Abstract
Designing effective anti-leukemic immunotherapy will require understanding mechanisms underlying tumor control or resistance. Here we report a mechanism of escape from immunologic targeting in an acute myeloid leukemia (AML) patient, who relapsed one year following immunotherapy with engineered T cells expressing a human leukocyte antigen A*02 (HLA-A2) restricted T cell receptor (TCR) specific for a Wilms’ Tumor Antigen 1 epitope, WT1126-134, (TTCR-C4). Resistance occurred despite persistence of functional therapeutic T cells and continuous expression of WT-1 and HLA-A2 by the patient’s AML cells. Analysis of the recurrent AML revealed expression of the standard proteasome, but limited expression of the immunoproteasome, specifically the beta-subunit 1i (β1i), which is required for presentation of WT1126-134. An analysis of a second patient treated with TTCR-C4 demonstrated specific loss of AML cells co-expressing β1i and WT1. To determine if the WT1 protein continued to be processed and presented in the absence of immunoproteasome processing, we identified and tested a TCR targeting an alternative, HLA-A2 restricted WT137-45 epitope, that was generated by immunoproteasome deficient cells, including WT1-expressing solid tumor lines. T cells expressing this TCR (TTCR37-45) killed the first patients’ relapsed AML resistant to WT1126-134 targeting, as well as other primary AML, in vitro. TTCR37-45 controlled solid tumor lines lacking immunoproteasome subunits both in vitro and in an NSG mouse model. As proteasome composition can vary in AML, defining and preferentially targeting such proteasome-independent epitopes may maximize therapeutic efficacy and potentially circumvent AML immune evasion by proteasome-related immunoediting.
One Sentence Summary:
Targeting the alternate Wilms Tumor-1 (WT1) epitope WT137-45 circumvents engineered T cell receptor gene therapy resistance in acute myeloid leukemia.
Introduction
Allogenic hematopoietic cell transplant (HCT) can cure some patients with high-risk acute myeloid leukemia (AML), which is largely a result of donor T cells eliminating leukemia by recognizing minor histocompatibility antigens or less commonly leukemia-associated antigens (1). This graft-versus-leukemia (GVL) effect is sporadic, entirely reliant on donor immune cells recognizing differences between host and donor cells, and is not predictable pre-HCT. Leukemic relapse post-HCT remains the leading cause of death, especially in patients who enter HCT with features of high-risk AML, in part due to the inability to ensure a GVL effect.
Adoptive therapy can provide T cells re-directed towards defined tumor-associated antigens. Most notably, T cells engineered with chimeric antigen receptors (CARs) targeting surface proteins, such as CD19 or B cell maturation antigen (BCMA), have shown stunning clinical results in B cell malignancies (2), but identifying AML surface proteins that are unique or safely targetable remains a challenge (3). By contrast, T cell receptors (TCRs) can recognize a broader set of peptides, which can be derived from intracellular and surface proteins (4). To achieve consistent antileukemic activity, we sought to target Wilms’ Tumor Antigen 1 (WT1), an intracellular transcription factor overexpressed in most leukemic cells but lowly expressed in normal adult tissues (5). Although not routinely present in peptide elution sets (6), suggesting that there may not be abundant presentation of the processed peptide, the WT1126-134 peptide was successfully eluted from a human leukocyte antigen (HLA)-A*02:01 leukemia, confirming presentation (7). T cells targeting WT1126-134 have contributed to maintenance of sustained complete responses in vaccine trials and exhibited a direct anti-leukemic effect in our group’s prior clinical study in which donor-derived WT1-specific CD8+ T cell clones were transferred into patients post-HCT (5, 8, 9).
Based on these results, we isolated a high affinity TCR (TCR-C4) restricted to HLA-A*02:01 and specific for the WT1126-134 epitope and cloned it into a clinically-validated lentiviral vector. Donor Epstein Barr Virus (EBV)-specific CD8+ T cells were selected as substrates to both minimize risk of graft-versus-host disease (GVHD) and enhance survival of therapeutic cells – EBV-specific CD8+ T cells were transduced with TCR-C4 (TTCR-C4) (10) (NCT01640301). Following prophylactic administration to patients with HLA-matched high-risk AML post-HCT, TTCR-C4 demonstrated in vivo persistence and sustained remissions were achieved in all patients despite risk of relapse that exceeded 50% (10). However, a formal assessment of direct TTCR-C4 anti-tumor activity was challenging as leukemia was undetectable at the time of TTCR-C4 transfer.
By re-directing T cell recognition towards a known tumor epitope, adoptive therapy trials offer the added benefit of limiting the tumor-T cell interaction to a defined target. Paradoxically, the requirements for a therapeutic response can often be better elucidated not in patients with complete sustained responses, but rather in patients whose tumors acquire resistance to the immune intervention. This can reveal critical response and evasion mechanisms (11).
Tumor escape after CAR T cell infusions has commonly reflected loss of the targeted antigen epitope from the cell surface (12). For TCR-mediated interventions, this is best paralleled by loss of expression of the targeted antigen or the HLA-restricting-allele (13). HLA loss has rarely been associated with post-HCT relapse in AML (13) and WT1 mutation was not a major contributor to immune escape in patients with AML who received WT1 peptide vaccination (14) or infusions of clonal WT1-specific T cells in our previous study (9). Instead, and specific to targeted TCR-T cell therapy, presentation of the targeted epitope can be impacted by proteasome processing (15). Indeed, the TCRC4-targeted epitope (WT1126-134) is reliant on the immunoproteasome (IP) (16), the dominant isoform in hemopoietic cells (17).
To investigate potential mechanisms of AML resistance to TTCR-C4, one patient was identified who relapsed after a second transplant. This patient experienced an apparent sustained remission after receiving TTCR-C4, but then relapsed one year later despite large numbers of persistent transferred TTCR-C4 cells. We performed high-dimensional analysis of the persistent T cells and recurrent AML, which confirmed TTCR-C4 were functional and that AML continued to express both restricting HLA Class I allele (HLA-A*02) and unmutated WT1. Further in-depth analysis and observations in a second patient who received TTCR-C4 with circulating WT1+ AML point to a previously undescribed mechanism of in vivo AML escape from a targeted T cell response, involving changes in antigen-processing of the targeted epitope. We then pursued an alternative HLA-A*02:01-restricted epitope, WT137-45, which is less susceptible to the observed mechanism of resistance and thus represents a promising TCR-T cell immunotherapy target. Our data underscore the necessity of elucidating acquired resistance mechanisms to inform rational design of more effective TCR-T cell therapies not only for AML, but for immunotherapy more broadly.
Results
An unusual clinical course warrants deeper investigation.
A 27-year-old patient developed recurrent AML with both extramedullary chloromas and minimal residual disease (MRD) in the bone marrow 7 months after receiving a second matched, unrelated donor HCT for relapsed refractory disease (Fig. 1A and Supplementary Methods). The patient was treated with local radiation and systemic salvage chemotherapy, which achieved remission as evidenced by no evaluable disease (NED), and then received an infusion of donor-derived TTCR-C4 on this study (10). Based on the history of relapsing after a second allo-HCT, he was at extremely high risk of early relapse and death, as AML relapse post-HCT is associated with a median survival of about 3 months (18). Surprisingly, the patient remained in remission without further therapy for 368 days before exhibiting recurrent AML with both a bladder chloroma and flow-cytometric evidence of MRD (0.04% AML cells) in the bone marrow. He then received reinduction chemotherapy followed by a second TTCR-C4 infusion on day 471 after the 1st infusion with T cells derived from the same pool of transduced cells originally infused, but the leukemia continued to relentlessly progress. This unusual clinical course with a long 1-year remission after multiple relapses led us to investigate potential immunologically related mechanisms of remission and relapse.
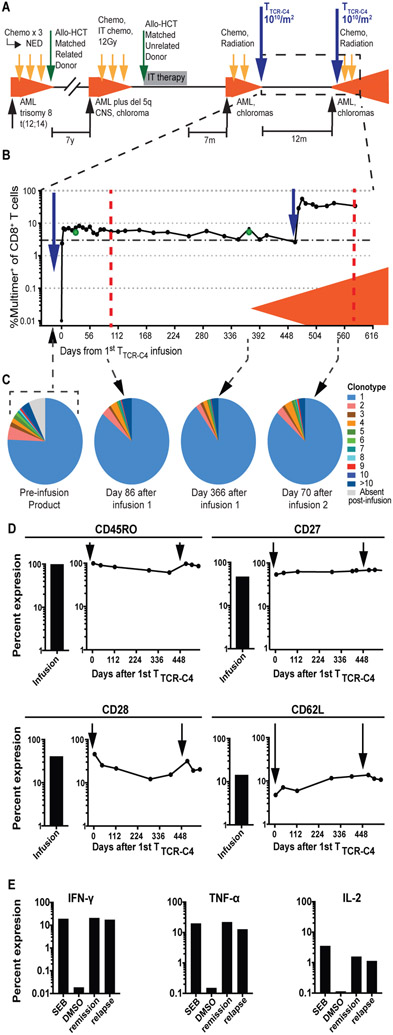
Fig. 1: TTCR-C4 persist and remain functional in the presence of relapsed AML.
(A) Timeline of patient’s treatment regimens. Chemo, chemotherapy; radiation, radiation therapy; IT, intrathecal; NED, no evidence of disease; CNS, central nervous system. (B) Percent multimer+ of CD8+ T cells in PBMCs (solid circles) and bone marrow (green circles) collected before and at defined timepoints after TTCR-C4 infusions are shown. The orange shaded area indicates presence of AML. Dotted red lines represent days 100 and 110 (or day 581 after the 1st) after 1st and 2nd infusion respectively. (C) Pie charts representing individual clonotypes composing the pre-infusion TTCR-C4 product (left) and at indicated timepoints (right) after both infusions are shown. (D) Percent expression of CD45RO, CD27, CD28 and CD62L on infusion products (bar graphs to the left) and at days 4, 42, 114, 289, 393, 478, 508 and 542 after the 1st infusion (graphs to the right) are shown. The three last timepoints are also days 7, 35 and 71 after the second infusion. Timing of infusions are indicated by black arrows. (E) Percent expression of IFN-γ, TNF-α and IL-2 (functional markers) in response to ex vivo stimulation WT1126-134 peptide (1nM) during remission (day 114 after 1st infusion) and relapse (day 581 after 1st infusion, day 110 after 2nd). Maximum and minimal cytokine expression following exposure to Staphylococcal enterotoxin B (SEB) and dimethylsufoxide (DMSO) are shown for each cytokine.
Phenotypically similar, functional TTCR-C4 persist through remission and relapse.
