Summary
Background:
Variants in multiple genetic loci modify the risk of non-alcoholic fatty liver disease (NAFLD) and cirrhosis but there are limited data on the quantitative impact of variant copies on liver fibrosis.
Aim:
To investigate the effect of PNPLA3, TM6SF2, MBOAT7, GCKR and HSD17B13 genotype on liver fibrosis assessed by magnetic resonance elastography (MRE), a reproducible, accurate, continuous biomarker of liver fibrosis.
Methods:
This is a cross-sectional analysis derived from a well-characterised cohort at risk for NAFLD who underwent genotyping and MRE assessment. Liver stiffness (LS) was estimated using MRE and advanced fibrosis was defined as liver stiffness ≥3.63 kilopascals (kPa). Univariable and multivariable linear and logistic regression analysis, were used to assess the association between genotype and MRE.
Results:
Two hundred sixty-four patients (63% women) with a mean age 53 (±17) years, and 31% Hispanic ethnicity with genotyping and MRE were included. The odds of advanced fibrosis were 3.1 (95% CI: 1.1–8.9, P = 0.04) for CG and 6.5 (95% CI: 2.2–18.9, P < 0.01) for GG compared to CC PNPLA3 genotype. Each PNPLA3 risk variant copy was associated with 0.40 kPa (95% CI: 0.19–0.61, P < 0.01) increase in LS on MRE in analysis adjusted for age, sex and BMI and there was significant genotype-age interaction (P < 0.01). Conversely, the protective TA allele in HSD17B13 was associated with a −0.41 kPa (95% CI: −0.76 to −0.05, P = 0.03) decrease in liver stiffness on MRE multivariable analysis.
Conclusion:
Knowledge of PNPLA3 and HSD17B13 genotype may assist in the non-invasive risk stratification of NAFLD with closer monitoring recommended for those with high genetic risk.
1 |. INTRODUCTION
Non-alcoholic fatty liver disease (NAFLD) is emerging as a leading cause of chronic liver disease globally, yet heterogeneity in disease severity and natural history have limited our ability to identify and treat the highest risk population.1,2 While NAFLD is closely tied to the presence of obesity and metabolic syndrome, approximately 20% of individuals with NAFLD have a body mass index (BMI) within the normal range.3 Furthermore, while the presence of steatohepatitis on liver biopsy identifies a high-risk population, it is impractical to scale to the estimated affected population of 1 billion. Therefore, there is a need to identify NAFLD phenotypes at the greatest risk for morbidity and mortality.
The strong genetic underpinnings of NAFLD are clearly demonstrated by familial clustering of advanced liver disease and racial/ethnic predisposition to disease.4,5 The most well-characterised common variant is the non-synonymous variant p.I148M in PNPLA3, which has increased prevalence in Hispanics6 and is associated with an increased risk of hepatic steatosis,6 non-alcoholic steatohepatitis (NASH),7,8 cirrhosis9 and hepatocellular carcinoma.10 However, despite the strong association, the PNPLA3 genotype has not been routinely incorporated into clinical practice. Additional loci including TM6SF2,11,12 MBOAT713 and GCKR14 have been associated with NAFLD risk and more recently a protective variant in HSD17B1315 was identified.
The most commonly used currently available non-invasive assessments do not incorporate genetic data to assess NAFLD severity and PNPLA3 genotype has performed poorly in discriminatory models identifying disease at a single time point. However, an unrealised value of genetic markers is to identify high-risk groups that may be at risk for an aggressive disease trajectory and future liver-related morbidity and mortality. The association between genotype and accurate, quantitative assessments of NAFLD activity is needed to better characterise the impact of genotype on NAFLD severity. While the liver biopsy is the reference standard, it evaluates only a small portion of the liver and utilises ordinal categories to stage fibrosis. In contrast, magnetic resonance elastography (MRE) is an accurate,16 reproducible,17 continuous measure of liver stiffness, which is associated with fibrosis and has been recently demonstrated to predict liver-related outcomes.18,19 Therefore, we hypothesised that variants in PNPLA3, TM6SF2, MBOAT7, GCKR and HSD17B13 would be associated with advanced fibrosis on MRE and that quantifying the magnitude of the association may assist in the non-invasive risk stratification of NAFLD patients. Using a well-characterised cohort of patients at risk for NAFLD, we evaluated the association between PNPLA3, TM6SF2, MBOAT7, GCKR and HSD17B13 genotype and advanced fibrosis on MRE as well as the quantitative impact of those variants on liver fibrosis on MRE.
2 |. MATERIALS AND METHODS
2.1 |. Study design
This is a cross-sectional analysis derived from a well-characterised prospective cohort of patients who had genotyping data and MRE assessment. This study included 264 uniquely phenotyped patients who underwent a standardised research visit: history, physical exam, PNPLA3 genotyping and MRE assessment between 2011 and 2017 at the University of California, San Diego NAFLD Research Center.20–24 All patients provided written informed consent prior to enrolling in the study and the study was approved by the UCSD Institutional Review Board.
2.2 |. Inclusion and exclusion criteria
Patients ≥18 years of age with written informed consent were included. Participants meeting any of the following criteria were excluded from the study: significant alcohol consumption (defined as ≥14 drinks/week for men or ≥7 drinks/week for women) within the previous 2-year period; evidence of active substance use; clinical or laboratory evidence of secondary causes or chronic conditions associated with hepatic steatosis including nutritional disorders, human immunodeficiency virus infection, and use of steatogenic drugs such as amiodarone, glucocorticoids, methotrexate, l-asparaginase and valproic acid; underlying liver disease other than NAFLD including viral hepatitis (assessed with serum hepatitis B surface antigen and hepatitis C RNA assays), haemochromatosis, Wilson’s disease, alpha-1 antitrypsin deficiency, glycogen storage disease, autoimmune hepatitis and cholestatic or vascular liver disease; major systemic illnesses; decompensated liver disease (defined as Child-Pugh score >7 points); contraindications to magnetic resonance imaging (MRI) including metallic implants, claustrophobia and body circumference exceeding the imaging chamber capacity; pregnancy or attempting to be pregnant; any other conditions believed by the principal investigator to affect patient’s competence or compliance to complete the study.
