Abstract
Acute respiratory infections due to viral or bacterial etiology can cause 60 deaths per one lakh population. Viral etiology is more common as compared to bacterial, but lack of definite diagnosis leads to increased usage of empirical antibiotics. During the first wave of the COVID-19 pandemic, there was a need to identify co-infections especially in severe acute respiratory illness (SARI) patients to identify it as one of the cofactors for increased severity of illness and to identify the causative agents in COVID-19 negative individuals. The SARS CoV-2 real time PCR was carried out using ICMR approved kits and the other respiratory viruses were detected using the multiplex commercially available real time kit. A total of 186 patients presenting with either SARI (89.8%) or influenza like illness (10.2%) were included in the study. Out of these, 43 (23.1%) were positive for SARS CoV-2 RNA and 2 (4.6%) patients with SARI showed concomitant infection with either human rhinovirus or human respiratory syncytial virus . Out of 143 patients negative for SARS CoV-2, 35 (24.5%) were positive for one or more microbial infections and 28 (19.6%) infected with other respiratory viral infection most common being human rhinovirus. The results suggest that viral coinfections are significantly higher among COVID-19 negative individuals (24.5% vs 4.6%) presenting with respiratory illness as compared to COVID-19 positive individuals possibly due to viral interference and competitive advantage of SARS-CoV-2 in modulating the host immunity. Further detailed research is required for the understanding of mechanisms of viral co-infection.
Keywords: Respiratory co-infection, SARI, ILI, First COVID-19 wave, Empirical antibiotic overuse
Introduction
According to the World Health Organization (WHO), acute respiratory infections (ARI) is defined as a sudden onset of respiratory symptoms like cough, coryza, fever and shortness of breath [1]. Due to ARI, there are 60 deaths per 1,00,000 population and viral etiology accounts for 30–70% of the cases [2]. The clinical differentiation of viral and bacterial illness can be difficult, hence there is a need for lab confirmation to identify the definitive etiology so that early antibiotics can be initiated for bacterial pneumonia while restricting their use in viral pneumonia.
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) which belongs to the group beta-coronavirus is a novel coronavirus which was declared a pandemic by WHO on 11th March 2020 [3]. Transmission of SARS-CoV-2 occurs primarily via contact with infected individuals, contact with respiratory droplets/fomites and airborne transmission by smaller droplets. Even though airborne transmission is not the primary route of transmission, this mode of spread can occur in enclosed spaces with poor ventilation and more than 30 min exposure with an infected person [4, 5]. The clinical manifestations due to this virus can range from asymptomatic infection seen in 85–90% cases to mild illness known as influenza like illness (ILI) to severe acute respiratory illness (SARI) requiring hospitalization [6].
The co-infection of other viral and bacterial agents in COVID-19 positive patients can lead to adverse outcomes in the form of increased mortality or prolonged hospitalization in these patients. Approximately 90% patients have been prescribed empirical antibiotics during the pandemic era [7]. The widespread and inappropriate use of antibiotics is likely to lead to antibiotic resistance (AMR) and also disturbance of microflora leading to antibiotic associated diarrhoea, which has a major impact on global health and the world economy [8, 9]. Early diagnosis and identification of causative agents will lead to better case management, reducing the cost of treatment and also reduce evolution of resistant pathogens in the hospital setup.
Multiplex real time RT-PCR is the gold standard method for the detection of co-infections [10] as the individual detection of these pathogens individually is time consuming and laborious [11]. During the pandemic era, there has been an increased focus on the presence of SARS-CoV-2, while the presence of co-infection in SARI/ILI patients with or without SARS-CoV-2 has been neglected. Some studies from Singapore have shown that only 8 (1.8%) out of 431 COVID positive patients had co-infection. Wang et al., from China have shown that 5.8% of SARS-CoV-2 infected patients and 18.4% of patients without COVID-19 had co-infection with other respiratory pathogens [12, 13]. However, to the best of our literature search, there is scarcity of data on the presence of co-infections in COVID-19 patients in the Indian scenario. Therefore, this study aimed to see the prevalence of co-infections among the COVID-19 suspected patients with SARI and ILI patients presenting to a tertiary care hospital in North India.