Both infusions of 1010 TTCR-C4/m2 (on days 0 and 471) were administered without prior lymphodepleting chemotherapy. TTCR-C4 in peripheral blood reached peak frequencies of 8.2% and 56% of CD8+ T cells after the 1st and 2nd infusion, respectively (Fig. 1B and Supplementary Methods). Overall, TTCR-C4 frequency remained above 3% with sustained, robust absolute TTCR-C4 counts (fig. S1A). Comparable high TTCR-C4 frequencies were observed in the bone marrow at days 28 and 368 (Fig. 1B, green circles). These high proportions of persistent transferred TTCR-C4 are analogous to results obtained in patients who received TTCR-C4 post-HCT prophylactically to prevent relapse (10). Both infusion products were expanded immediately prior to infusion from identical pre-infusion aliquots. Infusions primarily (>75%) consisted of a single expanded clonotype that also comprised majority of persistent TTCR-C4 after both infusions (Fig. 1C and fig. S1B). TTCR-C4 surface expression of memory-associated markers (CD45RO, CD27, CD28, and CD62L (19)) remained similar after both TTCR-C4 infusions (Fig. 1D, fig. S2A). Ex vivo analysis of recovered TTCR-C4 revealed the cells were functional, maintaining cytokine production in response to targets presenting the cognate WT1126-134 peptide during the period of apparent remission (day 114) and in the context of progressive AML (day 70 or day 541 after the 2nd or 1st infusion, respectively) (Fig. 1E, fig. S2B).
Single-cell RNA sequencing revealed that the TTCR-C4 transcriptome during remission was compatible with recent activation.
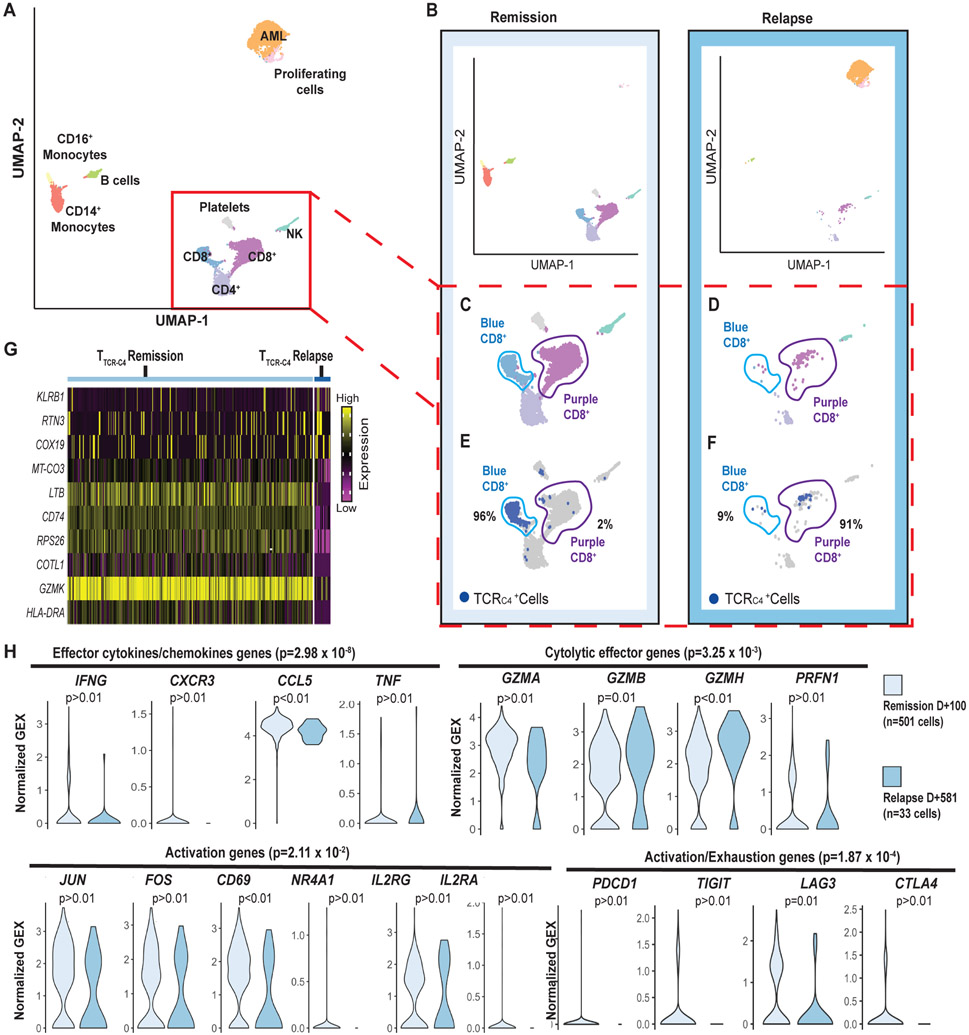
To investigate if transcriptomes of circulating TTCR-C4 differed between clinical remission and relapse, single-cell RNA sequencing (scRNAseq) (20) was performed on available peripheral blood mononuclear cells (PBMCs) 100 days after the 1st TTCR-C4 infusion (remission) and 110 days after the 2nd infusion (relapse), which represents 581 days after the 1st infusion (Fig. 1B, dotted red lines). Data were generated from a single sequencing run containing both samples, reads were aggregated together, and clusters were delineated (Fig. 2A, fig. S3) and then visualized separately, based on barcodes unique to the timepoint (Fig. 2B). Surprisingly, during clinical remission, CD8+ cells grouped into 2 separate and distinct clusters in an unsupervised transcriptional profile analysis (Fig. 2C, light blue and purple clusters), but the light blue cluster was nearly absent at relapse (Fig. 2D). During remission, 96% of TTCR-C4 as identified by TCR-C4 transgene expression, were grouped in the light blue cluster whereas endogenous CD8+ T cells grouped in the purple cluster (Fig. 2E). In contrast, during relapse in the presence of circulating blasts, 91% of TTCR-C4 grouped with the purple endogenous CD8+ cells (Fig. 2F). To probe for differences between TTCR-C4 at remission and relapse, an unsupervised differential gene expression analysis was performed. The top 10 significantly differentially expressed genes (p<0.05 and logFC >∣log2(1.5)∣) showed TTCR-C4 cells at remission had higher expression of genes associated with T cell activation, including lymphotoxin-β [LTB], granzyme K [GZMK], human leukocyte antigen (HLA)-DR [HLA-DR] and CD74 [CD74], which mediates assembly and trafficking of HLA class II complexes (Fig. 2G) (21-24). Gene set analysis of effector cytokine/chemokine genes (25) (interferon (IFN)-γ [IFNG], C-X-C motif chemokine receptor 1 [CX3CR1], C-C motif chemokine ligand 5 [CCL5] and tumor necrosis factor alpha (TNF-α) [TNF]), cytolytic effector genes (25) (granzymes A [GZMA], B [GZMB] and H [GZMH] and perforin [PRF1]), activation/proliferation genes (26) (Jun proto-oncogene [JUN], Fos proto-oncogene [FOS], CD69 [CD69], nuclear receptor subfamily 4 group A member 1 [NR4A1], interleukin (IL)-2 receptor subunit gamma [IL2RG], and IL-2 receptor subunit alpha [IL2RA]) and activation/exhaustion genes (programmed cell death receptor-1 [PDCD1], T-Cell immunoreceptor with Ig and ITIM domains, [TIGIT], lymphocyte activating 3 [LAG3], and cytotoxic T-lymphocyte associated protein 4, [CTLA4]) were significantly enriched in TTCR-C4 at remission versus relapse (p=2.98 x 10−8, p=3.25 x 10−3, p=2.11 x 10−2 and p=1.87 x 10−4 respectively) (Fig. 2H), suggesting repetitive stimulation or activation during remission. Although not every gene in each composite gene-set was individually statistically significant, each of the total gene-sets were significantly different. For example, expression of IFN-γ and TNF-α assessed individually did not achieve statistical significance (p >0.01, p >0.01 respectively) but were significant as part of the effector cytokine/chemokine gene-set (p =2.98 x 10−8), which may reflect both the known decline in expression of these genes after dissociation from the activating signal (27) and potential decline of polyfunctionality from repetitive activation events.
Fig. 2: TTCR-C4 present an activated transcriptional profile during remission but not during relapse.
(A) UMAP visualization of PBMCs from both the remission (100 days after 1st infusion) and relapse (110 days after the second infusion or 581 days after the 1st) (red dotted lines Fig. 1A) samples are visualized together (see Supplementary Methods). PBMCs (n=7704) clustered into populations as indicated by labels. Clustering biostatistical analysis is described in Supplementary Methods and representative marker genes are shown in fig. S3. (B) UMAP visualization of the separated PBMCs obtained at remission (left) and relapse (right). (C) Close-ups are shown for clusters containing T cells during remission with identification of two (light blue and purple) distinct CD8+ T cell clusters based on their transcriptional programs. (D) Close-ups are shown for clusters containing T cells during relapse revealing the CD8+ T cells are predominantly in the purple cluster. (E) Localization of TTCR-C4 (dark blue dots) cells during remission is shown. (F) Localization of TTCR-C4 (dark blue dots) cells during relapse is shown. Percentage of the total TTCR-C4 cells in each cluster are indicated. (G) The heat map shows the 10 most differentially expressed genes (DEG) comparing TTCR-C4 during remission (n=501) to those during relapse (n=33). (H) Analysis of gene-sets representing effector cytokines, cytolytic effector genes activation, and exhaustion genes were compared in TTCR-C4 during remission (light blue) and relapse (darker blue), with each transcript in the gene-set shown as a violin plot. The shape of the violin displays frequencies of values. Model-based Analysis of Single Cell Transcriptomics (MAST) was used to determine the significance shown above each plot. Significance thresholds were set a priori at a threshold of false discovery rate of 5% and positive or negative fold change > log2(1.5).
Furthermore, comparison of endogenous CD8+ T cells from the same sample to remission TTCR-C4 transcriptomes again revealed significant enrichment for the effector cytokine/chemokine, activation/proliferation, and activation/exhaustion gene-sets in TTCR-C4 (p=4.89 x 10−2, p=4.89 x 10−2 and p=0.007 respectively) (fig. S4), suggesting the profile represents antigen-specific events rather than global T cell activation in the host. As previously evidenced, TTCR-C4 remained functional at both timepoints (Fig. 1E), consistent with the T cells being in an activated but not exhausted state. The differences in gene expression (log2-fold change) observed at remission between TTCR-C4 and endogenous cells were lost at relapse (fig. S5).
As TTCR-C4 also contains an endogenous EBV-specific TCR, the patient was screened for evidence of EBV reactivation as a potential explanation for T cell activation. Very few copies of the EBV viral genome was detected immediately after the first TTCR-C4 infusion (10), but none were detected by day 7 or at serial timepoints thereafter (table S1). The results suggest TTCR-C4 may have lysed and eliminated an EBV reservoir shortly after infusion. The possibility of a low degree of EBV antigen presentation leading to signaling through the endogenous EBV-specific TCR cannot be eliminated and could explain T cell activation during the prolonged period of clinical remission. However, the latter appears unlikely, as TTCR-C4 no longer exhibited evidence of activation at relapse, a time when systemic illness might be more likely to lead to EBV reactivation. This further infers that TTCR-C4 at relapse were not responding to leukemia or a low degree of EBV antigen. Consequently, we next evaluated the antigenicity of the patient’s leukemia.
AML during relapse expressed both unmutated WT1 and HLA-A*02:01.