2.3 |. Clinical assessment and laboratory tests
All patients underwent a standardised clinical evaluation including a detailed history and a physical examination, which included vital signs, height, weight and anthropometric measurements, was performed by a trained clinical investigator. BMI was defined as the body weight (in kilograms) divided by height (in meters) squared. Alcohol consumption was documented outside clinical visits and confirmed in the research clinic using the Alcohol Use Disorders Identifications Test (AUDIT) and the Skinner questionnaire. Other causes of liver disease were systematically ruled out based on history and laboratory tests. Patients underwent the following biochemical tests: glucose, albumin, haemoglobin A1c, alanine aminotransferase, aspartate aminotransferase, total bilirubin, alkaline phosphatase, fasting lipid panel, platelets, insulin, international normalised ratio. FIB-4,25 NAFLD Fibrosis Score26 were calculated as described previously. Participants were instructed to fast for a minimum of 8 hours prior to collection of laboratory tests.
2.4 |. PNPLA3, TM6SF2, MBOAT7, GCKR and HSD17B13 genotyping
Whole-blood specimens collected during the research visit were used, and DNA was extracted using Qiagen’s DNeasy® Blood & Tissue Kit. PNPLA3, TM6SF2, MBOAT7, GCKR and HSD17B13 genotyping was performed in triplicate using single-nucleotide polymorphism assays from Applied Biosystems and analysed using Quant Studio.
2.5 |. Magnetic resonance imaging
Abdominal MRI was obtained on a single 3 Tesla magnetic resonance scanner (GE Signa EXCITE HDxt, GE Healthcare) at the UCSD MR3T Research Laboratory using previously described methods.27–31 MRI proton density fat fraction (MRI-PDFF) was used to measure hepatic steatosis and where NAFLD is defined as having an MRI-PDFF ≥5%. Liver stiffness was estimated using two-dimensional MRE, which is the most accurate biomarker for the quantitative assessment of liver stiffness as a surrogate for hepatic fibrosis.32–35 A passive driver was fitted around the body over the liver and connected to an acoustic active driver that delivered continuous vibrations at 60 hertz to produce shear waves in the liver, which were processed to generate elastograms depicting liver stiffness. Four slices were assessed, and co-localised regions of interest were manually specified as previously described.36
2.6 |. Definition of advanced fibrosis
Participants were considered to have advanced fibrosis if a liver stiffness of ≥3.63 kilopascals (kPa) was found on MRE. Previous studies have shown that ≥3.63 kPa on MRE provides an accuracy of 0.92 for the detection of advanced fibrosis, and it is the most accurate non-invasive test for the diagnosis of advanced fibrosis.32–34
2.7 |. Outcome measures
The primary outcome was the assessment of the association of PNPLA3 genotype with advanced fibrosis on MRE.
Secondary outcomes were assessing the association of TM6SF2, MBOAT7, GCKR and HSD17B13 with advanced fibrosis on MRE, PNPLA3, TM6SF2, MBOAT7, GCKR and HSD17B13 genotype on MRE as a continuous measure and the association between PNPLA3 genotype and MRI-PDFF.
2.8 |. Statistical analysis
We hypothesised that there would be an association between PNPLA3 risk variants and advanced fibrosis on MRE. Power analysis performed assuming that 60% of the cohort had one or two risk variants and that the presence of risk variants was associated with four times higher odds of advanced fibrosis (5% in CC and 20% in CG/GG) showed that a sample size of 190 would provide 80% power with a two-tailed alpha of 0.05 and therefore, we had adequate power to test and confirm our hypothesis. For patient characteristics, an ANOVA was performed on continuous variables presented as mean (SD), Kruskal-Wallis performed on those presented as median (IQR). Chi-square or Fisher’s exact test as appropriate on all categorical variables. Unadjusted logistic regression analyses were conducted to assess the odds ratio (OR) of various factors on the presence of advanced fibrosis (MRE ≥3.63). Moreover, the association between genotype and additional MRE cut points, 3.0 and 5.0 kPa was evaluated by logistic regression. In addition, logistic regression analyses, multivariable-adjusted for age, sex, BMI, diabetes mellitus (DM) and FIB-4, were conducted to assess the OR of five genetic loci on the presence of advanced fibrosis (MRE ≥3.63). Furthermore, linear regression was performed with MRE as a continuous measure as the outcome and each of the five genetic loci as the predictor. Interaction terms between age, BMI and PNPLA3 genotype were evaluated by adding cross-product terms to regression models. All statistical analyses were performed using SAS 9.4 (SAS Institute), and a P value less than 0.05 was considered statistically significant.