Materials and methods
Study participants
This study was a retrospective cross-sectional study which included nasopharyngeal swab samples routinely received in the Department of Virology, PGIMER, Chandigarh for COVID 19 testing. Detection of SARS-CoV-2 by RT-PCR was done for all the samples, which were processed in Biosafety cabinet class II A2 following all biosafety precautions. The RNA extraction was carried out by commercially available kits as per the manufacturer’s instructions and stored in – 80 °C in case of delay in testing. The RT-PCR was performed using the ICMR approved kits available during different time points following ICMR guidelines [14]. A total of 186 patients presented with respiratory illness (SARI/ILI) from April to September 2020 were included in the study. The study patients were categorized retrospectively as Group 1: SARI patients with COVID-19 (n = 28), Group 2: SARI patients without COVID-19 (n = 139) and Group 3: ILI patients with and without COVID-19 (n = 19).
Detection of co-infection by multiplex RT-PCR
The internal control (IC) containing the Equine Arteritis Virus (EAV) provided in the kit of FTD respiratory pathogen 21 kit (Fast-Tack Diagnostics, Europe) was used to spike the samples and negative control before extraction by commercial extraction kits. The FTD respiratory pathogen 21 kit uses TaqMan probe technology to detect 21 respiratory pathogens. These pathogens are Influenza A virus (IAV), Influenza B virus (IBV), Influenza A (H1N1) lineage virus, Human Coronaviruses (229E, NL63, OC43 and HKU1), Parainfluenza viruses (HPIV)-1,2,3 and 4, Human Metapneumovirus (HMPV) A & B, Human respiratory syncytial viruses (HRSV) A & B, Human Bocavirus (HBoV), Human Rhinovirus (HRV), Human Parechovirus (HPeV), Human Adenovirus (HAdV), Enterovirus (EV) and Mycoplasma pneumoniae. The Multiplex single tube RT-PCR was carried out using 12.5 µl of 2X RT PCR buffer, 1 µl of RT enzyme mix, 1.5 µl of five different primer probe mix individually added to mixture containing 10 µl RNA template. Reverse transcription at 50 °C for 15 min, initial denaturation at 94 °C for 1 min followed by 40 cycles of denaturation at 94 °C for 8 s and annealing at 60 °C for 1 min.
Statistical analysis
Mean age was calculated for all the groups and between the groups Pearson’s Chi square test was applied for categorical variable. p value of less than 0.05 was considered statistically significant.
Results
Demographic distribution and clinical presentation
Out of the 186 recruited patients, 82 (44.1%) belonged to Punjab, 49 (26.3%) were from Chandigarh, 35 (18.8%) from Haryana, 10 (5.3%) from Himachal Pradesh and 10 (5.3%) from other states.
SARI patients (n = 167)
This group included 28 (16.7%) SARI patients with COVID-19 (Group 1) and 139 (83.23%) SARI patients without COVID-19 (Group 2). Out of these, 57 (34.1%) patients were > 60 years of age and 32 (19.2%) were in the paediatric age group (≤ 15 years). The male: female ratio was 1.5:1.
The most common symptoms were breathlessness (79.6%), dry cough (71.2%) and fever (63.4%). Other less common symptoms were sore throat (26.9%), cough with expectoration (18.5%) and myalgia (14.9%). A total of 93 (55.6%) patients had underlying comorbid conditions which included hypertension (21.5%), followed by diabetes mellitus (17.3%) and cardiac disease (15.5%). Chronic renal disease and immunocompromised conditions were observed in 9.5% of SARI patients. The underlying comorbid conditions were more prevalent in SARI patients (Group 1 and 2) as compared to the other groups (p value-0.006). No significant difference was observed in the symptoms of SARI patients presenting with and without COVID-19 (Table 1).
Table 1.