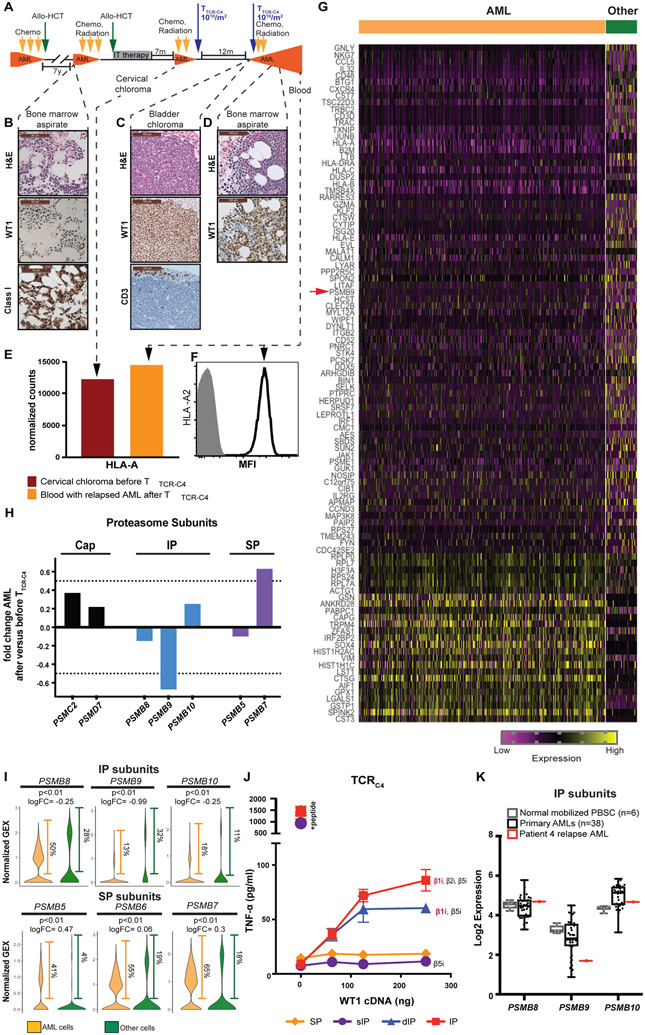
AML samples for immunohistochemistry (IHC) were obtained from a bone marrow aspirate during relapse after the first allo-HCT and prior to the first TTCR-C4 infusion (Fig. 3A and B), from a bladder chloroma at the time of relapse after the 1st TTCR-C4 infusion (Fig. 3A and C), and from a bone marrow aspirate 71 days after the 2nd TTCR-C4 infusion (Fig. 3A and D). All specimens showed abundant WT1 protein expression in the majority (>90%) of leukemic blasts (Fig. 3B to D). Clinical-grade next-generation sequencing performed at relapse after TTCR-C4 (day 58 after the 2nd TTCR-C4 infusion) found no WT1 sequence mutations (see Supplementary Methods). However, CD8+ T cells were not enriched in the chloroma (Fig. 3C), consistent with the lack of an effective T cell response at relapse. To assess potential differences in epitope presentation, HLA class I protein expression was evaluated. On AML prior to the second HCT, IHC could be performed only for total HLA class I expression, which was positive (Fig. 3B). HLA-A expression was evaluable in the cervical chloroma at relapse before any TTCR-C4 infusions, using bulk RNA sequencing on residual clinical material from an archival formalin fixed, paraffin embedded (FFPE) specimen. As a control, HLA-A was also analyzed from PBMC obtained 110 days after the 2nd TTCR-C4 infusion (the same sample on which scRNAseq was performed). HLA-A expression was detected in these samples (Fig. 3E), both of which contained greater than 89% AML. As a note, the NanoString platform utilized permitted only probing for HLA-A and not for a specific allele (see Supplementary Methods). However, HLA-A2 expression was directly evaluable on relapsed AML cells present in PBMCs obtained 110 days after the 2nd TTCR-C4 infusion (Fig. 3F). The presence of HLA class I at all time points, and specifically of HLA-A2 at the last relapse, suggests that HLA-A*02:01 expression was retained throughout the disease. The expression of both unmutated target protein and restricting HLA allele at late relapse excludes antigen loss or mutation as well as restricting HLA loss or downregulation as the cause of AML immune escape in the presence of abundant, potentially reactive, functional TTCR-C4.
Fig. 3: Reduced immunoproteasome expression is observed in relapsed AML.
(A) The timeline shows occurrence of bone marrow aspirate, myeloid chloroma, or blood obtained relative to the timeline of successive therapies. Presence of AML is indicated in orange. (B) Pre-TTCR-C4 AML Hematoxylin and Eosin (H&E) (top panel), immunohistochemistry (IHC) of WT1 expression (middle panel) and HLA class I expression (bottom panel) is shown. Scale bars are 50 μm. (C) H&E (top panel), WT1 IHC expression (middle panel) and CD3 infiltration (bottom panel) is shown for bladder chloroma obtained early post-TTCR-C4 relapse. Scale bars are 100 μm. (D) Post-TTCR-C4 AML H&E (top panel) and WT1 IHC (bottom panel) are shown. Scale bars are 100 μm. (E) Normalized counts (y-axis) of bulk RNA expression of HLA-A obtained from the pre-TTCR-C4 cervical chloroma and a post-TTCR-C4 PBMC sample are shown, both containing greater than 89% AML. (F) HLA-A2 protein expression was detected by flow cytometry of the post-TTCR-C4 blood. MFI, median fluorescence intensity. (G) The heat map shows the top 100 significantly differentially expressed genes (DEG) (p <0.05 and logFC>∣log2(1.5)∣) comparing AML (n=2,447) during relapse after T cell therapy to other cells in the sample (n=306). The red arrow indicates immunoproteasome (IP) subunit β1i [PSBM9] (H) The plot shows the fold change (y axis) of transcripts encoding proteasome cap ([PSMC2] and [PSMD7]; black bars), IP (β5i [PSMB8], β1i [PSMB9] and β2i [PSMB10]; blue bars) and standard proteasome (SP) (β5 [PSMB5], and β2 [PSMB7]; purple bars) sub-units comparing expression in the post-TTCR-C4 obtained from PBMCs to pre-TTCR-C4 AML obtained from a chloroma FFPE sample. β1 [PSMB6] was not available in the commercial RNA probe-set utilized. Bold dotted line indicates 50% change over baseline. (I) Expression of transcripts encoding genes constituting the active proteolytic site of the IP (β5i [PSMB8], β1i [PSMB9] and β2i [PSMB10]) and the SP (β5 [PSMB5], β1 [PSMB6] and β2 [PSMB7]) are shown at relapse comparing AML (n=2,447, orange) and other non-malignant cells (n=306, green) in the sample. Frequencies of cells with the detectable transcript are shown to the right of each violin plot. P values are indicated as well as the log2 fold change. Positive values indicate increased and negative decreased expression in AML. MAST was used to determine significance shown above each plot. Significance thresholds were set a priori at a threshold of false discovery rate of 5% and positive or negative fold change > log2(1.5)). (J) TNF-α production (pg/mL) was measured from T cells expressing TCRC4 cultured with cells expressing a specific proteasome isoform and transfected with varying amounts of WT1 cDNA (29) or pulsed with WT1126-134 peptide as a positive control. Data are presented as mean ± standard deviation. (K) RNAseq on viable leukemic blasts shows expression of genes constituting the active proteolytic site of the IP (β5i [PSMB8], β1i [PSMB9] and β2i [PSMB10]) in peripheral blood stem cells obtained from healthy donors (n=6, light gray), primary AML samples obtained from the Fred Hutch-University of Washington Hematopoietic Diseases Repository (black, n=38) and the patient (red, n=1). Box and whisker plot represents median, interquartile and range. Dots represent individual patient samples.
scRNAseq revealed relapsed AML after TTCR-C4 infusions had low expression of immunoproteasome subunits.
We next investigated antigen processing components in relapsed AML after TTCR-C4 infusions to account for the presumed failure of recognition. Evaluation by scRNAseq of the top 100 significant differentially expressed genes (p<0.05 and logFC>∣log2(1.5)∣) in AML versus non-AML hematopoietic cells (in the same sample) revealed that the AML cells had significantly (p<0.01 and logFC=−0.99) lower expression of the immunoproteasome subunit gene β1i [PSMB9] (Fig. 3G, red arrow), which was confirmed by very low to undetectable β1i protein incorporation in relapsed AML (fig. S6).
Due to the absence of a viably frozen pre-treatment AML sample, it was difficult to directly assess if this patient’s leukemia expressed the immunoproteasome before TTCR-C4 targeting WT1126-134 were initially infused. Therefore, we used residual clinical material from the cervical chloroma FFPE specimen (before the 1st TTCR-C4 administration) and PBMCs obtained 110 days after the 2nd TTCR-C4 infusion (Fig. 3E) to assess changes in proteasome subunit expression. Compared to pre-TTCR-C4, AML post-TTCR-C4 had more transcripts encoding proteins ubiquitously present in the caps of all 26S proteasomes (ATPase 2 [PSMC2] and non-ATPase 7 [PSMD7]) (Fig. 3H, black bars), suggesting comparatively increased total proteasome in AML post-TTCR-C4 (15). However, AML post-TTCR-C4 had less transcripts associated with the immunoproteasome subunits, β5i [PSMB8] (fold change −0.14) and β1i [PSMB9] (fold change −0.67), which would restrict immunoproteasome formation. Although β2i [PSMB10] was not decreased (fold change +0.25), its incorporation into an immunoproteasome requires β1i, as its presence alone cannot drive immunoproteasome formation (Fig. 3H, blue bars) (28). In contrast, although β1 was unavailable for assessment (see Supplementary Methods), an increase in β2 [PSMB7] (fold change +0.63) and minimal change in β5 [PSMB5] (fold change −0.1) was detected, suggesting that the AML may have preferentially formed the standard proteasome (Fig. 3H, purple bars). The decrease in β1i resulted in minimally detectable transcripts in the post-TTCR-C4 specimen (Fig. S6), confirmed by absent β1i protein expression (fig. S7). Furthermore, expression of transcripts encoding proteins constituting the active proteolytic site of the immunoproteasome (β5i [PSMB8], β1i [PSMB9], and β2i [PSMB10]) were significantly (p<0.01) decreased, whereas those constituting the active proteolytic site of the standard proteasome (β5 [PSMB5], β1 [PSMB6] and β2 [PSBM7]) were significantly increased (p<0.01) in the AML cells compared to other hematopoietic cells (Fig. 3I). Likewise, the frequency of cells expressing immunoproteasome subunits β1i [PSMB9], and β2i [PSMB10] were markedly lower in the relapsed AML, whereas the frequency of cells expressing standard proteasome subunits β5 [PSMB5], β1 [PSMB6] and β2 [PSBM7] was increased compared to other hematopoietic cells. Immunoproteasome subunit β5i [PSMB8], however, opposed this trend with 50% of relapsed AML cells expressing the transcript compared to 28% of other hematopoietic cells. Reduced [PSMB9] expression was not a result of gene mutation, as whole exome sequencing on the relapsed AML did not reveal any pathogenic mutations in PSMB9 (See Supplementary Methods). Moreover, the relapsed AML retained similar expression of IFN-γ response genes as other cells in the PBMCs, suggesting cytokine responsiveness remained intact (fig. S8).