3 |. RESULTS
3.1 |. Characteristics of the study population
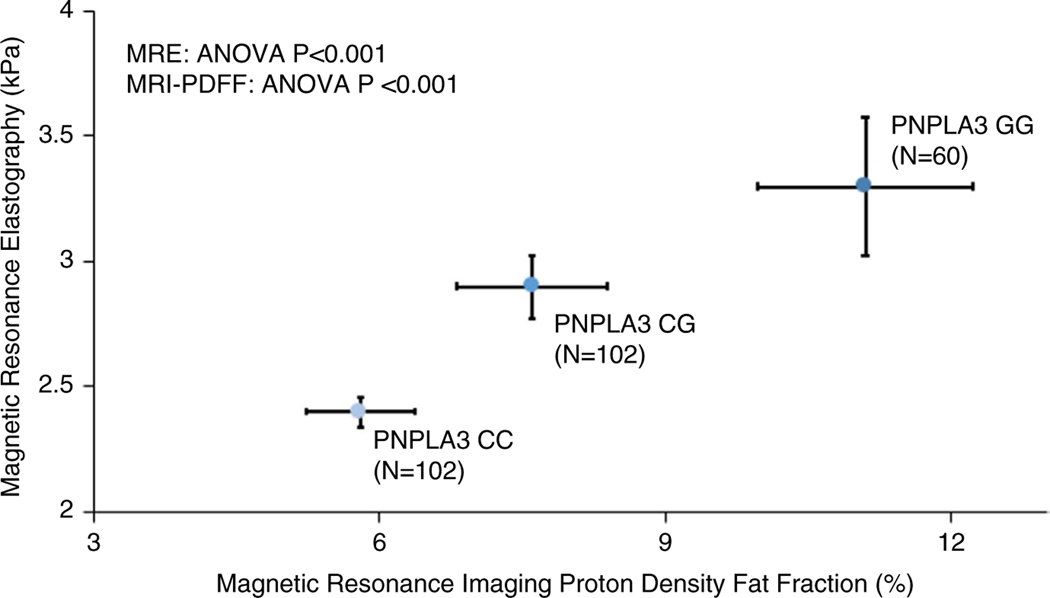
Two hundred sixty-four patients with genotype data and MRE were included. Participants had a mean age of 53 (±17) years and were predominantly female (63%); 55% white and 31% Hispanic ethnicity. The risk allele frequency for PNPLA3, TM6SF2, MBOAT7 and GCKR was 0.42 G allele, 0.06 T allele, 0.40 T allele and 0.43 T allele respectively. The protective TA allele frequency in HSD17B13 was 0.18. The mean BMI was 29 (±7) kg/m2. One hundred and twenty-two (46%) had NAFLD (MRI-PDFF ≥5%) and 34 (13%) had advanced fibrosis (MRE ≥3.63 kPa). (Table 1) A greater number of PNPLA3 risk alleles were associated with Hispanic ethnicity (G allele frequency 0.66 vs 0.30 in non-Hispanics P < 0.01), higher BMI (P = 0.01), increased family history of liver disease (P < 0.01) and higher alanine aminotransferase (ALT) (P = 0.02), aspartate aminotransferase (AST) (P = 0.01), alkaline phosphatase (P = 0.04), homeostatic model assessment method for insulin resistance (HOMA-IR) (P < 0.01), triglycerides (P = 0.0353) and high-d ensity lipoprotein cholesterol (HDL) (P = 0.0263) (Table 1). The mean (±SD) MRI-PDFF values for CC, CG and GG genotypes were 5.8% (±5.8), 7.6% (±7.9) and 11.1% (±8.8), respectively, P < 0.01, and the Mean (±SD) MRE values for CC, CG and GG genotypes were 2.4 kPa (±0.6), 2.9 kPa (±1.3) and 3.3 kPa (±2.2), respectively, P < 0.01 (Figure 1).
TABLE 1.
Clinical, demographic and imaging characteristics by PNPLA3 genotype
| PNPLA3 genotypes | Total N | C/C N = 102 | C/G N = 102 | G/G N = 60 | P |
|---|---|---|---|---|---|
| Demographic | |||||
| Age in years, mean (SD) | 263 | 52.8 (17.0) | 53.4 (17.2) | 53.2 (16.5) | 0.9727 |
| Male, n (%) | 264 | 41 (40.2%) | 34 (33.3%) | 22 (36.7%) | 0.5964 |
| BMI (kg/m2), mean (SD) | 256 | 28.9 (6.6) | 28.7 (7.9) | 32.0 (5.4) | 0.0103 |
| White, n (%) | 260 | 73 (71.6%) | 56 (56.0%) | 14 (24.1%) | <0.0001 |
| Diabetes, n (%) | 264 | 48 (47.1%) | 43 (42.2%) | 25 (41.7%) | 0.7188 |
| Hypertension, n (%) | 264 | 46 (45.1%) | 35 (34.3%) | 27 (45.0%) | 0.2241 |
| Hyperlipidaemia, n (%) | 176 | 11 (19.3%) | 15 (20.8%) | 11 (23.4%) | 0.8763 |
| Family history | |||||
| Liver disease, n (%) | 176 | 5 (8.8%) | 19 (26.4%) | 26 (55.3%) | <0.0001 |
| Biochemical profile | |||||
| HbA1c (%), median (IQR) | 259 | 6(1.3) | 5.9 (0.9) | 6 (1.4) | 0.5073 |
| AST (U/L), median (IQR) | 262 | 20.5 (8) | 24 (13) | 23 (20) | 0.0147 |
| ALT (U/L), median (IQR) | 261 | 19 (14) | 24.5 (19) | 26 (22) | 0.0201 |
| Alkaline phosphatase (U/L), median (IQR) | 262 | 71 (26) | 73 (37) | 79 (42) | 0.0433 |
| Total bilirubin (mg/dL), median (IQR) | 262 | 0.5 (0.3) | 0.4 (0.3) | 0.5 (0.3) | 0.0791 |
| Albumin (g/dL), median (IQR) | 262 | 4.5 (0.3) | 4.5 (0.4) | 4.4 (0.4) | 0.1288 |
| HOMA-IR median (IQR) | 248 | 2.7 (3) | 3.2 (4.3) | 5.5 (10.4) | <0.0001 |
| Triglycerides (mg/dL), median (IQR) | 260 | 96 (75) | 102 (103) | 125 (72.5) | 0.0353 |
| HDL (mg/dL), median (IQR) | 260 | 60 (27) | 55 (28) | 50 (18.5) | 0.0263 |
| LDL (mg/dL), median (IQR) | 259 | 99 (42) | 96 (43) | 106 (53) | 0.1444 |
| Platelet count (109/L), median (IQR) | 262 | 239.5 (84) | 236.5 (92.5) | 238.5 (117.5) | 0.8124 |
| INR, median (IQR) | 262 | 1 (0.1) | 1 (0.1) | 1 (0.1) | 0.2528 |
| Clinical prediction rule | |||||
| FIB-4, median (IQR) | 259 | 1 (0.7) | 1.1 (1) | 1.1 (1.2) | 0.5398 |
| NAFLD Fibrosis Score, median (IQR) | 252 | −1.3 (1.8) | −1.8 (2.6) | −1.2 (3.6) | 0.7150 |
| Imaging | |||||
| MRI-PDFF (%), mean (SD) | 264 | 5.8 (5.8) | 7.6 (7.9) | 11.1 (8.8) | <0.0001 |
| MRE (kPa), mean (SD) | 264 | 2.4 (0.6) | 2.9 (1.3) | 3.3 (2.2) | 0.0003 |
Note: ANOVA performed on continuous variables presented as mean (SD), Kruskal-Wallis performed on all other continuous variables. Chi-square or Fisher’s exact test as appropriate on all categorical variables.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; FIB-4, fibrosis index based on the 4 factor; HbA1c, haemoglobin A1c; HDL, high-density lipoprotein; HOMA-IR, homeostatic model assessment method for insulin resistance (calculated as (fasting insulin (μU/mL) * fasting glucose (mmol/L))/22.5); INR, International normalised ratio; IQR, interquartile range; LDL, low-density lipoprotein; MRE, magnetic resonance elastography; MRI-PDFF, magnetic resonance imaging proton density fat fraction; SD, standard deviation.
