17,18]. The conformational sampling afforded by computer simulations has additionally enabled the identification of promising druggable pockets in the DBD in a number of studies [6,8,19]. This includes cryptic pockets that are absent from the experimentally resolved structures but emerge in all-atom simulations, providing new therapeutic opportunities for p53 reactivation [8,19]. (Figure 2). Among these, the L1/S3 pocket is especially important as it exists across all mutant forms of p53 presenting a broad therapeutic opportunity [19].
Figure 2.
Druggable pockets in p53 DNA-binding domain. (a) L1/S3 pocket in an MD-generated open conformation [19], (b) Mutation-induced Y220C pocket from crystal structure (pdbID:3ZME, chain A), (c) L6 pocket in an MD-generated open conformation [8].
CTD
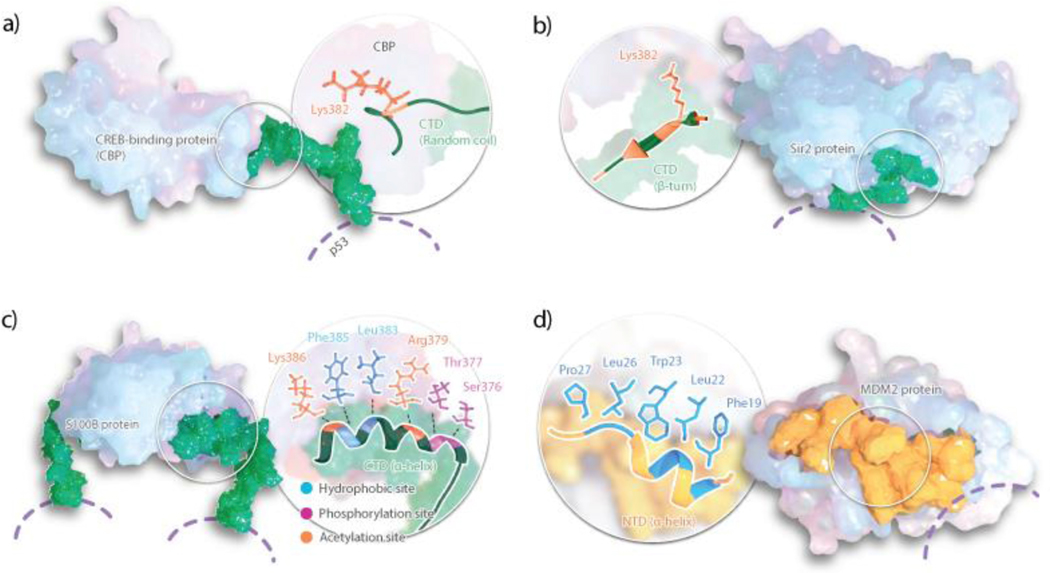
The p53 CTD exhibits properties of an intrinsically disordered protein, binding a number of proteins by adapting various conformations: an alpha-helix, a beta-strand, a beta-turn, or disordered structures [20–24] (Figure 3). Kumar et al. probed the CTD’s conformational behavior upon changing protein environment, finding a distinct and predictable switch of residues 380–388 between an α-helical and β-sheet secondary structures [25]. Via circular dichroism spectroscopy and FRET, they probed the importance of temperature and hydrophobic interactions with binding partners in the secondary structure of the CTD, finding that hydrophobicity allows the CTD to preferentially retain a more α-helical structure and higher temperatures allow for acquisition of secondary structure [25].
Figure 3.
Binding interface of p53 CTD and NTD with several protein interactors: (a) CREB-binding protein, (b) Sir2 protein, (c) S100B and (c) MDM2. CTD shown in green representation and NTD in yellow. Specific residues involved in the protein-protein interactions are highlighted.
Kannan et al. investigated the binding of the p53 CTD to 5 of its binding partners, namely S100B, cyclinA, CBP, sirtuin, and Set9 using equilibrium and non-equilibrium MD in explicit solvent [26]. The equilibrium MD demonstrated that the free p53 CTD peptide fluctuates between various conformations, including the conformations captured in crystal structures of its complexes [26]. Using non-equilibrium MD, the unbinding of the p53 CTD peptide was achieved, indicating that long-range electrostatic interactions (e.g., farther than 10 Å) results in formation of reactive conformations. This work further showed how peptide folding at or close to the binding interface steers the reactive conformations toward the binding partner [26].