To confirm that PSMB9 is required for WT1126-134 processing and TTCR-C4 recognition, TTCR-C4 were co-cultured with cells lines genetically modified to express the immunoproteasome, standard proteasome, an intermediate proteasome with a single immunoproteasome subunit (β5i, β1, β2, (sIP)), or an intermediate with two immunoproteasome subunits (β5i, β1i, β2, (dIP)) (29). Indeed, TTCR-C4 were found to only recognize cell lines expressing proteasome isoforms containing β1i (Fig. 3J).
To assess whether low PSMB9 may be present in other AML specimens, which would suggest this may be a more common phenomenon, we compared the relapsed leukemia to data from six normal-donor mobilized peripheral blood stem cell samples and 38 enriched primary AML blast samples obtained from the Fred Hutch-University of Washington Hematopoietic Diseases Repository. PSMB9 (β1i) RNA expression in this patient’s relapse AML was markedly lower compared to other AML specimens, but this reduction was not rare, as eight other AMLs also had low PSMB9 expression (Fig. 3K). With the caveat that adequate AML cells prior to the final relapse were unavailable for detailed analysis, these data suggests that the absence of β1i in this patient’s AML at relapse may have compromised recognition by TTCR-C4.
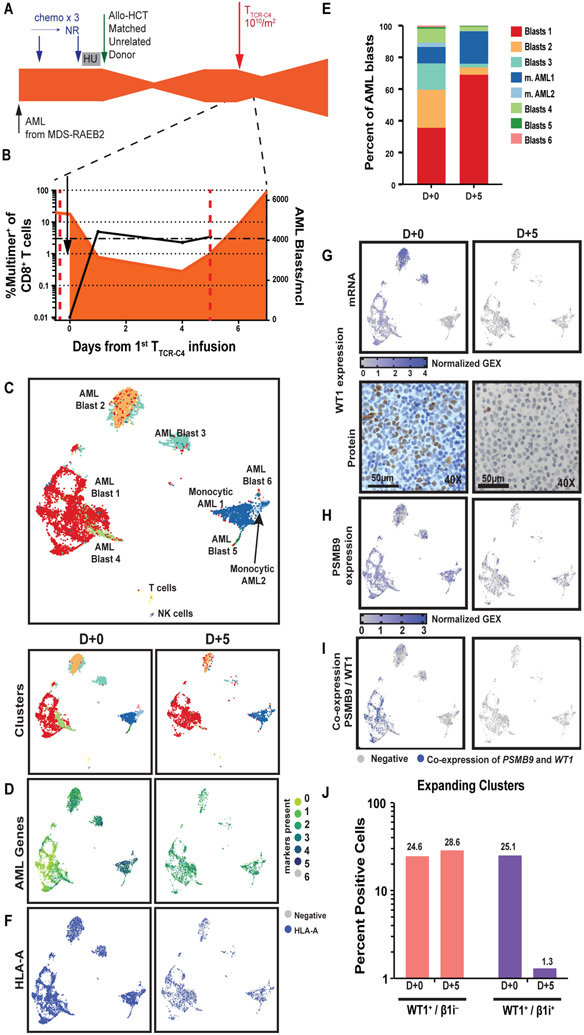
TTCR-C4 preferentially eliminates WT1+ AML blasts co-expressing PSMB9.
A second patient (Patient 2) on this trial was identified who received TTCR-C4 and had blasts available before and after T cell infusion (Fig. 4A and Supplementary Methods). This patient’s AML constituted 70.2% of PBMCs at the time of TTCR-C4 infusion. Although the absolute blast count moderately decreased immediately after infusion, the AML eventually progressed despite persistence of TTCR-C4 at greater than 3% of peripheral CD8+ T cells, and he succumbed to progressive disease 15 days after transfer (Fig. 4B, fig. S9). ScRNAseq on PBMCs obtained before TTCR-C4 infusion showed the AML blasts were heterogenous and formed seven leukemic clusters (Fig. 4C, fig. S10), all expressing one or more of the AML-associated genes CD34, CCND1, CDK6, FLT3, NPM1 and RUNX1(Fig. 4D, Fig. S11). Five days after infusion, clusters “AML Blasts 2” and “AML Blasts 3” decreased whereas clusters “AML Blasts 1” and “monocytic AML1” expanded (Fig. 4E). The expression of AML-associated genes was unchanged (fig. S11). HLA-A also remained expressed (Fig. 4F). However, the frequency of WT1-expressing cells decreased from 27% to 2.5% after TTCR-C4 exposure, with the most noticeable reduction in the two clusters that expressed the highest abundance of WT1 (Fig. 4G, top, corresponding to “AML Blasts 2” and “MAL Blasts 3” in Fig. 4C and E) WT1 loss was confirmed by IHC (Fig. 4G, bottom). PSMB9-expressing AML cells were detected after TTCR-C4 (Fig. 4H). In all blast populations expressing WT1 and β1i, but especially in the expanding clusters (“AML Blasts 1” and “monocytic AML1”), only the proportion of cells that co-expressed WT1 and β1i (PSMB9) was reduced after TTCR-C4 exposure (25.1% to 1.3% or a 19-fold reduction), whereas the proportion of cells that only expressed WT1 remained proportionately similar (24.6% to 28.6%) (Fig. 4I and J). These data support the hypothesis that TTCR-C4 exposure only efficiently eliminates WT1+ blasts that also express β1i.
Fig. 4: TTCR-C4 preferentially eliminates WT1+ AML co-expressing β1i.
(A) Timeline of patient’s treatment regimens. Chemo, chemotherapy; NR, no response; HU, hydroxyurea. (B) Percent multimer+ of CD8+ T cells is shown in peripheral blood collected before and at day +1, +4, and +5. The orange shaded area indicates presence of AML. Dotted red lines represent days +0 and +5 after TTCR-C4. (C) UMAP visualization is shown for scRNAseq on PBMCs from both before (D+0, n=7,381) TTCR-C4 infusion and 5 days post 1st infusion (D+5, n=3,319) (red dotted lines shown in Fig. 4B) aggregated into a single UMAP (top) or as individual time points (bottom, D+0 left and D+5 right). (D to I) UMAP of AML clusters on D+0 (left) or D+5 (right) are shown. (D) Expression of AML specific markers CD34, CCND1, CDK6, FLT3, NPM1, RUNX1 are shown. (E) Relative proportions of AML clusters at D+0 and D+5 are shown. (F) Expression of the gene encoding HLA-A is shown. (G) Expression of WT1 transcripts from peripheral AML (top panel) and immunohistochemistry (bottom panel) of WT1 protein performed on a bone marrow biopsy (D+0) and a PBMC cell pellet (D+5) are shown. (H) Expression of immunoproteasome gene β1i (PSMB9) is shown. (I) Co-expression of WT1 and PSMB9 is shown. (J) The bar graph shows the percent of cells that express WT1 only (pink bars) or WT1 and β1i ([PSMB9]) (purple bars) before (D+0) and after (D+5) TTCR-C4 infusion. Percent values are indicated above each column.
Targeting WT137-45 can overcome an absence of WT1 immunoproteasome processing and expand therapeutic potential to solid tumors.
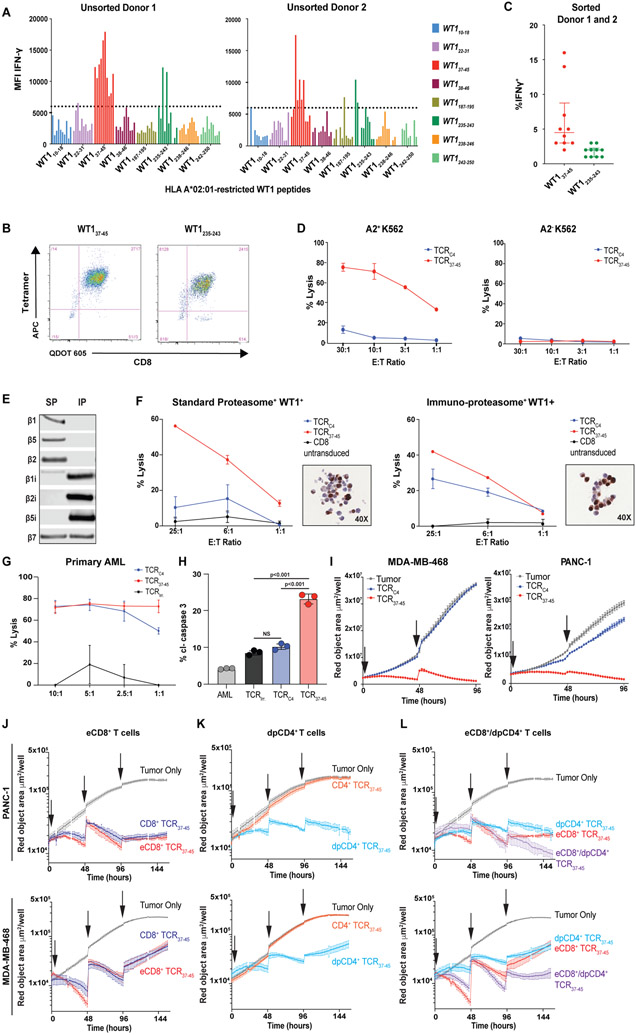
Identifying epitopes not requiring a specific proteasome isoform might avoid potential immunotherapy resistance and thereby broaden the clinical translation of targeting WT1. We therefore investigated whether targeting alternative HLA-A*02:01-restricted WT1 epitopes could have a therapeutic potential towards AML with low proteasome expression, presumed resistant to TTCR-C4, or solid tumors that generally lack immunoproteasome processing. We generated T cell lines from 15 healthy donors (two shown) recognizing eight previously described HLA-A*02:01-restricted WT1 epitopes (Fig. 5A) (30). Our screen identified WT137-45 (also identified by Ruggiero et al.(31)) and WT1235-243 as promising immunogenic candidates. Cell lines targeting WT137-45 and WT1235-243 were sorted on tetramer positivity (Fig. 5B) and assessed for reactivity towards the HLA-A*02:01 transfected, WT1+ K562 cell line which primarily expresses the standard proteasome and is not lysed by T cells targeting WT1126-134 (Fig. 5C and D) (16). A high affinity TCR reactive to WT137-45 (TCR37-45) was isolated from the cell lines and inserted into a lentiviral backbone (see Supplementary Methods). CD8+ T cells expressing TCR37-45 (TTCR37-45) recognized (fig. S12) and lysed HLA-A2- and WT1-transduced HEK-293 cell lines (endogenous standard proteasome expression) engineered or not to express the immunoproteasome (Fig. 5E and F, see Supplementary Methods) (29). Of note, WT1 expression was not uniform in the cells from the WT1-transduced immunoproteasome and standard proteasome cell lines (Fig. 5F), which limited the maximal lytic activity achieved in this assay. CD8+ T cells expressing either TCRC4 or TCR37-45 lysed HLA-A2+WT1+primary AML cells (Fig. 5G), but only TCR37-45 induced caspase 3 cleavage in the first patient’s relapsed, TTCR-C4-resistant AML (Fig. 5H). Furthermore, only TCR37-45 reduced growth of the WT1 expressing solid tumor cell lines PANC-1 (32) and HLA-A*02:01-transduced MDA-MD-683 (33) in a live tumor visualization assay (Fig. 5I, see Supplementary Methods). Exposing the PANC-1 cell line to IFN-γ for 48 hours increased β1i expression (fig. S13A) and enhanced recognition by TCRC4 (fig. S13B).