FIGURE 1.
PNPLA3 risk alleles increase both mean liver stiffness of magnetic resonance elastography and liver fat on magnetic resonance imaging proton density fat fraction shown with standard error bars
3.2 |. Factors associated with advanced fibrosis on MRE
Higher AST (OR = 1.05 [95% CI: 1.02–1.07, P < 0.01]), ALT (OR = 1.01 [95% CI: 1.00–1.03, P = 0.02]), alkaline phosphatase (OR = 1.02 [95% CI: 1.01–1.04, P < 0.01]), total bilirubin (OR = 6.60 [95% CI: 2.9–15.0, P < 0.01]) and HOMA-IR (OR = 1.07 [95% CI: 1.03–1.12, <0.01]) were associated with advanced fibrosis. Lower albumin (OR = 0.09 [95% CI: 0.03–0.29, P < 0.01]), HDL (OR = 0.96 [95% CI: 0.94–0.99, P < 0.01]), LDL-cholesterol (OR = 0.98 [95% CI: 0.963–0.990, P < 0.01]) and platelet count (OR = 0.98 [95% CI: 0.97–0.99, P < 0.01]) were associated with advanced fibrosis (Table 2).
TABLE 2.
Factors associated with advanced fibrosis on MRE
| Advanced fibrosis on MRE (≥3.63) OR (95% CI) | P-value | |
|---|---|---|
| PNPLA3 genotype | ||
| CC (ref) | 1 | |
| CG | 3.086 (1.068–8.918) | 0.0374 |
| GG | 6.467 (2.214–18.892) | 0.0006 |
| Demographic & biochemical | ||
| BMI (kg/m2) | 1.030 (0.981–1.081) | 0.2398 |
| HbA1c (%) | 1.134 (0.888–1.446) | 0.3135 |
| AST (U/L) | 1.046 (1.024–1.068) | <0.0001 |
| ALT (U/L) | 1.014 (1.002–1.026) | 0.0247 |
| Alkaline phosphatase (U/L) | 1.024 (1.012–1.036) | <0.0001 |
| Total bilirubin (mg/dL) | 6.604 (2.917–14.951) | <0.0001 |
| Albumin (g/dL) | 0.094 (0.031–0.286) | <0.0001 |
| HOMA-IR | 1.071 (1.026–1.118) | 0.0016 |
| Triglycerides (mg/dL) | 1.003 (0.999–1.007) | 0.1405 |
| HDL (mg/dL) | 0.960 (0.935–0.986) | 0.0024 |
| LDL (mg/dL) | 0.977 (0.963–0.990) | 0.0007 |
| Platelet count (109/L) | 0.977 (0.970–0.985) | <0.0001 |
| Clinical prediction rules | ||
| FIB-4 | 1.619 (1.291–2.031) | <0.0001 |
| NAFLD Fibrosis Score | 2.287 (1.725–3.032) | <0.0001 |
| Imaging | ||
| MRI-PDFF | 0.992 (0.944–1.042) | 0.7357 |
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; FIB-4, fibrosis index based on the 4 factor; HbA1c, haemoglobin A1c; HDL, high-density lipoprotein; HOMA-IR, homeostatic model assessment method for insulin resistance (calculated as (fasting insulin (μU/mL) * fasting glucose (mmol/L))/22.5); INR, international normalised ratio; LDL, low-density lipoprotein; MRI-PDFF, magnetic resonance imaging proton density fat fraction.
In addition, a one-unit increase in FIB-4 (OR = 1.62 [95% CI: 1.3–2.0, P < 0.01]) and NAFLD Fibrosis Score (OR = 2.29 [95% CI: 1.7–3.0, P < 0.01]) was associated with higher odds of advanced fibrosis. The number of PNPLA3 risk variants was strongly associated with advanced fibrosis on MRE, (OR = 3.1 [95% CI: 1.1–8.9, P = 0.04]) for CG and (OR = 6.5 [95% CI: 2.2–18.9, P < 0.01]) for GG compared to CC. The association between PNPLA3 risk variants and advanced fibrosis on MRE persisted in models for age and sex (OR = 3.1 [95% CI: 1.1–9.4, P = 0.04]) for CG and (OR = 7.4 [95% CI: 2.4–22.7, P < 0.01]) for GG compared to CC. Furthermore, PNPLA3 genotype was associated with advanced fibrosis despite adjustment for age, sex, BMI and FIB-4 score (OR = 2.94 [95% CI: 1.06–8.14, P = 0.04]) (Table 3). In addition, adjusted analysis PNPLA3 risk alleles remained associated with advanced fibrosis in a model adjusted for age, sex and DM (OR = 3.21 [95% CI: 3.21–9.61, P = 0.04]) for CG and (OR = 7.56 [95% CI: 2.45–23.3, P < 0.01]) for GG compared to CC. Finally, on analysis stratified by Hispanic ethnicity the association between PNPLA3 genotype and liver stiffness on MRE in the Hispanic population (N = 81) did not meet the threshold for statistical significance with wide confidence intervals, however, CG/GG genotype was associated with increased liver stiffness compared to CC in an analysis restricted to the non-Hispanic population (Table S1).
TABLE 3.
Multivariable models assessing the association of PNPLA3 genotype with advanced fibrosis
| Advanced fibrosis on MRE (≥3.63) OR (95% CI) | P-value | |
|---|---|---|
| Model 1 | ||
| CC (ref) | 1 | |
| CG | 3.120 (1.051–9.383) | 0.0405 |
| GG | 7.380 (2.405–22.650) | 0.0005 |
| Model 2: Model 1 + BMI | ||
| CC (ref) | 1 | |
| CG | 3.154 (1.059–9.395) | 0.0391 |
| GG | 6.829 (2.176–21.428) | 0.0010 |
| Model 3: Model 2 + FIB 4 | ||
| CC (ref) | 1 | |
| CG | 2.417 (0.783–7.456) | 0.1247 |
| GG | 5.257 (1.616–17.104) | 0.0058 |
| Model 4: Model 1 + DM | ||
| CC (ref) | 1 | |
| CG | 3.205 (1.068–9.614) | 0.0377 |
| GG | 7.564 (2.452–23.334) | 0.0004 |
Note: Model 1: Adjusted for age and sex.