Demographic parameters in SARI patients with and without COVID-19
| Demographic parameter | Total (n = 167) |
SARI patients with COVID-19 (Group 1, n = 28) |
SARI patients without Covid-19 (Group 2, n = 139) |
p value |
|---|---|---|---|---|
| Age (years) | ||||
| < 12 | 23 (13.7%) | 2 (7.1%) | 21 (15.1%) | 0.309 |
| 12–40 | 43 (25.7%) | 5 (17.8%) | 38 (27.3%) | |
| 41–60 | 45 (26.9%) | 8 (28.5%) | 37 (26.6%) | |
| ≥ 61 | 55 (32.9%) | 13 (46.4%) | 43 (30.9%) | |
| Gender | ||||
| Male | 101 (60.4%) | 17 (60.7%) | 84 (60.4%) | 0.978 |
| Female | 66 (39.5%) | 11 (39.28%) | 55 (39.5%) | |
| Symptoms | ||||
| Cough | 119 (71.2%) | 20 (71.4%) | 99 (71.2%) | 0.206 |
| Fever | 106 (63.4%) | 17 (60.7%) | 89 (64.0%) | |
| Sore throat | 45 (26.9%) | 9 (32.14%) | 36 (25.8%) | |
| Chest pain | 8 (4.7%) | 1 (3.5%) | 7 (5.0%) | |
| Abdominal pain | 15 (8.9%) | 1 (3.5%) | 14 (10.0%) | |
| Diarrhoea | 10 (5.9%) | 2 (7.1%) | 8 (5.7%) | |
| Nausea | 7 (4.1%) | 2 (7.1%) | 5 (3.5%) | |
| Body ache | 25 (14.9%) | 5 (17.8%) | 20 (14.3%) | |
| Sputum | 31 (18.5%) | 2 (7.1%) | 29 (20.3%) | |
| Vomiting | 7 (4.1%) | 0 (0%) | 7 (5.0%) | |
| Haemoptysis | 4 (2.3%) | 2 (7.1%) | 2 (1.4%) | |
| Nasal discharge | 8 (4.7%) | 1 (3.5%) | 7 (5.9%) | |
| Breathlessness | 133 (79.6%) | 23 (82.14%) | 110 (79.1%) | |
*p value < 0.05-Significant
ILI patients (n = 19)
Out of these ILI patient group, 15 (78.9%) were found to be COVID-19 positive. The median age group was 32 years (range 0.2–75 years). The male: female ratio was 1.1:1. Among these patients, the underlying comorbid conditions included chronic kidney disease (CKD), heart disease, hypertension and immunocompromised in 15.7% each and diabetes mellitus in 5.5% patients. The details of these patients are summarized in Table 2. The presence of sore throat was significantly higher in ILI patients positive for COVID-19 as compared to ILI without COVID-19.
Table 2.
Clinical presentation in ILI patients (Group 3)
| Demographic parameter | Total (n = 19) |
ILI patients with COVID-19 (n = 15) |