Fig. 5: Identification of an alternate HLA-A*02:01-restricted WT1 peptide (WT137-45 ) which is not dependent on immunoproteasome processing.
(A) Ten CD8+ T cell lines from 2 HLA-A*02:01-expressing healthy donors were generated against 8 candidate HLA-A*02:01-binding WT1 peptides (x-axis). Each cell line’s ability to produce IFN-γ following stimulation with the immunoproteasome-deficient but WT1+ HLA-A*02:01-transduced K562 cell line at an E:T of 1:1 was assessed. Peptides were considered to have elicited high reactivity from donor T cells as measured by MFI (above the arbitrary threshold of 6000) for the IFN-γ positive cells. (B) Representative flow plots show CD8+ T cell lines targeting WT137-45 or WT1235-243 that were sorted using the corresponding tetramers. (C) Sorted T cell lines targeting WT137-45 had the highest comparative reactivity, assessed by IFN-γ MFI, when cultured with HLA A*02:01-tansduced K562 cells. TCRs targeting WT137-45 were isolated from T cells in these lines. Data are presented as median with the interquartile range. All data points are shown. (D) Lysis of K562 ± expression of HLA-A*02:01 by CD8+ T cells from a healthy donor transduced with TCRC4 or a TCR targeting WT137-45 (TCRWT1-37-45 #1) confirmed that expression of the WT137-45 peptide was not dependent on IP processing. Error bars show standard deviation of triplicate wells. (E) The immunoblot shows protein expression of catalytic proteasome subunits in HEK-293 (endogenous SP expression) ± engineering to express the IP. (F) Percent lysis (y-axis) is shown for IP/SP-skewed HEK-293 cells lines by CD8+ T cells transduced to express either TCRC4 (blue lines), TCR37-45 (red lines) and untransduced CD8+ T cells (black lines) at indicated effector to target (E:T) ratios (x-axis). Inset depicts WT1 IHC of IP/SP HEK-293 cells transduced with WT1-and HLA-A*02:01. IP/SP cell lines demonstrate heterogeneity of WT1 expression. Error bars show standard deviation of triplicate wells. (G) Percent lysis (y-axis) of a primary leukemia by CD8+ T cells transduced to express either an irrelevant TCR (TCRirr.; black lines), TCRC4 (blue lines) or TCR37-45 (red lines) is shown. (H) Percent AML cells expressing cleaved caspase 3 (cl-caspase 3, y-axis) is shown for Patient 1’s relapsed AML either alone (gray) or cultured with CD8+ T cells with the endogenous TCR removed using CRISPR-Cas9 and transduced to express TCRirr. (black), TCRC4 (blue), or TCR37-45 (red). Error bars show standard deviation of triplicate wells. (I) Growth kinetics are shown for WT1+ cell lines PANC-1 and HLA-A*02:01-transduced MDA-MD-468 in live tumor-visualization assay in the absence (gray lines), or presence of CD8+ T cells transduced to express either TCRC4 (blue lines) or TCR37-45 (red lines). Effector to target (E:T) ratio of 10:1 was used and arrows indicate addition of tumor cells to culture. Standard error of triplicate wells are shown. (J to L) Growth kinetics are shown for WT1+ cell lines PANC-1 and HLA-A*02:01-transduced MDA-MB-468 in live tumor-visualization assay in the absence (gray lines), or presence of TTCR37-45 cells. (J) CD8+ T cells were transduced with only TCR37-45 (light-blue line) or a poly-cistronic construct containing TCR37-45 and CD8αβ (red line, eCD8+). (K) CD4+ T cells were transduced with only TCR37-45 (blue line) or a poly-cistronic construct containing TCR37-45 and CD8αβ (green line, dpCD4+). (L) A comparison of eCD8+ alone (red line), dpCD4+ alone (green line) or eCD8+/dpCD4+ in combination (1:1 ratio, purple line) is shown. For all conditions, arrows indicate addition of tumor cells to culture. Standard error of triplicate wells are shown. 4:1 E:T ratio, total T cells was kept consistent for all conditions.
In anticipation of targeting WT137-45 for future clinical translation in both solid and liquid tumors, we sought to maximize the clinical efficacy by also modeling the engagement of both CD4+ and CD8+ T cells, as has proven efficient for CAR T cells approaches with a 1:1 ratio CD8+:CD4+ (34). Antigen-specific CD4+ T cells, restricted by HLA class II, enhance CD8+ T cell survival, proliferation and function (35). As HLA class II expression is not always expressed by tumors (36), an alternative is to impart tumor recognition to CD4+ T cells through expression of a class I-restricted TCR that would co-localize engineered CD8+ and CD4+ T cells on the same tumor target without requiring HLA class II expression. Although high affinity TCRs can engage CD4+ T cells without CD8αβ binding, TCRs with such high affinity are rarely detectable, especially for targeting self-proteins such as WT1 (37). To engineer functional CD4+ T cells targeting WT137-45 we incorporated CD8αβ along with TCR37-45 into a lentiviral vector (see Supplementary Methods). Transduction with this vector generated cells with additional CD8 on the surface yielding ‘enhanced’ CD8+ T cells (eCD8+) and CD4+ T cells co-expressing CD8 and CD4 (double positive CD4+, dpCD4+). We next assessed the relative contribution of the CD8αβ to the function of CD8+ and CD4+ T cells individually and in combination by intentionally decreasing the effector to target ratio from 10:1 to 4:1 and repetitively adding tumor cells to “stress” the engineered T cells. Increased expression of CD8αβ did not substantially improve tumor control by eCD8+ cells compared to TCR-transduced CD8+ T cells (Fig. 5J), but dpCD4+ did mediate a sustained antitumor response, whereas CD4+ T cells transduced with the TCR alone were ineffective (Fig 5K). Furthermore, a 1:1 mixture of eCD8+/dpCD4+ TTCR37-45 cells more effectively controlled both solid tumor cell lines, PANC-1 and HLA-A*02:01-transduced MDA-MB-468, than either eCD8+ or dpCD4+ TTCR37-45 cells alone (Fig. 5L). Of note, none of these combinations were effective in clearing PANC-1 targets when CD8αβ was added to TCRC4 in CD4+ and CD8+ T cells, suggesting the addition of CD8αβ does not rescue reactivity in the context of limited antigen processing (fig. S14). DpCD4+ T cells, when compared to eCD8+ T cells alone, generally secreted abundant inflammatory cytokines (IFN-γ, TNF-α, IL-2), cytokines associated with a helper function (IL-6, IL-17, and IL-21) or a Th2 response (IL-4, IL-13) and limited IL-10 associated with regulatory function (38). Although co-cultures of eCD8+/dpCD4+ resulted in less abundant cytokines compared to dpCD4+ T cells alone, all cytokines, except for IL-6, were significantly (p <0.01) greater for eCD8+/dpCD4+ than for eCD8+ T cells alone suggesting cytokines secreted by dpCD4+ in co-culture may have provided support for the effector function of eCD8+ T cells (fig. S15). Together, these in vitro data suggest TCRs targeting WT137-45 might be efficient against AMLs and solid tumors with limited immunoproteasome expression (15).
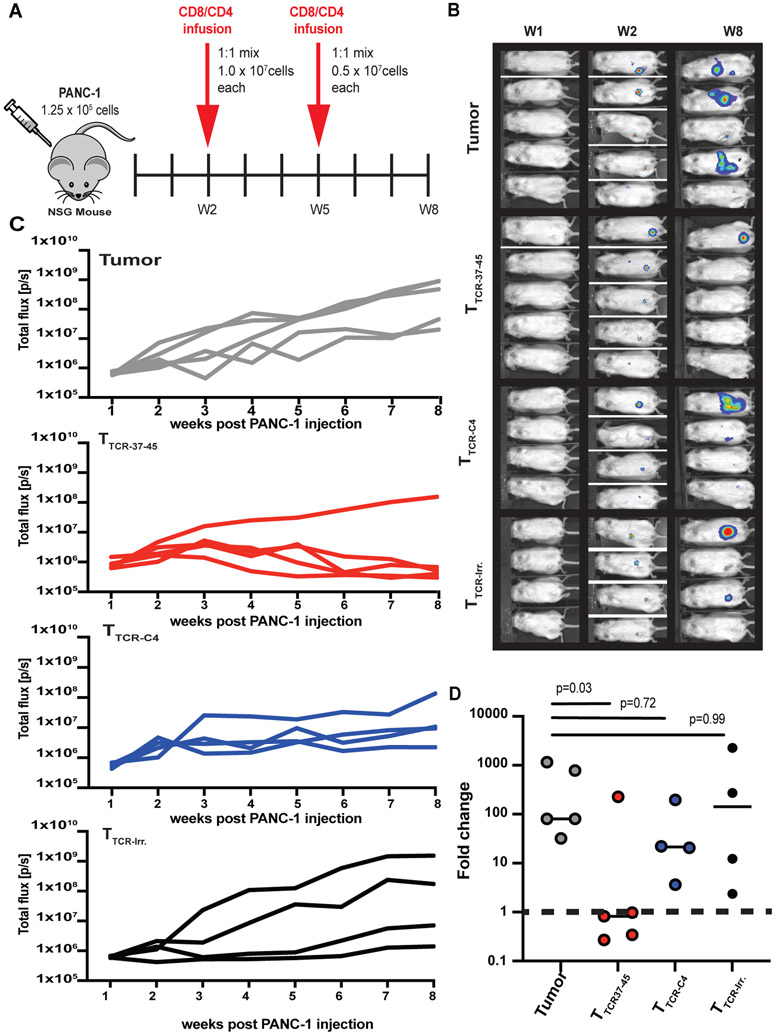
In a murine model, TTCR37-45 cells but not TTCR-C4 cells reduced the tumor burden of PANC-1, a solid tumor with limited β1i.
We next investigated whether eCD8+/dpCD4+ TTCR37-45 could reduce the burden of established PANC-1 tumors, which incorporate limited β1i into the proteasome without prolonged exposure to IFN-γ (fig. S13A). We used a NOD-scid IL2Rgammanull (NSG) mouse model with established PANC-1 tumors to compare the activity of eCD8+/dpCD4+ with TTCR37-45 or TTCR-C4 or irrelevant control (TTCR-irr) (Fig. 6A). Indeed, eCD8+/dpCD4+ TTCR37-45 effectively reduced detectable tumor in 4 of 5 mice, but TTCR-C4 and TTCR-Irr did not (Fig. 6B and C). Thus, TTCR37-45 was the only transduced T cell that significantly (p =0.03) reduced PANC-1 tumor compared to tumor alone (Fig. 6D). Moreover, eCD8+/dpCD4+ TTCR-37-45 T cells also controlled PANC-1 tumor outgrowth (P<0.0001) in a ‘preventive’ NSG mouse model (fig. S16).