Two recent separate studies on the p53 CTD peptide investigated secondary structure formation called molecular recognition features (MoRFs) with microsecond-timescale conventional MD or nanosecond-timescale replica-exchange MD [25,27]. Iida et al. focused on the interaction of the CTD peptide with S100B using virtual-system coupled multicanonical MD, a generalized ensemble MD simulation for enhanced conformational sampling [28]. They concluded that the CTD peptide adopts various conformations upon binding S100B rather than the single α-helical conformation observed in the NMR structure of the p53CTD-S100B complex. Analyzing the multimodal structural distribution of the complex, they found the conformation observed in the NMR model is the most probable orientation in the ensemble. They also found that the entropy of p53 CTD peptide in the S100B-bound state is higher than that of the free state, indicating conformational entropy may not affect p53CTD-S100B complex formation.
NTD
In the intrinsically disordered NTD, residues 10 to 40 are particularly important as they are found in a stable α-helix while in complex with MDM2, an important negative regulator of p53 [29] (Figure 3). The interface of the p53 NTD and MDM2 complex is an important drug target; as such, NTD structure and dynamics has received the attention of many researchers. Herrero-Nieto et al. combined MD at the millisecond timescale with MSMs to explore the ensemble of conformations of the p53 NTD peptide in isolation [30]. They found multiple states enriched in secondary structure elements including an α-helix as well as β-sheets in the conformational landscape of p53 NTD corresponding to about 40% of the equilibrium population. Apart from these structurally and kinetically diverse ordered states, the remaining 60% was completely heterogeneous and lacked any secondary structure. Zhao et al. coupled replica exchange molecular dynamics (REMD) simulations with MSMs to investigate the effect of dual phosphorylation at Ser46 and Thr55 on conformational ensemble of the p53 NTD peptide and found a slightly larger α-helical content in the ensemble after phosphorylation [31].
p53 and MDM2 engage in a negative autoregulatory loop, where upregulation of p53 leads to increased expression of MDM2, which in turn binds and targets p53 for ubiquitin-dependent degradation. Many tumors exploit this negative feedback loop by introducing MDM2 mutations to increase p53 degradation in cancer cells. Due to the co-dependence of these two proteins, there have been great strides in utilizing computational methods to understand this protein-protein interaction. For example, metadynamics simulations [32], potential of mean force (PMF) studies [33], parallel cascade selection molecular dynamics (PaCS-MD) [34], MSMs [35–37], and modeling employing limited data (MELD)-accelerated MD [38] have been used to understand the binding/unbinding process of p53 and MDM2 at a molecular level. These methods were able to accurately predict the thermodynamic and kinetics of p53/MDM2 binding and reveal states along the p53/MDM2 binding trajectory, which can all be used as models in p53/MDM2 structure-based drug design.
Our enhanced understanding of the p53/MDM2 interaction aids in designing inhibitors to disrupt this protein-protein interaction. This is evidenced by the number of drug discovery programs targeting p53/MDM2 as discussed in a recent review by Miller, Gaiddon, and Storr [4]. The Amgen compound, AMG232 (currently in Phase I/II clinical trials), is one of the most potent p53/MDM2 inhibitors to date exhibiting sub-nanomolar binding, inducing significant p53 upregulation, and demonstrating clinically suitable PK properties. Aileron Therapeutics developed a stapled peptide, ALRN-6924, a highly potent dual inhibitor of both MDM2 and MDMX that has been shown to restore p53 activity in vitro and in vivo and is undergoing Phase I clinical studies. MDMX is also a negative regulator of p53 that binds its transactivation domain to inhibit activation of p53 transcription factors; in particular, MDMX binds MDM2 to prevent its auto-ubiquitination, which stabilizes p53 ubiquitin-dependent degradation [39,40]. Therefore, dual targeting of both MDM2 and MDMX has provided an interesting avenue in significantly enhancing anticancer activity.