Abbreviations: BMI, body mass index; DM, diabetes mellitus; FIB 4, fibrosis index based on the 4 factor; MRE, magnetic resonance elastography.
Risk variants in TM6SF2, MBOAT7 and GCKR did not have a statistically significant association with advanced fibrosis (Table S2), but the point estimates were greater than 1 compared when compared to no risk variants. The protective HSD17B13:TA variant was protective against advanced fibrosis (OR = 0.337 [95% CI: 0.124–0.915], P = 0.0329) compared to the referent genotype TT. Fifty-nine and nineteen patients had a liver stiffness ≥3.0 and 5.0 kPa respectively. In sensitivity analyses for lower (3.0 kPa) and higher cut points (5.0 kPa), indicating moderate fibrosis and cirrhosis with a high risk of decompensation, respectively, PNPLA3 GG genotype remained associated with moderate fibrosis at the 3.0 kPa cut-point compared to CC, however, other risk variants did not meet the threshold of statistical significance (Table S3). When evaluating the higher cut-point of 5.0 kPa, no PNPLA3 CC (referent) patients had liver stiffness ≥5.0 kPa limiting the analysis for that genotype. The HSD17B13 TA variant remained significantly protective, albeit with only one patient with the TA allele with liver stiffness ≥5.0 kPa (Table S4).
3.3 |. Quantitative impact of PNPLA3 risk variants on hepatic steatosis and fibrosis on MRI-PDFF and MRE
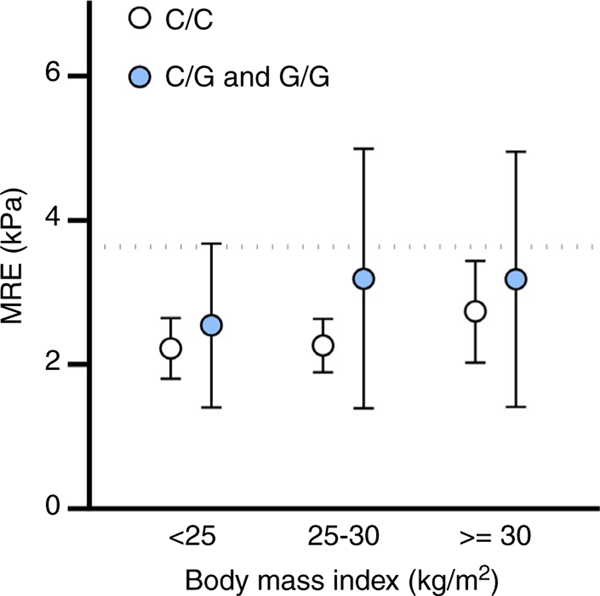
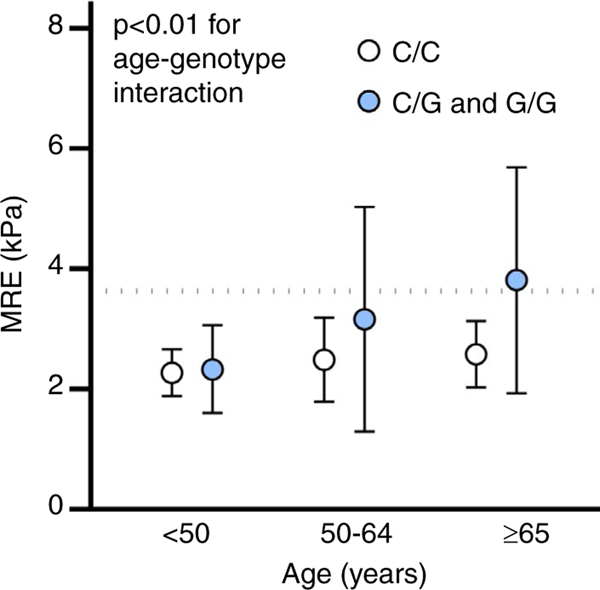
Each PNPLA3 risk variant copy was associated a 2.56% (95% CI: 1.39–3.72, P < 0.01) increase in hepatic steatosis on MRI-PDFF on linear regression and remained similar in multivariable analysis adjusted for age, sex and BMI, 2.11% (95% CI: 1.02–3.21, P < 0.01). Each PNPLA3 risk variant copy was associated a 0.45 kPa (95% CI: 0.23–0.66, P < 0.01) increase in liver stiffness on MRE on linear regression and remained similar in multivariable analysis adjusted for age, sex and BMI, 0.40 kPa (95% CI: 0.19–0.61, P < 0.01). Graphical representation of the interaction between PNPLA3 genotype and BMI suggested demonstrated that higher BMI category combined with increased risk variants increased liver stiffness (Figure 2), however, the P-value for an interaction term between BMI and PNPLA3 genotype was not statistically significant, (P = 0.40). An interaction for age and PNPLA3 genotype was significant, (P = 0.002) and demonstrates similar liver stiffness in patients <50 regardless of PNPLA3 genotype. However, in populations age 50–64 years and ≥65 years PNPLA3 genotype is strongly associated with increased liver stiffness (Figure 3) demonstrating the value of incorporating genotype into the frequency of monitoring particularly as patients age.
FIGURE 2.
Liver stiffness on magnetic resonance elastography by PNPLA3 genotype and body mass index
FIGURE 3.
Liver stiffness on magnetic resonance elastography by PNPLA3 genotype and age
3.4 |. Quantitative impact of TM6SF2, MBOAT7, GCKR and HSD17B13 variants on fibrosis on MRE
Risk variants in TM6SF2, MBOAT7 and GCKR did not have a statistically significant association with liver stiffness on MRE on linear regression, however, the point estimates were greater than 0; 0.12 (95% CI: −0.45 to 0.69, P = 0.6705), 0.14 (95% CI: −0.1 to 0.39, P = 0.2557) and 0.14 (95% CI: −0.11 to 0.39, P = 0.282). On multivariable analysis adjusted for age, sex and BMI, each TM6SF2, MBOAT7 and GCKR risk variant copy did not have a statistically significant association with liver stiffness on MRE; 0.11 (95% CI: −0.44 to 0.66, P = 0.6814), 0.14 (95% CI: −0.1 to 0.38, P = 0.2495) and 0.21 (95% CI: −0.03 to 0.45, P = 0.092) respectively. The protective TA allele in HSD17B13 was less common in Hispanic compared to non-Hispanic populations (minor allele frequency 0.09 vs 0.23, P < 0.01) and was associated with a −0.40 (95% CI: −0.77 to −0.02, P = 0.0387) decrease in liver stiffness on MRE on unadjusted linear regression and on multivariable analysis adjusted for age, sex and BMI was associated with a −0.41 (95% CI: −0.76 to −0.05, P = 0.0263) decrease in liver stiffness on MRE.