Fig 6: TTCR37-45 cells reduce PANC-1 solid tumor burden in an NSG mouse model.
(A) Schematic for NSG experimental treatment model. (N=2) Data from one of two biological replicates is shown. (B) Bioluminescence images of mice are shown at weeks 1, 2, and 8 post PANC-1leu injection. All mice had detectable tumors at the time of adoptive transfer. (C) Total flux (p/s) for each treatment group was measured over time: Tumor (No T cells, n=5); TTCR37-45 (n=5); TTCR-C4 (n=4) TTCR-Irr. (n=4). (D) Fold change relative to baseline (Week 1) at week 8 is shown for each treatment group. Data in (D) were analyzed by a Kruskal-Wallis test for non-parametric data. n.s., not significant. Horizontal bars indicate the mean.
Discussion
Here, we show evidence pointing to a previously undescribed mechanism of AML resistance to immunologic targeting that is proteasome-dependent. Proteasomes are responsible for the degradation of proteins into peptides subsequently presented by HLA class I molecules, and the enzymatic composition of the proteasome can determine precisely which peptides are presented (39). In our first clinical case, escape under the immunologic pressure of a focused antigen-specific T cell response may have resulted in selection of leukemic cells with altered protein processing machinery that abrogated presentation of the targeted epitope.
The standard proteasome is expressed in all somatic cells and contains a catalytic core composed of three enzymatic subunits (β1, β2, and β5). The immunoproteasome, by contrast, is expressed in hematopoietic cells and can be induced in most somatic cells by IFN-γ. It has a different catalytic core comprised of distinct enzymatic β-subunits (β1i, β2i, and β5i). The immunoproteasome and standard proteasome each produce a broad spectrum of peptides, including many identical peptides, but the immunoproteasome produces a more diverse set of peptides that are generally considered more immunogenic (39). Proteasomes assemble in a cooperative fashion. Subunits β2i and β1i are mutually incorporated into the pro-form of the proteasome and promote subsequent incorporation of β5i to form a functional immunoproteasome (28). In the absence of β2i and β1i, β2 is incorporated which inhibits β2i incorporation and prompts standard proteasome formation through incorporation of β5 then β1 (40). Formation of immunoproteasome is more efficient and takes precedence over standard proteasome formation, which is evident by the dominant presence of intact immunoproteasomes in cells that express the catalytic components of both proteasome types (40, 41). Unlike solid tumors that preferentially express the standard proteasome, AML and other hematopoietic cells generally express the immunoproteasome predominantly, and thus largely display peptides that have been processed by the immunoproteasome (42). As the WT126-134 peptide appears to be exquisitely dependent on the presence of β1i as an integral part of the immunoproteasome, processing of this epitope would be severely hampered following loss or downregulation of β1i
The proteolytic machinery of tumor cells has been shown to impact the outcome of epitope-specific immunotherapies, and immunoproteasome deficiency is a feature of limited response to immunotherapy in solid tumors (43, 44). For example, a MAGE-A3 HLA-B40 restricted peptide was previously shown to be exclusively presented by immunoproteasome-expressing tumor cells (45), and the targeted WT1126-134 epitope was shown to be poorly or not presented by solid tumor lines (16). By contrast, efficient presentation of the melanoma-antigen 1 (MART1) epitope, MART126-35, and of a renal cell carcinoma antigen, depends on standard proteasome processing and is obstructed by induction of the immunoproteasome, which no longer produces these epitopes and thus paradoxically can allow tumors to evade elimination by antigen-specific T cells that are producing IFN-γ following target recognition (46). Alternative processing defects can also be responsible for tumor evasion. In mice that lack expression of the endoplasmic reticulum aminopeptidase 1 (ERAP1), a component of the antigen processing machinery that trims peptides in the ER to fit in the binding groove of HLA class I molecules, HLA class I-positive, IFN-γ-responsive tumors progressed despite the presence of functional, antigen-specific cytotoxic T lymphocytes due to failure to present trimming-dependent epitopes (47). Consistent with our results, these findings highlight how changes in a tumor cell’s antigen processing machinery can disrupt immune recognition, particularly if a single epitope is targeted.
Our study has limitations and raises questions that warrant future elucidation. We only observed one such ‘resistant’ case in this small initial trial and no additional HLA-A2+, WT1+, PSMB9low primary AMLs could be obtained from our repository. Thus, no conclusions can be derived as to the frequency of this mechanism either under natural immunologic pressure or as a result of immunotherapy. The infused TTCR-C4 also contained their endogenous EBV-specific TCR, and signaling through this receptor cannot be formally excluded as an explanation for the activation signature observed at remission. The timing of the clinical sampling for the first patient and the unavailable ‘fresh’ leukemia prior to TTCR-C4 infusion, hampered the ability to unequivocally demonstrate the recurrence was due to β1i loss. Although we observed a roughly 60% reduction in PSMB9 transcripts post TTCR-C4 (Fig. 3H, Supplementary Methods), the unavailability of ‘fresh’ leukemia before TTCR-C4 infusion did not allow for a direct comparison of incorporated proteasome subunits before and after TTCR-C4 by immunoblot. The presence of other leukemias with low β1i and the progression of the second patient’s AML with reduced WT1/β1i co-expression after TTCR-C4 suggests there are many other potential obstacles to efficacy with engineered T cells. Establishing a murine model that can recapitulate loss of β1i as a result of immunological pressure or more generally the study of AML in the presence of T cell infusions to investigate escape mechanisms is warranted, especially as TCR gene therapy becomes more common.
The results underscore the importance of epitope selection for T cell therapy or vaccination strategies, in which the goal is to enhance the magnitude of T cell responses to selected epitopes (48). In particular, epitopes commonly identified in public databases reflect responses to epitopes expressed by target cells in inflammatory conditions or presented by professional antigen-presenting cells, both of which likely express the immunoproteasome, which can lead to selection of potentially sub-optimal targets. In the face of tumor heterogeneity, strategies targeting multiple proteins or epitopes using engineered T cells will likely be necessary. Targeting epitopes efficiently processed by both the standard proteasome and immunoproteasome may also increase the chances of circumventing proteasome-dependent immune-evasion (15). We pursued the latter strategy in searching for an epitope not exclusively dependent on the immunoproteasome. Our results suggest that selecting T cells/TCRs recognizing WT137-45 (See also, Ruggiero et al.(31)) rather than WT1126-134 should assure that changes in the relative immunoproteasome expression cannot abrogate presentation of WT1-expressing cells and allow for immune evasion, thus promoting successful clinical translation both for vaccines and TCR-engineered T cell therapies.
Materials and Methods:
Study Design.
The goal of this study was to uncover the molecular mechanisms underlying observed clinical relapse for two patients treated with TTCR-C4 in arm two of this clinical trial (NCT1640301) (10). Both patients presented with circulating AML in the presence of persisting therapeutic TTCR-C4 T cells. First, scRNAseq was performed on both patients at two time points relevant to their respective clinical courses (as materials allowed). This facilitated simultaneous investigation of immune cells and leukemic cells for potential mechanisms of evasion in an unbiased fashion. Patient 1 showed a marked decrease in PSMB9 expression, a subunit of the immunoproteasome complex known to be necessary for generation of the WT1126-134 peptide suggesting a probable mechanism of immune-evasion. Subsequent experiments were aimed at validating this hypothesis and determining which immunoproteasome β-subunits were critical for generating WT1126-134. We then sought to find an alternative TCR not reliant on immunoproteasome subunits for presentation. Additional peptides were screened for immunogenicity from fifteen donors and candidate TCRs were tested for ability to lyse the immunoproteasome deficient, WT1+ leukemia line, K562. The resulting TCR recognized WT137-45 (TTCR37-45). Next, we tested the ability of TTCR37-45 to recognize Patient 1’s relapsed leukemia. Finally, we investigated the efficacy of TTCR37-45 in an immunoproteasome deficient solid tumor model, in vitro and in vivo. Female, NSG (Jackson Laboratory, 005557) mice six to eight weeks of age were used in our in vivo studies. Mice in our preventative model (n = 10, n= 5 in each group) were randomly assigned a treatment group prior to TTCR37-45 infusion, which was administered by researchers in our group. Mice enrolled in our treatment study (n = 32) were randomly assigned a treatment group after tumor was detected. Our pre-established exclusion criteria excluded mice without detectable tumor leaving 18 mice included in our study, which were randomly divided in to four groups. Tumor burden was measured in each group prior to infusion to confirm there was no significant difference between groups (p ≥ 0.64) and assessed each week thereafter. T cell infusions were administrated by core facility in this blinded study. Fold change in tumor was calculated by dividing the total flux detected on week eight by the total flux detected on week two immediately prior to T cell infusion. ARRIVE guidelines were followed for in vivo studies.
Statistical Analysis.
A variety of statistical methods were used in this investigation requiring a specific test depending on the data type. Each test is described in the respective section of the supplementary materials where the technique was used. Briefly, all non-sequencing statistical analyses were done using GraphPad Prism. For in vitro comparisons of TCRs (TTCR-C4, TTCR37-45, TTCR-Irr. versus tumor/AML alone), a one-way analysis of variance (ANOVA) with Tukey multiple comparisons was used after normal data distribution was determined by a Shapiro-Wilk test. For our mouse model in vivo comparisons of TCRs (TTCR-C4, TTCR37-45, TTCR-Irr. versus tumor/AML alone), a one-way ANOVA with Tukey’s multiple comparison test was used to confirm there was no significant difference between the groups prior to infusion and, post-therapy, a Kruskal-Wallis test for non-parametric data was used. For comparisons of TTCR37-45 to tumor alone, a two-tailed t test was used. For scRNAseq, significance thresholds were set a priori at a threshold of false discovery rate of 5% and positive or negative fold change > log2(1.5).
Data and materials availability:
All data associated with this paper are in the paper or supplementary materials and the clinical protocol is available upon request. All requests for raw data, code, and materials are promptly reviewed by the Fred Hutchinson cancer Research Center to verify if the request is subject to any intellectual property confidential obligations. WT1-specific TCRs are available from Fred Hutchinson Cancer Research Center under a material transfer agreement for preclinical research purposes only. Patient-related data not included in the paper were generated as part of a clinical trial and may be subject to patient confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. PBMC scRNAseq is available at the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) Accession number: GSE190501. R code used to generate the UMAP plots and perform data analysis was performed with Seurat software packages; this is described and referenced in the corresponding supplementary materials and methods.
Supplementary Material
Acknowledgments:
We thank the Fred Hutch Good Manufacturing Practice Cell Processing Facility for generating TTCR-C4, the Immune Monitoring Laboratory for generating tetramers, the Flow Cytometry Facility for providing instruments and assistance in flow cytometry assays, the Genomics Core for sequencing and data processing, the Program in Immunology, Immunotherapy Integrated Research Center, Seattle Cancer Care Alliance Immunotherapy Clinic staff, The Fred Hutch/University of Washington Hematopoietic Disease Repository for providing primary AML samples, and patients and their families.