Modeling the full-length p53
Structural characterization of full-length proteins that contain folded and disordered domains is a major challenge. Thus, experimental and computational approaches generally analyze various fragments of such multidomain IDPs in isolation. However, folded or disordered components of multidomain IDPs do not function as isolated entities: instead, all components of the entire protein act in synergy. In support of this, Wright et al. recently showed the NTD’s importance in DNA binding accuracy using NMR and inteins, evidencing that the NTD reduces DBD binding to nonspecific p53 DNA by five-fold. This suggests competition between NTD and non-specific DNA in binding to the DBD while leaving response-element DNA binding uninhibited [41]. Furthermore, He et al. probed the NTD-DBD interactions via NMR, showing small chemical shifts suggesting weak interactions, perhaps regulating binding between the DBD and response element DNA by nucleic acid mimicry or electrostatic screening [42].
Although there are high-resolution structures available for different domains of the full-length protein (fl-p53), there is no x-ray crystallographic or NMR structure of the entire fl-p53 [43]. Characterizing the structural ensembles and long timescale dynamics of the fl-p53 structure, or at least of large multi-domain fragments, while in complex with DNA or other protein interactors in atomic detail would be invaluable for understanding the p53-activated tumor suppression pathways, as well as how its behavior changes with p53 cancer mutations, post-translational modifications, or binding of small molecules or protein interactors. A first glimpse into the fl-p53 monomer concerted dynamics has been provided by MD simulations of Chillemi et al. [44]. These simulations revealed correlated motions between the transactivation domain 1 (TAD1) and the proline-rich region of the NTD, the tetramerization region in the CTD, and Lys120 in the DBD.
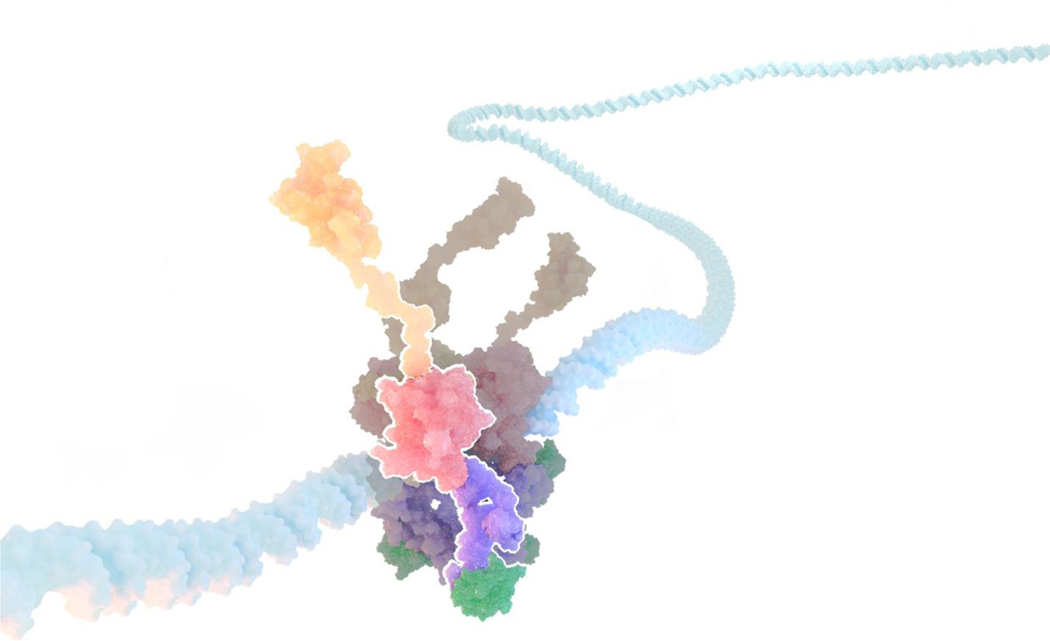
Advances in computing architectures and software performance have made possible simulations of the explicitly solvated full-length DNA-complexed p53 at sufficient timescales that can reveal domain dynamics and allosteric communication in the fl-p53 tetramer context. MD simulations of our all-atom integrative fl-p53 tetramer models bound to three different DNA sequences, namely a p21 response element, a PUMA response element and a nonspecific DNA sequence, yielded final structures consistent with electron microscopy maps and, for the first time, showed the direct interactions of the p53 CTD with DNA at the atomic level [45]. (Figure 4). Solvent mapping analyses of these nearly 1 μs-long simulations revealed multiple potential druggable pockets in p53 and a collective principal component analysis identified sequence-dependent differential quaternary binding modes of the p53 tetramer to DNA [45]. In a subsequent study including wild-type p53 and the R175H mutant in the fl-p53 tetramer context, the symmetric quaternary DNA binding mode observed for the wild-type DBDs was found absent in the R175H mutant system, similar to the case of wild-type p53 DBDs binding to nonspecific DNA in the previous study [46]. These findings prove the requirement of functional p53 and an optimal DNA sequence for a productive binding mode of DBD and DNA at the molecular level.