4 |. DISCUSSION
Using a diverse, well-characterised cohort of patients with PNPLA3 genotype and MRE assessment, we demonstrate that PNPLA3 risk alleles are strongly associated with advanced fibrosis and each PNPLA3 risk allele is associated with a 0.40 kPa increase in liver stiffness on MRE adjusting for age, sex and BMI. This strong association quantifies the impact of PNPLA3 genotype on liver fibrosis in patients at risk for NAFLD. Conversely, the protective TA variant in HSD17B13 is associated with a −0.41 kPa decrease in liver stiffness on MRE on adjusted analysis. In the context of previously described cut-points the difference between CC and GG genotype could translate to the difference between minimal fibrosis (stage 0–1) and advanced fibrosis (stage 3–4). Importantly, a strong interaction between age and PNPLA3 genotype reveals the potential clinical utility of genotype on identifying patients at risk for a more aggressive disease trajectory. Currently, liver biopsy or non-invasive tests, including liver stiffness measurements, are used to determine the frequency of monitoring and choice of intervention in patients with NAFLD. However, we demonstrate that the presence of PNPLA3 risk variants is associated with a divergent and more aggressive disease trajectory in patients 50 years or older, which may warrant closer monitoring or early intervention.
4.1 |. In context with published literature
The severity of fibrosis is the strongest predictor of outcomes in patients with NAFLD.37 Generally, clinical prediction rules are used for the high negative predictive value to identify patients at low risk for advanced fibrosis who require only limited monitoring,38 however, a subset of patients can progress rapidly from minimal to advanced fibrosis.39 Importantly paired biopsy studies have demonstrated that 40%−50% of patients with NAFLD without NASH can progress to NASH on a subsequent biopsy, which is associated with progression to significant fibrosis.40 The underlying factors associated with a transition to more severe disease and the optimal strategy to monitor these patients is undefined.
The p.I148M-variant in PNPLA3 is the most well-characterised common variant in NAFLD and is associated with the full spectrum of disease in NAFLD including hepatic steatosis,6 NASH,8,41 advanced fibrosis7 and hepatocellular carcinoma.10 Furthermore, a population-based study of the National Health and Nutrition Examination Survey demonstrated that homozygosity with the risk allele is associated with a >8 fold increased hazard ratio for liver-related death.42 Stender and colleagues evaluated the quantitative impact of PNPLA3 risk variants on hepatic steatosis using magnetic resonance spectroscopy and described an important interaction between adiposity and genotype that impacts hepatic steatosis and the risk of cirrhosis.43 While previous studies demonstrated an association with fibrosis, an accurate continuous measure of liver fibrosis is required to quantify the impact of risk variants on liver fibrosis. Krawczyk et al, demonstrated an association between PNPLA3 genotype and liver stiffness on vibration-controlled transient elastography (VCTE), however, VCTE has inferior diagnostic accuracy and repeatability44 when compared to MRE, limiting its precision to quantify the impact of risk variants on liver fibrosis. Furthermore, their study cohort primarily consisted of patients with viral hepatitis with only 7.1% with NAFLD. This study utilised MRE, the most accurate, non-invasive, continuous marker of liver fibrosis in NAFLD. Furthermore, we demonstrated that the association between genotype and advanced fibrosis persisted despite adjustment for FIB-4 and quantified the association between genotype and hepatic steatosis using MRI-PDFF. Importantly, we demonstrated a strong interaction between age, PNPLA3 risk variants and fibrosis on MRE that began to diverge at age 50. Moreover, all patients with a very high liver stiffness, ≥5.0 kPa, carried at least one copy of the PNPLA3 risk variant, further underscoring its importance as a major risk factor for progressive liver fibrosis. Taken together, closer monitoring may be warranted for patients with PNPLA3 risk allele, even if they are low-risk based on non-invasive biomarker assessment, particularly when age 50 years or older.
Furthermore, we also evaluated additional genetic loci associated with NAFLD severity including TM6SF2, MBOAT7, GCKR and HSD17B13. Each risk allele had a point estimate consistent with a minor, non-significant impact on liver stiffness on MRE. However, the protective TA variant in HSD17B13 had a strong protective association on liver stiffness on MRE of similar magnitude to PNPLA3. To date, studies have focused on assessing the protective effect of HSD17B13 on aminotransferases15,45 or semi-quantitative measures on liver biopsy.46 This study demonstrates the magnitude of the variant on well-phenotyped patients using the most accurate, continuous biomarker of liver fibrosis, which can be used in future polygenic assessments of fibrosis risk.
4.2 |. Strengths and limitations
Although this study evaluates a diverse, prospectively recruited well-phenotyped cohort at risk for NAFLD, certain limitations merit acknowledgment. First, this is a cross-sectional study and to better evaluate the impact of genotype on transitioning to higher risk NAFLD, longitudinal studies will be required. However, the association between risk alleles and liver fibrosis persisted despite multivariable adjustment and we demonstrated a strong interaction between PNPLA3 genotype and age. Second, this is a single-centre study, and our sample size limited the ability to evaluate for interactions with obesity. However, despite being a single-centre study, our population was diverse with approximately one-third of patients of Hispanic ethnicity. Finally, we evaluated five loci with prior studies demonstrating a strong association with NAFLD. Additional, loci including NCAN,47 PPP1R3B48 and LYPLAL149 were not assessed. Despite this, few studies have evaluated the five included loci in patients with a detailed NAFLD assessment. Further, multi-centre studies utilising MRE may be required to better evaluate if a significant interaction between age, obesity, genetic risk and liver fibrosis in NAFLD and quantify a smaller impact of risk alleles in TM6SF2, MBOAT7 and GCKR.