ILI patients without Covid-19 (n = 4) |
p value |
|---|---|---|---|---|
| Age (years) | ||||
| Median (range) | 32 (0.2–75 years) | 30 (0.5–60 years) | 53 (0.2–75 years) | 0.073 |
| Gender | ||||
| Male | 10 (52.6%) | 8 (53.3%) | 2 (50%) | 0.906 |
| Female | 9 (47.3%) | 7 (46.6%) | 2 (50%) | |
| Symptoms | ||||
| Cough | 17 (89.4%) | 13 (86.6%) | 4 (100%) | 0.44 |
| Fever | 15 (78.9%) | 11 (73.3%) | 4 (100%) | 0.245 |
| Sore throat | 13 (68.4%) | 13 (86.6%) | 0 (0%) | 0.001* |
| Body ache | 2 (10.5%) | 1 (6.6%) | 1 (25%) | 0.288 |
| Sputum | 1 (5.2%) | 1 (6.6%) | 0 (0%) | 0.59 |
| Haemoptysis | 1 (5.2%) | 1 (6.6%) | 0 (0%) | 0.596 |
*p value < 0.05-Significant
Detection of viral and bacterial respiratory pathogens
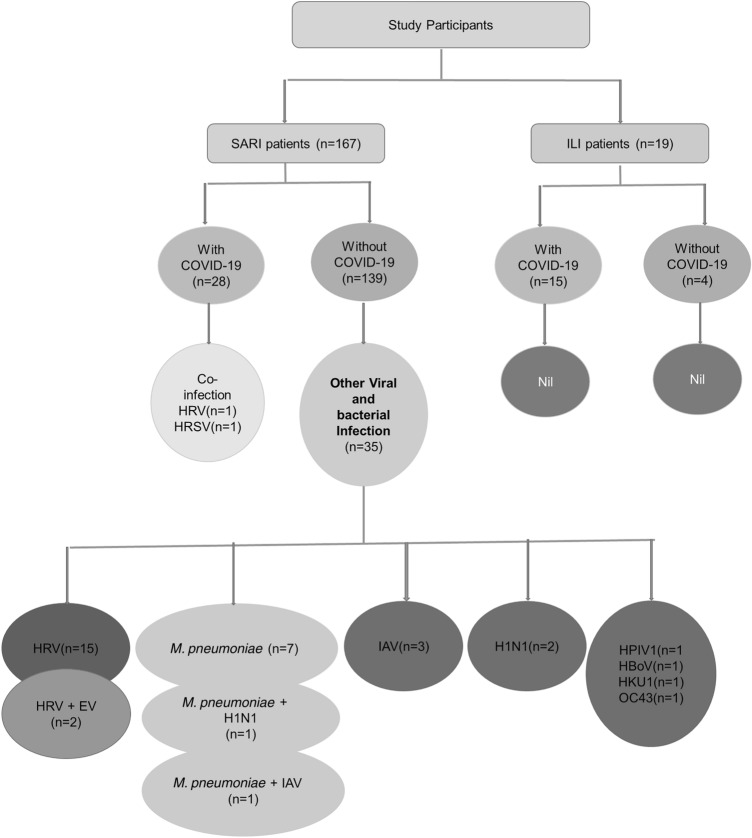
Out of 186 patients, 37 (19.8%) were found to be infected with respiratory pathogens. Among these, all (n = 37,100%) patients presented with SARI. No ILI patients showed the presence of any associated infection. Out of 37 patients, 33 (89.2%) were found to have infection with a single pathogen and 4 (10.8%) showed dual infection.
Among 33 patients showing the presence of single infection of respiratory pathogens, HRV infection accounted for 16 (48.5%, 16/33) followed by detection of Mycoplasma pneumoniae in 7 (21.2%), IAV in 3 (9.1%), Influenza A/ H1N1 in 2 (6.1%) and one (3%) each of HKU1, OC43, HPIV-1, HRSV and HBoV as represented in Fig. 1. One patient with HRV infection and one with HRSV were concomitantly infected with SARS-CoV-2 and both belonged to SARI category. Four patients with dual infection: two patient had HRV and EV, Mycoplasama pneumoniae with H1N1 and Mycoplasma pneumoniae with IAV (other than H1N1) in one each. The data was shown in Fig. 1. Of 143 SARS-CoV-2 negative patients, 19.6% (n = 28) had respiratory viral infection and Mycoplasma pneumoniae accounted for 6.3% (n = 9). The cycle threshold (Ct value) for these respiratory pathogens was found to be between 18 and 33 with proper sigmoid curves.
Fig. 1.