Figure 4.
Full-length p53 tetramer model in complex with DNA. DNA is depicted in light blue surface representation while NTD, DBD, TET and CTD domains of one fl-p53 monomer are highlighted in orange, pink, purple and green surface representation, respectively.
Methods that can identify correlated motions and allosteric communication in protein structures can be extremely helpful for the interpretation of simulations of dynamic multidomain proteins such as full-length p53. A recent development of the dynamical network analysis method by Melo et al. provides exciting opportunities for the efficient and automated identification of communication pathways in biomolecular structures, including visualization in VMD [47]. The methodology is optimized enough to efficiently process large multi-subunit protein simulations, making it exceptionally poised for p53 and other complex proteins analysis. Another method developed by Porter et al. looks for cooperative changes in solvent exposure as indication of functionally relevant conformational changes and led to the identification of not only allosteric signaling but also cryptic pockets in tested systems [48]. Furthermore, connecting dynamical network analysis with MSMs is likely to indicate how residue-level communication networks shift in different states. Multiscale methods that intersect computational approaches operating at different scales are tools capable of addressing longer-range structure, dynamics and function questions in this and other systems [49].
Conclusions & Outlook
In this review we discuss recent efforts that have provided hitherto unseen insights into the structure, dynamics, function, and druggable new pockets of p53 through integrating cutting-edge computational and experimental biophysical approaches. Techniques that drill down into one p53 domain have provide value in their own right, but structure based drug design has great potential to now also use such integrated methods on the full-length p53 structure as well as its complex with DNA. We posit that the larger-scale approaches to p53 reactivation will be particularly useful in drug design, especially in the way that they provide a more complete understanding of how dynamics affects function, including insights on longer-range allosteric effects.
Taking a higher level view, the field of computational chemistry has made significant strides in developing methods to understand protein complexes, with p53 commonly used as an appropriate example. A related field that stands to benefit from such approaches is PROTAC design. PROTACs are a promising therapeutic modality, which selectively targets proteins for degradation through exploitation of the intracellular ubiquitin-proteasome machinery [50]. These hetero-bifunctional molecules consist of a target protein ligand, an E3 ligase ligand, and an often flexible linker. This new approach in drug design shows great promise in selectively and rapidly degrading target proteins, thereby inhibiting downstream signals associated with disease pathways. Indeed, PROTAC design may be a worthwhile approach in targeting p53-associated tumors. For example, Li et al. developed MD-224, the first MDM2 PROTAC that rapidly degrades MDM2 at concentrations < 1 nM in human leukemia cells and thereby prevents p53 degradation by MDM2 [51]. Similar computational approaches, as we and others have demonstrated for p53, can be used to learn the optimal ternary complex for PROTACS, as well as understand multidomain dynamics and communication in IDPs.
Similar to p53, many multi-domain hub proteins have long IDP regions which are used for mediating the interactions with their protein partners. Impairment of such interactions are often significant and disease associated. The computational approaches used to explore fl-p53 dynamics can be adopted to investigate dynamics and long-range communication of large multi-domain proteins and their complexes. With larger and larger structural information emerging from recent advances in single particle cryoelectron microscopy and cryoelectron tomography, computational approaches that can explore such realistic complexes without losing atomic detail will become an asset.
Acknowledgements
This work was supported by the National Institutes of Health grant number 1R01GM132826.
Footnotes
Conflicts of interest statement
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Joerger AC, Fersht AR: The p53 Pathway: Origins, Inactivation in Cancer, and Emerging Therapeutic Approaches. Annu Rev Biochem 2016, 85:375–404. [DOI] [PubMed] [Google Scholar]
- 2.Bykov VJN, Eriksson SE, Bianchi J, Wiman KG: Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 2018, 18:89–102. [DOI] [PubMed] [Google Scholar]
- 3. Tan YS, Mhoumadi Y, Verma CS: Roles of computational modelling in understanding p53 structure, biology, and its therapeutic targeting. J Mol Cell Biol 2019, 11:306–316. *Presents a comprehensive account of computational contributions to the p53 field.