4.3 |. Implications for future research
This study quantifies the impact of PNPLA3 genotype on liver fibrosis and demonstrates a strong association that persists despite adjustment for FIB-4 score. The clinical utility of genotyping for common variants, including PNPLA3, will be to identify patients at risk for a more aggressive disease trajectory. Rather than basing ongoing monitoring and interventions on a clinical prediction rule or liver stiffness measurement, consideration of genetic risk may focus interventions on patients with mild disease who are at risk for progression, particularly as they age beyond 50 years. HSD17B13 genotype may ameliorate the genetic risk of liver fibrosis with a similar magnitude as PNPLA3 risk variants increase risk and should be incorporated into a genetic risk assessment. In addition, emerging evidence suggests that PNPLA3 genotype may be associated with a differential response to certain treatments and PNPLA3 expression may represent a target for future pharmacotherapy of affected individuals.50
Currently, clinical trials of therapeutic agents in NAFLD must account for the heterogeneous trajectory of NASH including an up to 30% placebo response rate, which mandates larger sample sizes with substantially higher associated costs. Through the assessment of genetic risk, clinical trials can enrich populations at greatest risk for disease progression. By utilising an accurate, continuous biomarker of fibrosis, this study quantifies the effect of risk and protective variants and provides regression coefficients that may be incorporated into the calculation of polygenic risk. For example an individual who is PNPLA3 G/G, HSD17B13 T/TA may be intermediate risk and comparable to PNPLA3 C/G, HSD17B13 T/T. As regulatory bodies in Europe and the United States have emphasised improvement in fibrosis as a key endpoint, assessment of polygenic risk for fibrosis may substantially improve the trial design.
Future studies will need to incorporate polygenic risk and the interaction with lifestyle factors to better classify the risk of liver-related and all-cause mortality in patients at risk for NAFLD. In conclusion, our study demonstrates that PNPLA3 genotype is associated with liver fibrosis assessed on MRE and that the impact of being homozygous for the risk allele compared to wild-type could translate into the difference between minimal/no fibrosis and advanced fibrosis. Importantly, the impact of PNPLA3 risk alleles becomes pronounced in patients 50 years or older. These data require further validation in longitudinal multicentre studies to identify if PNPLA3 genotype can predict patients at risk for a more aggressive disease trajectory and if protective variants in HSD17B13 minimise that risk.
Supplementary Material
ACKNOWLEDGEMENT
Declaration of personal interests: Potential conflict of interest for Rohit Loomba: Dr Loomba serves as a consultant or advisory board member for Bird Rock Bio, Celgene, Enanta, GRI Bio, Madrigal, Metacrine, NGM, Sanofi, Arrowhead Research, Galmed, NGM, GNI, NovoNordisk, Merck, Siemens, Pfizer, Gilead. Glympsebio, In addition, his institution has received grant support from Allergan, BMS, BI, Daiichi-Sankyo Inc, Eli-Lilly, Galectin, Galmed, GE, Genfit, Intercept, Janssen Inc, Madrigal, Merck, NGM, Pfizer, Prometheus, Siemens, and Sirius. He is also co-founder of Liponexus Inc
Funding information
RL receives additional funding support from NIEHS (5P42ES010337), NCATS (5UL1TR001442), and NIDDK (R01DK106419 and P30DK120515) and DOD PRCRP (CA170674P2). VA is supported by NIDDK (K23DK119460). DD was supported by an ACTRI/NIH grant KL2TR001444, the San Diego Digestive Diseases Research Center (NIH DK120515) and the Southern California Research Center for ALPD and Cirrhosis funded by the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health under Award Number 5P50AA011999.
Footnotes
DATA AVAILABILITY STATEMENT
The summary statistics of data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
SUPPORTING INFORMATION
Additional supporting information will be found online in the Supporting Information section.
REFERENCES
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- 2.Loomba R, Wong R, Fraysse J, et al. Nonalcoholic fatty liver disease progression rates to cirrhosis and progression of cirrhosis to decompensation and mortality: a real world analysis of Medicare data. Aliment Pharmacol Ther. 2020;51:1149–1159. [DOI] [PubMed] [Google Scholar]
- 3.Ye Q, Zou B, Yeo YH, et al. Global prevalence, incidence, and outcomes of non-obese or lean non-alcoholic fatty liver disease: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2020;5:739–752. [DOI] [PubMed] [Google Scholar]
- 4.Caussy C, Soni M, Cui J, et al. Nonalcoholic fatty liver disease with cirrhosis increases familial risk for advanced fibrosis. J Clin Invest. 2017;127:2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. [DOI] [PubMed] [Google Scholar]
- 6.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51:1209–1217. [DOI] [PubMed] [Google Scholar]
- 8.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. [DOI] [PubMed] [Google Scholar]
- 9.Emdin CA, Haas M, Ajmera V, et al. Association of genetic variation with cirrhosis: a multi-trait genome-wide association and gene-environment interaction study. Gastroenterology. 2021; 160:1620–1633.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YL, Patman GL, Leathart JB, et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2014;61:75–81. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y-L, Reeves HL, Burt AD, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46:352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mancina RM, Dongiovanni P, Petta S, et al. The MBOAT7-TMC4 Variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zain SM, Mohamed Z, Mohamed R. Common variant in the glucokinase regulatory gene rs780094 and risk of nonalcoholic fatty liver disease: a meta-analysis. J Gastroenterol Hepatol. 2015;30:21–27. [DOI] [PubMed] [Google Scholar]
- 15.Abul-Husn NS, Cheng X, Li AH, et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu C, Caussy C, Imajo K, et al. Magnetic resonance vs transient elastography analysis of patients with nonalcoholic fatty liver disease: a systematic review and pooled analysis of individual participants. Clin Gastroenterol Hepatol. 2019;17:630–637.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venkatesh SK, Wang G, Teo LL, Ang BW. Magnetic resonance elastography of liver in healthy Asians: normal liver stiffness quantification and reproducibility assessment. J Magn Reson Imaging. 2014;39:1–8. [DOI] [PubMed] [Google Scholar]
- 18.Han MAT, Vipani A, Noureddin N, et al. MR elastography-based liver fibrosis correlates with liver events in nonalcoholic fatty liver patients: a multicenter study. Liver Int. 2020;40:2242–2251. [DOI] [PubMed] [Google Scholar]