Funding:
This work was supported by P01CA18029-41 (to PDG), SITC-Merck Immunotherapy Fellowship (to KGP), National Institutes of Health T32CA080416 (to MCL), National Institutes of Health 5K08CA169485 (to AGC), Immunotherapy Integrated Research Center at FHCRC (to AGC), Damon Runyon (to AGC), Guillot Family Zach Attacks Leukemia Foundation (to AGC and PDG), Juno Therapeutics (to PDG), and the Ludwig Institute for Cancer Research (to BJVdE).
Footnotes
Competing interests: AGC and PDG have received support from Juno Therapeutics. PDG was a consultant, has received support from, and had ownership interest in Juno Therapeutics. He consults and has ownership interest in RAPT Bio., Elpiscience, Celsius, and Nextech. PDG and TMS are inventors on patent application PCT/US2015/042986 held/submitted by Fred Hutchinson Cancer Research Center that covers WT1 p126-specific TCRs that has been licensed to Juno Therapeutics, a Bristol-Myers Squibb Company. PDG,TMS and AGC are inventors on patent application PCT/US2016/068556 submitted by Fred Hutchinson Cancer Research Center and Adaptive Biotechnologies Corporation that covers WT1 p37-specific TCRs. Fred Hutchinson Cancer Research Center has licensed its interest to Juno Therapeutics, a Bristol-Myers Squibb Company. AGC and KP have received reagents from 10X Genomics. RG has received consulting income from Illumina, Juno Therapeutics, Takeda, Infotech Soft, Celgene, Merck. RG has received research support from Janssen Pharmaceuticals and Juno Therapeutics. RG declares ownership in Ozette technologies, Modulus Therapeutics, CellSpace Biosciences and minor stock ownership in 10X Genomics. BJVdE has ownership interests in iTeos Therapeutics and Oncorus. BJVdE is a consultant for iTeos therapeutics, Amgen, Oncorus, and Vaccitech. CCY is a consultant for Merck, Celgene, Twinstrands, Eli Lilly (loxo), Blueprint, and OBI. The other authors declare no other competing interests. MB is now employed by BMS who has licensed the aforementioned patents.
References and Notes:
- 1.Dickinson AM, Norden J, Li S, Hromadnikova I, Schmid C, Schmetzer H, Jochem-Kolb H, Graft-versus-Leukemia Effect Following Hematopoietic Stem Cell Transplantation for Leukemia. Front Immunol 8, 496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim F, Ang SO, Emerging CAR landscape for cancer immunotherapy. Biochem Pharmacol 178, 114051 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Perna F, Berman SH, Soni RK, Mansilla-Soto J, Eyquem J, Hamieh M, Hendrickson RC, Brennan CW, Sadelain M, Integrating Proteomics and Transcriptomics for Systematic Combinatorial Chimeric Antigen Receptor Therapy of AML. Cancer Cell 32, 506–519 e505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone JD, Kranz DM, Role of T cell receptor affinity in the efficacy and specificity of adoptive T cell therapies. Frontiers in immunology 4, 244 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugiyama H, WT1 (Wilms' tumor gene 1): biology and cancer immunotherapy. Jpn J Clin Oncol 40, 377–387 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Narayan R, Olsson N, Wagar LE, Medeiros BC, Meyer E, Czerwinski D, Khodadoust MS, Zhang L, Schultz L, Davis MM, Elias JE, Levy R, Acute myeloid leukemia immunopeptidome reveals HLA presentation of mutated nucleophosmin. PLoS One 14, e0219547 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berlin C, Kowalewski DJ, Schuster H, Mirza N, Walz S, Handel M, Schmid-Horch B, Salih HR, Kanz L, Rammensee HG, Stevanovic S, Stickel JS, Mapping the HLA ligandome landscape of acute myeloid leukemia: a targeted approach toward peptide-based immunotherapy. Leukemia 29, 647–659 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Ochsenreither S, Fusi A, Busse A, Bauer S, Scheibenbogen C, Stather D, Thiel E, Keilholz U, Letsch A, "Wilms Tumor Protein 1" (WT1) peptide vaccination-induced complete remission in a patient with acute myeloid leukemia is accompanied by the emergence of a predominant T-cell clone both in blood and bone marrow. J Immunother 34, 85–91 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Chapuis AG, Ragnarsson GB, Nguyen HN, Chaney CN, Pufnock JS, Schmitt TM, Duerkopp N, Roberts IM, Pogosov GL, Ho WY, Ochsenreither S, Wolfl M, Bar M, Radich JP, Yee C, Greenberg PD, Transferred WT1-Reactive CD8+ T Cells Can Mediate Antileukemic Activity and Persist in Post-Transplant Patients. Science translational medicine 5, 174ra127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapuis AG, Egan DN, Bar M, Schmitt TM, McAfee MS, Paulson KG, Voillet V, Gottardo R, Ragnarsson GB, Bleakley M, Yeung CC, Muhlhauser P, Nguyen HN, Kropp LA, Castelli L, Wagener F, Hunter D, Lindberg M, Cohen K, Seese A, McElrath MJ, Duerkopp N, Gooley TA, Greenberg PD, T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat Med 25, 1064–1072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A, Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruella M, Maus MV, Catch me if you can: Leukemia Escape after CD19-Directed T Cell Immunotherapies. Comput Struct Biotechnol J 14, 357–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterhouse M, Pfeifer D, Pantic M, Emmerich F, Bertz H, Finke J, Genome-wide profiling in AML patients relapsing after allogeneic hematopoietic cell transplantation. Biol Blood Marrow Transplant 17, 1450–1459 e1451 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Busse A, Letsch A, Scheibenbogen C, Nonnenmacher A, Ochsenreither S, Thiel E, Keilholz U, Mutation or loss of Wilms' tumor gene 1 (WT1) are not major reasons for immune escape in patients with AML receiving WT1 peptide vaccination. J Transl Med 8, 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigneron N, Abi Habib J, Van den Eynde BJ, Learning from the Proteasome How To Fine-Tune Cancer Immunotherapy. Trends Cancer 3, 726–741 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Jaigirdar A, Rosenberg SA, Parkhurst M, A High-avidity WT1-reactive T-Cell Receptor Mediates Recognition of Peptide and Processed Antigen but not Naturally Occurring WT1-positive Tumor Cells. J Immunother 39, 105–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn DJ, Orlowski RZ, The immunoproteasome as a target in hematologic malignancies. Semin Hematol 49, 258–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moukalled NM, Kharfan-Dabaja MA, What is the role of a second allogeneic hematopoietic cell transplant in relapsed acute myeloid leukemia? Bone Marrow Transplant 55, 325–331 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Restifo NP, Gattinoni L, Lineage relationship of effector and memory T cells. Curr Opin Immunol 25, 556–563 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papalexi E, Satija R, Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 18, 35–45 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Gommerman JL, Browning JL, Ware CF, The Lymphotoxin Network: orchestrating a type I interferon response to optimize adaptive immunity. Cytokine Growth Factor Rev 25, 139–145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo Y, Chen J, Zhao T, Fan Z, Granzyme K degrades the redox/DNA repair enzyme Ape1 to trigger oxidative stress of target cells leading to cytotoxicity. Molecular immunology 45, 2225–2235 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Peterlin BM, Transcriptional regulation of HLA-DRA gene. Res Immunol 142, 393–399 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Schroder B, The multifaceted roles of the invariant chain CD74--More than just a chaperone. Biochim Biophys Acta 1863, 1269–1281 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Greenough TC, Straubhaar JR, Kamga L, Weiss ER, Brody RM, McManus MM, Lambrecht LK, Somasundaran M, Luzuriaga KF, A Gene Expression Signature That Correlates with CD8+ T Cell Expansion in Acute EBV Infection. J Immunol 195, 4185–4197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CY, Forman LW, Faller DV, Calcium-dependent immediate-early gene induction in lymphocytes is negatively regulated by p21Ha-ras. Mol Cell Biol 16, 6582–6592 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slifka MK, Rodriguez F, Whitton JL, Rapid on/off cycling of cytokine production by virus-specific CD8+ T cells. Nature 401, 76–79 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Groettrup M, Standera S, Stohwasser R, Kloetzel PM, The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc Natl Acad Sci U S A 94, 8970–8975 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abi Habib J, De Plaen E, Stroobant V, Zivkovic D, Bousquet MP, Guillaume B, Wahni K, Messens J, Busse A, Vigneron N, Van den Eynde BJ, Efficiency of the four proteasome subtypes to degrade ubiquitinated or oxidized proteins. Sci Rep 10, 15765 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doubrovina E, Carpenter T, Pankov D, Selvakumar A, Hasan A, O'Reilly RJ, Mapping of novel peptides of WT-1 and presenting HLA alleles that induce epitope-specific HLA-restricted T cells with cytotoxic activity against WT-1(+) leukemias. Blood 120, 1633–1646 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. E. Ruggiero E., Prodeus A, Magnani ZI, Camisa B, Merelli I,, S. L. Politano C, Potenza A, Cianciotti BC, Manfredi F, Di Bono M, Vago L,, M. S. Tassara M, Ponzoni M, Sanvito F, Liu D, Balwani I, Galli R, Genua ORM, O’Connell D, Dutta I, Yazinski SA, McKee M, Arredouani M,, C. F. Schultes B, Bonini C, CRISPR-based gene disruption and integration of high-avidity, WT1-specific T cell receptors improve anti-tumor T cell function. Science of Translational Medicine, (2021). [DOI] [PubMed] [Google Scholar]
- 32.Glienke W, Maute L, Wicht J, Bergmann L, Wilms' tumour gene 1 (WT1) as a target in curcumin treatment of pancreatic cancer cells. Eur J Cancer 45, 874–880 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Loeb DM, Evron E, Patel CB, Sharma PM, Niranjan B, Buluwela L, Weitzman SA, Korz D, Sukumar S, Wilms' tumor suppressor gene (WT1) is expressed in primary breast tumors despite tumor-specific promoter methylation. Cancer Res 61, 921–925 (2001). [PubMed] [Google Scholar]