- 4.Miller JJ, Gaiddon C, Storr T: A balancing act: using small molecules for therapeutic intervention of the p53 pathway in cancer. Chem Soc Rev 2020, 49:6995–7014. [DOI] [PubMed] [Google Scholar]
- 5.Loh SN: Follow the mutations: Toward class-specific, small-molecule reactivation of p53. Biomolecules 2020, 10: 10.3390/biom10020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pradhan MR, Siau JW, Kannan S, Nguyen MN, Ouaray Z, Kwoh CK, Lane DP, Ghadessy F, Verma CS: Simulations of mutant p53 DNA binding domains reveal a novel druggable pocket. Nucleic Acids Res 2019, 47:1637–1652. *Applies MD, umbrella sampling and benzene mapping simulations to propose the S6/S7 loop destabilization as an initiating event towards p53 structural destabilization and aggregation. The conformational perturbations also suggest a pocket with druggable characteristics which bound FDA approved drugs in computational screening.
- 7.Liu X, Tian W, Cheng J, Li D, Liu T, Zhang L: Microsecond molecular dynamics simulations reveal the allosteric regulatory mechanism of p53 R249S mutation in p53-associated liver cancer. Comput Biol Chem 2020, 84:107194. [DOI] [PubMed] [Google Scholar]
- 8. Barros EP, Demir Ö, Soto J, Cocco MJ, Amaro RE: Markov State Models and NMR Uncover an Overlooked Allosteric Loop in p53. bioRxiv 2020, **Presents unprecedentedly extensive sampling of p53 DBD for wild-type and the Y220C mutant and identifies an overlooked allosteric loop (loop L6) through the use of Markov State Models.
- 9.Bromley D, Daggett V: Tumorigenic p53 mutants undergo common structural disruptions including conversion to α-sheet structure. Protein Sci 2020, 29:1983–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bej A, Rasquinha JA, Mukherjee S: Conformational Entropy as a Determinant of the Thermodynamic Stability of the p53 Core Domain. Biochemistry 2018, 57:6265–6269. [DOI] [PubMed] [Google Scholar]
- 11.Lima I, Navalkar A, Maji SK, Silva JL, De Oliveira GAP, Cino EA: Biophysical characterization of p53 core domain aggregates. Biochem J 2020, 477:111–120. [DOI] [PubMed] [Google Scholar]
- 12.Garg A, Hazra JP, Sannigrahi MK, Rakshit S, Sinha S: Variable Mutations at the p53-R273 Oncogenic Hotspot Position Leads to Altered Properties. Biophys J 2020, 118:720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang GZ, Fersht AR: Multisite aggregation of p53 and implications for drug rescue. Proc Natl Acad Sci U S A 2017, 114:E2634–E2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bykov VJN, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G: Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med 2002, 8:282–288. [DOI] [PubMed] [Google Scholar]
- 15.Bykov VJN, Issaeva N, Zache N, Shilov A, Hultcrantz M, Bergman J, Selivanova G, Wiman KG: Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem 2005, 280:30384–30391. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann S, Bykov VJN, Ali D, Andreń O, Cherif H, Tidefelt U, Uggla B, Yachnin J, Juliusson G, Moshfegh A, et al. : Targeting p53 in vivo: A first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol 2012, 30:3633–3639. [DOI] [PubMed] [Google Scholar]
- 17.Bauer MR, Jones RN, Tareque RK, Springett B, Dingler FA, Verduci L, Patel KJ, Fersht AR, Joerger AC, Spencer J: A structure-guided molecular chaperone approach for restoring the transcriptional activity of the p53 cancer mutant Y220C. Futur Med Chem 2019, 11:2491–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bauer MR, Krämer A, Settanni G, Jones RN, Ni X, Khan Tareque R, Fersht AR, Spencer J, Joerger AC: Targeting Cavity-Creating p53 Cancer Mutations with Small-Molecule Stabilizers: The Y220X Paradigm. ACS Chem Biol 2020, 15:657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wassman CD, Baronio R, Demir Ö, Wallentine BD, Chen C-K, Hall LV, Salehi F, Lin D, Chung BP, Hatfield GW, et al. : Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53 ¨. Nat Commun 2013, 4:1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rustandi RR, Baldisseri DM, Weber DJ: Structure of the negative regulatory domain of p53 bound to S100B(ββ). Nat Struct Biol 2000, 7:570–574. [DOI] [PubMed] [Google Scholar]