- 19.Gidener T, Ahmed OT, Larson JJ, et al. Liver stiffness by magnetic resonance elastography predicts future cirrhosis, decompensation, and death in NAFLD. Clin Gastroenterol Hepatol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le T-A, Chen J, Changchien C, et al. Effect of colesevelam on liver fat quantified by magnetic resonance in nonalcoholic steatohepatitis: a randomized controlled trial. Hepatology. 2012;56:922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loomba R, Sirlin CB, Ang B, et al. Ezetimibe for the treatment of nonalcoholic steatohepatitis: assessment by novel magnetic resonance imaging and magnetic resonance elastography in a randomized trial (MOZART trial). Hepatology. 2015;61:1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomba R, Schork N, Chen C-H, et al. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015;149:1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doycheva I, Cui J, Nguyen P, et al. Non-invasive screening of diabetics in primary care for NAFLD and advanced fibrosis by MRI and MRE. Aliment Pharmacol Ther. 2016;43:83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loomba R, Cui J, Wolfson T, et al. Novel 3D magnetic resonance elastography for the noninvasive diagnosis of advanced fibrosis in NAFLD: a prospective study. Am J Gastroenterol. 2016;111: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sterling RK, Lissen E, Clumeck N, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. [DOI] [PubMed] [Google Scholar]
- 26.Angulo P, Hui JM, Marchesini G, et al. The NAFLD fibrosis score: a noninvasive system that identifies liver fibrosis in patients with NAFLD. Hepatology. 2007;45:846–854. [DOI] [PubMed] [Google Scholar]
- 27.Tang AN, Desai A, Hamilton G, et al. Accuracy of MR imaging-estimated proton density fat fraction for classification of dichotomized histologic steatosis grades in nonalcoholic fatty liver disease. Radiology. 2015;274:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang AN, Tan J, Sun M, et al. Nonalcoholic fatty liver disease: MR imaging of liver proton density fat fraction to assess hepatic steatosis. Radiology. 2013;267:422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeder SB, Robson PM, Yu H, et al. Quantification of hepatic steatosis with MRI: the effects of accurate fat spectral modeling. J Magn Reson Imaging. 2009;29:1332–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hines CDG, Frydrychowicz A, Hamilton G, et al. T(1) independent, T(2) (*) corrected chemical shift based fat-water separation with multi-peak fat spectral modeling is an accurate and precise measure of hepatic steatosis. J Magn Reson Imaging. 2011;33:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Permutt Z, Le TA, Peterson MR, et al. Correlation between liver histology and novel magnetic resonance imaging in adult patients with non-alcoholic fatty liver disease -MRI accurately quantifies hepatic steatosis in NAFLD. Aliment Pharmacol Ther. 2012;36:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park CC, Nguyen P, Hernandez C, et al. Magnetic resonance elastography vs transient elastography in detection of fibrosis and noninvasive measurement of steatosis in patients with biopsy-proven nonalcoholic fatty liver disease. Gastroenterology. 2017;152:598–607.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imajo K, Kessoku T, Honda Y, et al. Magnetic resonance imaging more accurately classifies steatosis and fibrosis in patients with nonalcoholic fatty liver disease than transient elastography. Gastroenterology. 2016;150:626–637.e7. [DOI] [PubMed] [Google Scholar]
- 34.Hsu E, Feghali-Bostwick CA. Insulin-like growth factor-II is increased in systemic sclerosis-associated pulmonary fibrosis and contributes to the fibrotic process via Jun N-terminal kinase-and phosphatidylinositol-3 kinase-dependent pathways. Am J Pathol. 2008;172:1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung J, Loomba RR, Imajo K, et al. MRE combined with FIB-4 (MEFIB) index in detection of candidates for pharmacological treatment of NASH-related fibrosis. Gut. 2020;0:1–8. https://gut.bmj.com/content/early/2020/11/18/gutjnl-2020-322976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dulai PS, Sirlin CB, Loomba R. MRI and MRE for non-invasive quantitative assessment of hepatic steatosis and fibrosis in NAFLD and NASH: clinical trials to clinical practice. J Hepatol. 2016;65:1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulai PS, Singh S, Patel J, et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: systematic review and meta-analysis. Hepatology. 2017;65:1557–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castera L, Friedrich-Rust M, Loomba R. Noninvasive assessment of liver disease in patients with nonalcoholic fatty liver disease. Gastroenterology. 2019;156:1264–1281.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh S, Allen AM, Wang Z, Prokop LJ, Murad MH, Loomba R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: a systematic review and meta-analysis of paired-biopsy studies. Clin Gastroenterol Hepatol. 2015;13:643–654.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleiner DE, Brunt EM, Wilson LA, et al. Association of histologic disease activity with progression of nonalcoholic fatty liver disease. JAMA Netw Open. 2019;2:e1912565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52:894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unalp-Arida A, Ruhl CE. Patatin-like phospholipase domain-containing protein 3 I148M and liver fat and fibrosis scores predict liver disease mortality in the U.S. population. Hepatology. 2020;71:820–834. [DOI] [PubMed] [Google Scholar]
- 43.Stender S, Julia Kozlitina G, Nordestgaard B, Tybjærg-Hansen A, Hobbs HH, Cohen JC. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat Genet. 2017;49:842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loomba R. Role of imaging-based biomarkers in NAFLD: recent advances in clinical application and future research directions. J Hepatol. 2018;68:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gellert-Kristensen H, Nordestgaard BG, Tybjaerg-Hansen A, Stender S. High risk of fatty liver disease amplifies the alanine transaminase-lowering effect of a HSD17B13 variant. Hepatology. 2020;71:56–66. [DOI] [PubMed] [Google Scholar]
- 46.Pirola CJ, Garaycoechea M, Flichman D, et al. Splice variant rs72613567 prevents worst histologic outcomes in patients with nonalcoholic fatty liver disease. J Lipid Res. 2019;60:176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gorden A, Yang R, Yerges-Armstrong LM, et al. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dongiovanni P, Meroni M, Mancina RM, et al. Protein phosphatase 1 regulatory subunit 3B gene variation protects against hepatic fat accumulation and fibrosis in individuals at high risk of nonalcoholic fatty liver disease. Hepatol Commun. 2018;2:666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlsson B, Lindén D, Brolén G, et al. Review article: the emerging role of genetics in precision medicine for patients with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2020;51:1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.