Flowchart showing the co-infections in study participants
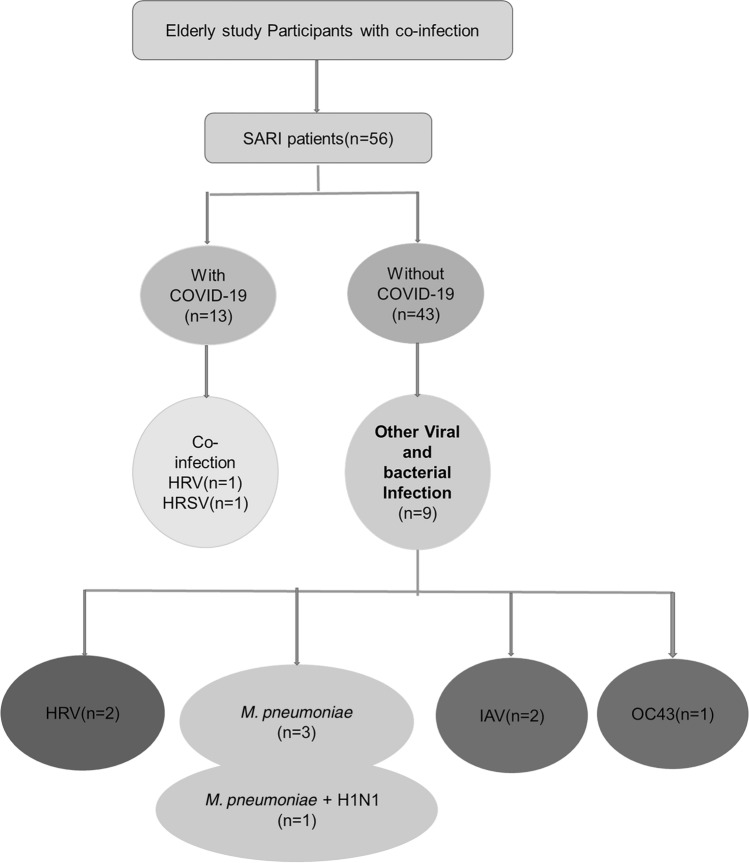
Higher rate of other microbial infection was seen in elderly SARI patients without COVID-19 as compared to those with COVID-19 (9/43, 20.9% vs 2/13, 15.4%) and details are shown in Fig. 2.
Fig. 2.
Flowchart of elderly SARI patients with co-infections
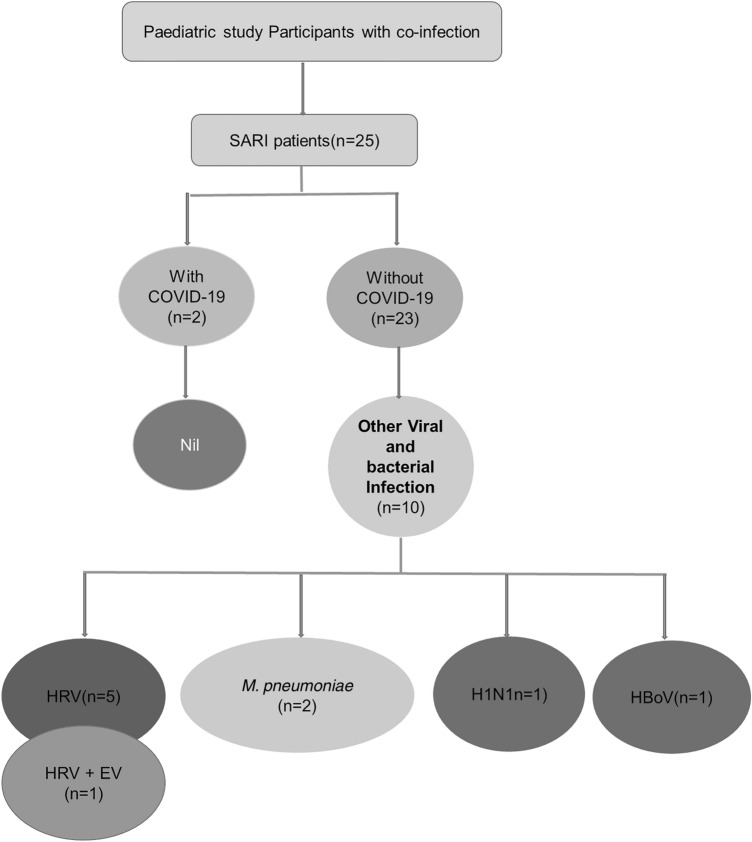
Similarly, higher rate of other microbial infection was seen in paediatric SARI patients without COVID-19 as compared to those with COVID-19 (10/23, 43.5% vs 0/2, 0%) and details are shown in Fig. 3. Three paediatric patients positive for HRV were found to be immunocompromised or had underlying cardiac disease.
Fig. 3.
Flowchart of paediatric SARI patients with co-infections
Discussion
The most common viruses implicated in ARI includes Human respiratory syncytial virus (HRSV), influenza virus (IAV, H1N1, IBV), parainfluenza virus (HPIV), human Bocavirus (HBoV), human metapneumovirus (HMPV), adenovirus (HAdV), rhinovirus (HRV), enterovirus (EV) and coronaviruses. [2, 15]. Early diagnosis is of utmost importance for timely management and initiation of specific antimicrobial/antiviral therapy. However, due to non-availability of rapid diagnostic techniques in most diagnostic laboratories, empirical broad spectrum antibiotic therapy is started based upon radiological or clinical findings.