- 21.Lowe ED, Tews I, Cheng KY, Brown NR, Gul S, Noble MEM, Gamblin SJ, Johnson LN: Specificity determinants of recruitment peptides bound to phosphoCDK2/cyclin A. Biochemistry 2002, 41:15625–15634. [DOI] [PubMed] [Google Scholar]
- 22.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C: Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell 2002, 10:523–535. [DOI] [PubMed] [Google Scholar]
- 23.Mujtaba S, HeY, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM: Structural Mechanism of the Bromodomain of the Coactivator CBP in p53 Transcriptional Activation. Mol Cell 2004, 13:251–263. [DOI] [PubMed] [Google Scholar]
- 24.Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, et al. : Regulation of p53 activity through lysine methylation. Nature 2004, 432:353–360. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A, Kumar P, Kumari S, Uversky VN, Giri R: Folding and structural polymorphism of p53 C-terminal domain: One peptide with many conformations. Arch Biochem Biophys 2020, 684:108342. [DOI] [PubMed] [Google Scholar]
- 26.Kannan S, Lane DP, Verma CS: Long range recognition and selection in IDPs: The interactions of the C-terminus of p53. Sci Rep 2016, 6:23750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fadda E, Nixon MG: The transient manifold structure of the p53 extreme C-terminal domain: Insight into disorder, recognition, and binding promiscuity by molecular dynamics simulations. Phys Chem Chem Phys 2017, 19:21287–21296. [DOI] [PubMed] [Google Scholar]
- 28.Iida S, Kawabata T, Kasahara K, Nakamura H, Higo J: Multimodal Structural Distribution of the p53 C-Terminal Domain upon Binding to S100B via a Generalized Ensemble Method: From Disorder to Extradisorder. J Chem Theory Comput 2019, 15:2597–2607. [DOI] [PubMed] [Google Scholar]
- 29.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP: Structure of the MDM2 oncoprotein bound to the p53 tumor supressor transactivation domain. Science (80- ) 1996, 274:948–953. [DOI] [PubMed] [Google Scholar]
- 30. Herrera-Nieto P, Pérez A, De Fabritiis G: Characterization of partially ordered states in the intrinsically disordered N-terminal domain of p53 using millisecond molecular dynamics simulations. Sci Rep 2020, 10:12402. *Provides a detailed structural and kinetic description of the conformational landscape of p53 NTD using millisecond-long MD simulations in combination with Markov State Models.
- 31.Zhao L, Ouyang Y, Li Q, Zhang Z: Modulation of p53 N-terminal transactivation domain 2 conformation ensemble and kinetics by phosphorylation. J Biomol Struct Dyn 2020, 38:2613–2623. [DOI] [PubMed] [Google Scholar]
- 32.Zou R, Zhou Y, Wang Y, Kuang G, Ågren H, Wu J, Tu Y: Free Energy Profile and Kinetics of Coupled Folding and Binding of the Intrinsically Disordered Protein p53 with MDM2. J Chem Inf Model 2020, 60:1551–1558. [DOI] [PubMed] [Google Scholar]
- 33.Das P, Mattaparthi VSK: Computational Investigation on the p53-MDM2 Interaction Using the Potential of Mean Force Study. ACS Omega 2020, 5:8449–8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tran DP, Kitao A: Dissociation Process of a MDM2/p53 Complex Investigated by Parallel Cascade Selection Molecular Dynamics and the Markov State Model. J Phys Chem B 2019, 123:2469–2478. [DOI] [PubMed] [Google Scholar]
- 35.Tra DP, Kita A: Kinetic Selection and Relaxation of the Intrinsically Disordered Region of a Protein upon Binding. J Chem Theory Comput 2020, 16:2835–2845. [DOI] [PubMed] [Google Scholar]