- 34.Turtle CJ, Hanafi LA, Berger C, Gooley TA, Cherian S, Hudecek M, Sommermeyer D, Melville K, Pender B, Budiarto TM, Robinson E, Steevens NN, Chaney C, Soma L, Chen X, Yeung C, Wood B, Li D, Cao J, Heimfeld S, Jensen MC, Riddell SR, Maloney DG, CD19 CAR-T cells of defined CD4+:CD8+ composition in adult B cell ALL patients. J Clin Invest 126, 2123–2138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tay RE, Richardson EK, Toh HC, Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thibodeau J, Bourgeois-Daigneault MC, Lapointe R, Targeting the MHC Class II antigen presentation pathway in cancer immunotherapy. Oncoimmunology 1, 908–916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ecsedi M, McAfee MS, Chapuis AG, The Anticancer Potential of T Cell Receptor-Engineered T Cells. Trends Cancer 7, 48–56 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raphael I, Nalawade S, Eagar TN, Forsthuber TG, T cell subsets and their signature cytokines in autoimmune and inflammatory diseases. Cytokine 74, 5–17 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spaapen RM, Neefjes J, Immuno-waste exposure and further management. Nat Immunol 13, 109–111 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Bai M, Zhao X, Sahara K, Ohte Y, Hirano Y, Kaneko T, Yashiroda H, Murata S, Assembly mechanisms of specialized core particles of the proteasome. Biomolecules 4, 662–677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan S, van den Broek M, Schwarz K, de Giuli R, Diener PA, Groettrup M, Immunoproteasomes largely replace constitutive proteasomes during an antiviral and antibacterial immune response in the liver. J Immunol 167, 6859–6868 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Rouette A, Trofimov A, Haberl D, Boucher G, Lavallee VP, D'Angelo G, Hebert J, Sauvageau G, Lemieux S, Perreault C, Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci Rep 6, 34019 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T, Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 14, 135–146 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Tripathi SC, Peters HL, Taguchi A, Katayama H, Wang H, Momin A, Jolly MK, Celiktas M, Rodriguez-Canales J, Liu H, Behrens C, Wistuba II, Ben-Jacob E, Levine H, Molldrem JJ, Hanash SM, Ostrin EJ, Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc Natl Acad Sci U S A 113, E1555–1564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz ES, Chapiro J, Lurquin C, Claverol S, Burlet-Schiltz O, Warnier G, Russo V, Morel S, Levy F, Boon T, Van den Eynde BJ, van der Bruggen P, The production of a new MAGE-3 peptide presented to cytolytic T lymphocytes by HLA-B40 requires the immunoproteasome. J Exp Med 195, 391–399 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keller M, Ebstein F, Burger E, Textoris-Taube K, Gorny X, Urban S, Zhao F, Dannenberg T, Sucker A, Keller C, Saveanu L, Kruger E, Rothkotter HJ, Dahlmann B, Henklein P, Voigt A, Kuckelkorn U, Paschen A, Kloetzel PM, Seifert U, The proteasome immunosubunits, PA28 and ER-aminopeptidase 1 protect melanoma cells from efficient MART-126-35 -specific T-cell recognition. Eur J Immunol 45, 3257–3268 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Textor A, Schmidt K, Kloetzel PM, Weissbrich B, Perez C, Charo J, Anders K, Sidney J, Sette A, Schumacher TN, Keller C, Busch DH, Seifert U, Blankenstein T, Preventing tumor escape by targeting a post-proteasomal trimming independent epitope. J Exp Med 213, 2333–2348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krishna S, Anderson KS, T-Cell Epitope Discovery for Therapeutic Cancer Vaccines. Methods Mol Biol 1403, 779–796 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Horowitz MM, High-resolution typing for unrelated donor transplantation: how far do we go? Best Pract Res Clin Haematol 22, 537–541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, Dombret H, Ebert BL, Fenaux P, Larson RA, Levine RL, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz M, Sierra J, Tallman MS, Tien HF, Wei AH, Lowenberg B, Bloomfield CD, Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129, 424–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Araki D, Wood BL, Othus M, Radich JP, Halpern AB, Zhou Y, Mielcarek M, Estey EH, Appelbaum FR, Walter RB, Allogeneic Hematopoietic Cell Transplantation for Acute Myeloid Leukemia: Time to Move Toward a Minimal Residual Disease-Based Definition of Complete Remission? J Clin Oncol 34, 329–336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armand P, Kim HT, Logan BR, Wang Z, Alyea EP, Kalaycio ME, Maziarz RT, Antin JH, Soiffer RJ, Weisdorf DJ, Rizzo JD, Horowitz MM, Saber W, Validation and refinement of the Disease Risk Index for allogeneic stem cell transplantation. Blood 123, 3664–3671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, Wood BL, Methods of Detection of Measurable Residual Disease in AML. Current hematologic malignancy reports 12, 557–567 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Festuccia M, Deeg HJ, Gooley TA, Baker K, Wood BL, Fang M, Sandmaier BM, Scott BL, Minimal Identifiable Disease and the Role of Conditioning Intensity in Hematopoietic Cell Transplantation for Myelodysplastic Syndrome and Acute Myelogenous Leukemia Evolving from Myelodysplastic Syndrome. Biol Blood Marrow Transplant 22, 1227–1233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang M, Storer B, Wood B, Gyurkocza B, Sandmaier BM, Appelbaum FR, Prognostic impact of discordant results from cytogenetics and flow cytometry in patients with acute myeloid leukemia undergoing hematopoietic cell transplantation. Cancer 118, 2411–2419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones S, Peng PD, Yang S, Hsu C, Cohen CJ, Zhao Y, Abad J, Zheng Z, Rosenberg SA, Morgan RA, Lentiviral vector design for optimal T cell receptor gene expression in the transduction of peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Hum Gene Ther 20, 630–640 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapuis AG, Thompson JA, Margolin KA, Rodmyre R, Lai IP, Dowdy K, Farrar EA, Bhatia S, Sabath DE, Cao J, Li Y, Yee C, Transferred melanoma-specific CD8+ T cells persist, mediate tumor regression, and acquire central memory phenotype. Proc Natl Acad Sci U S A, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chapuis AG, Desmarais C, Emerson R, Schmitt TM, Shibuya K, Lai I, Wagener F, Chou J, Roberts IM, Coffey DG, Warren E, Robbins H, Greenberg PD, Yee C, Tracking the Fate and Origin of Clinically Relevant Adoptively Transferred CD8+ T Cells In Vivo. Sci Immunol 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J, Dudley ME, Rosenberg SA, Robbins PF, Persistence of multiple tumor-specific T-cell clones is associated with complete tumor regression in a melanoma patient receiving adoptive cell transfer therapy. J Immunother 28, 53–62 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A, PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horton H, Thomas EP, Stucky JA, Frank I, Moodie Z, Huang Y, Chiu YL, McElrath MJ, De Rosa SC, Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. Journal of immunological methods 323, 39–54 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moncunill G, Han H, Dobano C, McElrath MJ, De Rosa SC, OMIP-024: pan-leukocyte immunophenotypic characterization of PBMC subsets in human samples. Cytometry. Part A : the journal of the International Society for Analytical Cytology 85, 995–998 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Limaye AP, Huang ML, Atienza EE, Ferrenberg JM, Corey L, Detection of Epstein-Barr virus DNA in sera from transplant recipients with lymphoproliferative disorders. J Clin Microbiol 37, 1113–1116 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boeckh M, Huang M, Ferrenberg J, Stevens-Ayers T, Stensland L, Nichols WG, Corey L, Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J Clin Microbiol 42, 1142–1148 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH, Massively parallel digital transcriptional profiling of single cells. Nature communications 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902 e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ilicic T, Kim JK, Kolodziejczyk AA, Bagger FO, McCarthy DJ, Marioni JC, Teichmann SA, Classification of low quality cells from single-cell RNA-seq data. Genome Biol 17, 29 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R, MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hafemeister C, Satija R, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, Wappett M, Sharpe A, Dymond M, Barrett JC, Harrington EA, Marshall G, Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res 75, 2587–2593 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Pritchard CC, Salipante SJ, Koehler K, Smith C, Scroggins S, Wood B, Wu D, Lee MK, Dintzis S, Adey A, Liu Y, Eaton KD, Martins R, Stricker K, Margolin KA, Hoffman N, Churpek JE, Tait JF, King MC, Walsh T, Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn 16, 56–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JE, Ao L, Miller Z, Kim K, Wu Y, Jang ER, Lee EY, Kim KB, Lee W, PSMB9 codon 60 polymorphisms have no impact on the activity of the immunoproteasome catalytic subunit B1i expressed in multiple types of solid cancer. PLoS One 8, e73732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ho WY, Nguyen HN, Wolfl M, Kuball J, Greenberg PD, In vitro methods for generating CD8+ T-cell clones for immunotherapy from the naive repertoire. J Immunol Methods 310, 40–52 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, Van den Eynde BJ, Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 107, 18599–18604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vigneron N, Ooms A, Morel S, Ma W, Degiovanni G, Van den Eynde BJ, A peptide derived from melanocytic protein gp100 and presented by HLA-B35 is recognized by autologous cytolytic T lymphocytes on melanoma cells. Tissue Antigens 65, 156–162 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Pogosova-Agadjanyan EL, Moseley A, Othus M, Appelbaum FR, Chauncey TR, Chen IL, Erba HP, Godwin JE, Fang M, Kopecky KJ, List AF, Pogosov GL, Radich JP, Willman CL, Wood BL, Meshinchi S, Stirewalt DL, Impact of Specimen Heterogeneity on Biomarkers in Repository Samples from Patients with Acute Myeloid Leukemia: A SWOG Report. Biopreserv Biobank 16, 42–52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, Wang S, Li W, RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 80.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 81.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, Barnes I, Berry A, Bignell A, Carbonell Sala S, Chrast J, Cunningham F, Di Domenico T, Donaldson S, Fiddes IT, Garcia Giron C, Gonzalez JM, Grego T, Hardy M, Hourlier T, Hunt T, Izuogu OG, Lagarde J, Martin FJ, Martinez L, Mohanan S, Muir P, Navarro FCP, Parker A, Pei B, Pozo F, Ruffier M, Schmitt BM, Stapleton E, Suner MM, Sycheva I, Uszczynska-Ratajczak B, Xu J, Yates A, Zerbino D, Zhang Y, Aken B, Choudhary JS, Gerstein M, Guigo R, Hubbard TJP, Kellis M, Paten B, Reymond A, Tress ML, Flicek P, GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, D766–D773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robinson MD, Oshlack A, A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guillaume B, Chapiro J, Stroobant V, Colau D, Van Holle B, Parvizi G, Bousquet-Dubouch MP, Theate I, Parmentier N, Van den Eynde BJ, Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci U S A 107, 18599–18604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bandaranayake AD, Correnti C, Ryu BY, Brault M, Strong RK, Rawlings DJ, Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res 39, e143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Findlay L, Eastwood D, Stebbings R, Sharp G, Mistry Y, Ball C, Hood J, Thorpe R, Poole S, Improved in vitro methods to predict the in vivo toxicity in man of therapeutic monoclonal antibodies including TGN1412. J Immunol Methods 352, 1–12 (2010). [DOI] [PubMed] [Google Scholar]
- 87.Riddell SR, Watanabe KS, Goodrich JM, Li CR, Agha ME, Greenberg PD, Restoration of viral immunity in immunodeficient humans by the adoptive transfer of T cell clones. Science 257, 238–241 (1992). [DOI] [PubMed] [Google Scholar]
- 88.Jedema I, van der Werff NM, Barge RM, Willemze R, Falkenburg JH, New CFSE-based assay to determine susceptibility to lysis by cytotoxic T cells of leukemic precursor cells within a heterogeneous target cell population. Blood 103, 2677–2682 (2004). [DOI] [PubMed] [Google Scholar]
- 89.Nguyen DN, Roth TL, Li PJ, Chen PA, Apathy R, Mamedov MR, Vo LT, Tobin VR, Goodman D, Shifrut E, Bluestone JA, Puck JM, Szoka FC, Marson A, Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat Biotechnol 38, 44–49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cao X, Shores EW, Hu-Li J, Anver MR, Kelsall BL, Russell SM, Drago J, Noguchi M, Grinberg A, Bloom ET, et al. , Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity 2, 223–238 (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.