Since the start of COVID-19 pandemic, a large number of SARI/ILI patients are being suspected and investigated mainly for SARS-CoV-2 infection by Real time PCR/Rapid antigen test (RAT) than any other viral infections. The disease severity and increased mortality has been reported among elderly individuals and/ or with comorbid conditions such as diabetes mellitus, hypertension, chronic kidney disease, coronary artery disease, patients on immunosuppression etc. Though molecular techniques have been established almost all across the nation, the detection of multiple pathogens by uniplex PCR is time consuming and laborious. Therefore, the present study was planned to understand the prevalence of different infectious agents among SARI/ILI patients during a COVID-19 pandemic situation by using commercially available multiplex Real time PCR kits (FTD respiratory pathogen 21 kit, Fast-Tack Diagnostics, Europe).
The majority 167 (89.8%) of the study patients included in the present study were SARI patients as they were more likely to get admitted in the hospital and to have concomitant infections due the disease severity. Among SARI, male patients were 1.5 times more affected as compared to females and similar gender predominance was seen among COVID-19 patients [16]. The predominant symptoms among SARI patients were breathlessness (79.6%) followed by cough (71.2%) and fever (63.4%). On the other hand, ILI patients had cough (89.4%), fever (78.9%) and sore throat (68.4%) as their symptoms. In a previous study from our Centre, mortality rates among COVID-19 SARI and non-COVID-19 SARI were 31.6% and 28.2% respectively with no statistically significant difference (p = 0.593) according to Pannu et al. study [17].
In the COVID-19 pandemic era, the antibiotic use for hospitalized ARI patients is about 74.5% according to Cox et al., 2020 [18]. The results of the present study showed that co-infection with other respiratory pathogen was not seen among the majority of the SARI COVID-19 positive patients which is in concordance with study by Nowak et al., 2020 and Contou et al., 2020 [19, 20]. The possible reason would be the competitive advantage of SARS-COV-2 in modulating the host immunity [19]. Respiratory pathogens other than SARS-CoV-2 were detected among 35/143, (24.5%) of SARI patients who were negative for COVID-19. Among the SARI with COVID-19 positive patients, two (7.1%, 2/28) had infection with HRV and HRSV. The results of the present study are also in concordance with the study conducted by Wang et al., from Wuhan, China which showed; out of 613 patients tested for the presence of 13 respiratory pathogens only 5.8% of SARS-CoV 2 infected patients and 18.4% of patients without COVID-19 had co-infection [13]. On the contrary, the study conducted by Kim et al., in the US showed that out of 1217 patients there was co-infection in 19.8% of SARS CoV 2 infected patients and 26.5% of patients without COVID-19 and hence there was no significant difference in co-infection among these patients [21].
Identification of pathogens among SARI patients without COVID-19 is crucial. Timely establishment of diagnosis and better patient management is necessary among critically ill patients. Among the 143 patients without SARS-CoV-2, about 28 (19.6%) infected with other respiratory viral infection and Mycoplasma pneumoniae accounted for 9 (6.3%). Among the viral infections, HRV and IAV were the most common respiratory pathogens. Viral infections being more common, our study findings do not support the use of broad spectrum antibacterial as the first line therapy, which may further increase the chance of emergence of resistant bacteria. Explicit testing of viral pathogen may be able to strengthen antimicrobial stewardship by rationalizing antibiotic policy as well as saving on the unnecessary use of antibiotics considering the cost–benefit ratio. However, azithromycin is a broad spectrum antibiotic was widely used in pandemic scenario [22].