- 36.Zhou G, Pantelopulos GA, Mukherjee S, Voelz VA: Bridging Microscopic and Macroscopic Mechanisms of p53-MDM2 Binding with Kinetic Network Models. Biophys J 2017, 113:785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul F, Wehmeyer C, Abualrous ET, Wu H, Crabtree MD, Schöneberg J, Clarke J, Freund C, Weikl TR, Noé F: Protein-peptide association kinetics beyond the seconds timescale from atomistic simulations. Nat Commun 2017, 8:1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrone JA, Perez A, MacCallum J, Dill KA: Computed Binding of Peptides to Proteins with MELD-Accelerated Molecular Dynamics. J Chem Theory Comput 2017, 13:870–876. [DOI] [PubMed] [Google Scholar]
- 39.Sharp DA, Kratowicz SA, Sank MJ, George DL: Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 1999, 274:38189–38196. [DOI] [PubMed] [Google Scholar]
- 40.Stad R, Ramos YFM, Little N, Grivell S, Attema J, Van der Eb AJ, Jochemsen AG: Hdmx stabilizes Mdm2 and p53. J Biol Chem 2000, 275:28039–28044. [DOI] [PubMed] [Google Scholar]
- 41.Krois AS, Jane Dyson H, Wright PE: Long-range regulation of p53 DNA binding by its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci U S A 2018, 115:E11302–E11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He F, Borcherds W, Song T, Wei X, Das M, Chen L, Daughdrill GW, Chen J: Interaction between p53 N terminus and core domain regulates specific and nonspecific DNA binding. Proc Natl Acad Sci U S A 2019, 116:8859–8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emamzadah S, Tropia L, Halazonetis TD: Crystal Structure of a Multidomain Human p53 Tetramer Bound to the Natural CDKN1A (p21) p53-Response Element. Mol Cancer Res 2011, 9:1493–1500. [DOI] [PubMed] [Google Scholar]
- 44.Chillemi G, Davidovich P, D’Abramo M, Mametnabiev T, Garabadzhiu AV, Desideri A, Melino G: Molecular dynamics of the full-length p53 monomer. Cell Cycle 2013, 12:3098–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Demir Ö, Ieong PU, Amaro RE: Full-length p53 tetramer bound to DNA and its quaternary dynamics. Oncogene 2017, 36:1451–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Offutt TL, Ieong PU, Demir Ö, Amaro RE: Dynamics and Molecular Mechanisms of p53 Transcriptional Activation. Biochemistry 2018, 57:6528–6537. **Provides a fully integrated view of the full length p53 in complex with various DNA; explores wild type and mutant forms using conventional all-atom MD and shows how the R175H p53 mutant exhibits a different DNA binding mode
- 47. Melo MCR, Bernardi RC, De La Fuente-Nunez C, Luthey-Schulten Z: Generalized correlation-based dynamical network analysis: A new high-performance approach for identifying allosteric communications in molecular dynamics trajectories. J Chem Phys 2020, 153:134104. **Provides optimization of the dynamical network analysis method for efficient identification of information pathways and dynamic networks in proteins, including large protein complexes, through automated analysis via Jupyter notebooks and visualizations in VMD.
- 48. Porter JR, Moeder KE, Sibbald CA, Zimmerman MI, Hart KM, Greenberg MJ, Bowman GR: Cooperative Changes in Solvent Exposure Identify Cryptic Pockets, Switches, and Allosteric Coupling. Biophys J 2019, 116:818–830. *Presents the concept of solvent exposed cryptic pockets as ‘exposons’ and demonstrates a novel methodology for combining MSMs to discover and target these newly revealed regions
- 49.Amaro RE, Mulholland AJ: Multiscale methods in drug design bridge chemical and biological complexity in the search for cures. Nat Rev Chem 2018, 2:0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersson M, Crews CM: PROteolysis TArgeting Chimeras (PROTACs) — Past, present and future. Drug Discov Today Technol 2019, 31:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. LY, YanJ, AguilaA, McEacherD, PrzybranowskS, LiL, YanY, WanM, HaX, WanS: Discovery of MD-224 as a First-in-Class, Highly Potent, and Efficacious Proteolysis Targeting Chimera Murine Double Minute 2 Degrader Capable of Achieving Complete and Durable Tumor Regression. J Med Chem 2019, 62:448–466. *Presents the first example of using an emerging therapeutic modality (PROTAC) in a potential treatment for p53-associated tumors.