Co-infection among the SARS-CoV-2 positive SARI patients was found in 1% (2/167) of the study participants. Similarly, results were found in Wee et al. study, which shows the co-infection rate between SARS-CoV-2 and other respiratory viruses was found to be low, at 1.4% [12]. The findings signifies that co-infections were uncommon among SARI COVID-19 patients during the first wave of the pandemic. Paediatric population is vulnerable to respiratory infections and 19.2% of the study participants belonged to the paediatric age group. Among the paediatric population, SARI without COVID-19, had higher rate of microbial infection compared to SARI with COVID-19 and the most common respiratory pathogen was HRV (24%). These results are consistent with Canela et al. study where the common pathogens were influenza H1N1 (43.1%), EV/HRV (41.4%), HRSV (12.1%), HMPV (12.1%), HAdV (6.9%), and HBoV (3.5%) with viral co-infection seen in 22.4% of the paediatric cases [23].
Several studies and reports have highlighted the presence of co-infection of Influenza virus with SARS-CoV-2 [24–28]. Hence, IAV, H1N1 virus, IBV and SARS-CoV-2 should be available as a combo surveillance kit for detect most common pathogens in a single multiplex kit which would help in early diagnosis and detection of circulating strains among patients presenting with ARI symptoms. This will help the clinician in establishing the diagnosis in a cost effective manner in a short span of time. Moreover, additional studies are required to establish whether simultaneous viral infection in SARS-CoV-2 patients could potentially drive viral interference or influence disease outcome. There is also a need to study the co-infection among severe COVID-19 patients admitted in ICU patients as emergence of lymphocytopenia in COVID-19 patient results in a state of immunodeficiency, which makes it easier to be co-infected with other respiratory pathogens. The results of the present study showed that co-infections with other respiratory pathogens were uncommon among COVID-19 patients in the tertiary care hospital of north India. The major limitation of the present study was the small sample size, variation in the number of included study participants and inability to find patient’s disease outcome because it is a lab based retrospective study.
In conclusion, significantly higher viral co-infection was found among COVID-19 negative individuals (24.5%) in comparison with COVID-19 positive individuals (4.6%). HRV and IAV being most common among the viral infections and establishment of definitive diagnosis would decrease the unnecessary antibiotic use among these patients thereby, reducing the evolution of superbugs which cause nosocomial infection. Management of patients with early diagnosis and conclusive treatment is the requirement in this era for better patient care hence multiplexing assays are the need of the hour.
Acknowledgements
This work was carried out under the activity of Regional VRDL at PGIMER, Chandigarh. The support from PGIMER and ICMR is duly acknowledged.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or non-profit sectors.
Declarations
Conflict of interest
Authors declare that they have no conflict of interest.
Consent for publication
All authors have read the manuscript and approved the final manuscript.
Ethical approval
This study was approved by Institutional ethical committee vide no. NK/7067/Study/159 dated 13.02.2021.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Monika Sapra and Sangeetha Kirubanandhan share combined first author.
References
- 1.Identification of ILI/SARI/ARI cases As per WHO case definition (Referred and Systematic collected samples). 2021. https://dhr.gov.in/sites/default/files/Influenza%20Algorithm.pdf. Accessed 10 Aug 2021.
- 2.Hatem A, Mohamed S, Elhassan UEA, Ismael EAM, Rizk MS, El-Kholy A, et al. Clinical characteristics and outcomes of patients with severe acute respiratory infections (SARI): results from the Egyptian surveillance study 2010–2014. Multidiscip Respir Med. 2019;14(1):1–12. doi: 10.1186/s40248-019-0174-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medicine TLR. COVID-19 transmission—up in the air. Lancet Respir Med. 2020;8(12):1159. doi: 10.1016/S2213-2600(20)30514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations. 2021. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations. Accessed 20 Jul 2021.
- 6.Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 7.Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. 2018;11:1645. doi: 10.2147/IDR.S173867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahony JB. Detection of respiratory viruses by molecular methods. Clin Microbiol Rev. 2008;21(4):716–747. doi: 10.1128/CMR.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malhotra B, Swamy MA, Reddy PVJ, Kumar N, Tiwari JK. Evaluation of custom multiplex real-time RT-PCR in comparison to fast-track diagnostics respiratory 21 pathogens kit for detection of multiple respiratory viruses. Virol J. 2016;13(1):1–7. doi: 10.1186/s12985-016-0549-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wee LE, Ko KKK, Ho WQ, Kwek GTC, Tan TT, Wijaya L. Community-acquired viral respiratory infections amongst hospitalized inpatients during a COVID-19 outbreak in Singapore: co-infection and clinical outcomes. J Clin Virol. 2020;128:104436. doi: 10.1016/j.jcv.2020.104436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Wu Q, Xu W, Qiao B, Wang J, Zheng H, et al. Clinical diagnosis of 8274 samples with 2019-novel coronavirus in Wuhan. MedRxiv. 2020 doi: 10.1101/2020.02.12.20022327. [DOI] [Google Scholar]
- 14.ICMR Guidelines for COVID-19 testing in Private Laboratories. 2020. https://www.icmr.gov.in/pdf/covid/labs/Notification_ICMR_Guidelines_Private_Laboratories.pdf. Accessed 25 Jul 2021.
- 15.Cimolai N. Complicating infections associated with common endemic human respiratory coronaviruses. Health Secur. 2021;19(2):195–208. doi: 10.1089/hs.2020.0067. [DOI] [PubMed] [Google Scholar]
- 16.Bwire GM. Coronavirus: why men are more vulnerable to Covid-19 than women? SN Compr Clin Med. 2020;2(7):874–876. doi: 10.1007/s42399-020-00341-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pannu AK, Kumar M, Singh P, Shaji A, Ghosh A, Behera A, et al. Severe acute respiratory infection surveillance during the initial phase of the COVID-19 outbreak in North India: a comparison of COVID-19 to other SARI causes. Indian J Crit Care Med. 2021;25(7):761–767. doi: 10.5005/jp-journals-10071-23882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox MJ, Loman N, Bogaert D, O'Grady J. Co-infections: potentially lethal and unexplored in COVID-19. Lancet Microbe. 2020;1(1):e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowak MD, Sordillo EM, Gitman MR, Mondolfi AEP. Co-infection in SARS-CoV-2 infected patients: where are influenza virus and rhinovirus/enterovirus? J Med Virol. 2020;92(10):1699–1700. doi: 10.1002/jmv.25953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Contou D, Claudinon A, Pajot O, Micaëlo M, Flandre PL, Dubert M, et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann Intensive Care. 2020;10(1):1–9. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim D, Quinn J, Pinsky B, Shah NH, Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020;323(20):2085–2086. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldenburg CE, Doan T. Azithromycin for severe COVID-19. Lancet. 2020;396(10256):936–937. doi: 10.1016/S0140-6736(20)31863-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canela LNP, Magalhães-Barbosa MCd, Raymundo CE, Carney S, Siqueira MM, Prata-Barbosa A, et al. Viral detection profile in children with severe acute respiratory infection. Braz J Infect Dis. 2018;22:402–411. doi: 10.1016/j.bjid.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miatech JL, Tarte NN, Katragadda S, Polman J, Robichaux SB. A case series of coinfection with SARS-CoV-2 and influenza virus in Louisiana. Respir Med Case Rep. 2020;31:101214. doi: 10.1016/j.rmcr.2020.101214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashemi SA, Safamanesh S, Ghafouri M, Taghavi MR, Heydari MSMZ, Ahmadabad HN, et al. Co-infection with COVID-19 and influenza A virus in two died patients with acute respiratory syndrome, Bojnurd, Iran. J Med Virol. 2020 doi: 10.1002/jmv.26014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Lai X, Chen Z, Tu S, Qin K. Clinical characteristics of critically ill patients co-infected with SARS-CoV-2 and the influenza virus in Wuhan, China. Int J Infect Dis. 2020;96:683–687. doi: 10.1016/j.ijid.2020.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue H, Zhang M, Xing L, Wang K, Rao X, Liu H, et al. The epidemiology and clinical characteristics of co-infection of SARS-CoV-2 and influenza viruses in patients during COVID-19 outbreak. J Med Virol. 2020;92(11):2870–2873. doi: 10.1002/jmv.26163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng Y, Ma J, Wang H, Wang X, Hu Z, Li H, et al. Co-infection of influenza A virus and SARS-CoV-2: A retrospective cohort study. J Med Virol. 2021;93(5):2947–2954. doi: 10.1002/jmv.26817. [DOI] [PMC free article] [PubMed] [Google Scholar]