Abstract
Uveal melanoma is the most common intraocular cancer in the adult eye. R183 and Q209 were found to be mutational hotspots in exon 4 and exon 5 of GNAQ and GNA11 in Caucasians. However, only a few studies have reported somatic mutations in GNAQ or GNA11 in uveal melanoma in Chinese. We extracted somatic DNA from paraffin-embedded biopsies of 63 Chinese uveal melanoma samples and sequenced the entire coding regions of exons 4 and 5 in GNAQ and GNA11. The results showed that 33% of Chinese uveal melanoma samples carried Q209 mutations while none had R183 mutation in GNAQ or GNA11. In addition, seven novel missense somatic mutations in GNAQ (Y192C, F194L, P170S, D236N, L232F, V230A, and M227I) and four novel missense somatic mutations in GNA11 (R166C, I200T, S225F, and V206M) were found in our study. The high mutation frequency of Q209 and the novel missense mutations detected in this study suggest that GNAQ and GNA11 are common targets for somatic mutations in Chinese uveal melanoma.
Keywords: uveal melanoma, GNAQ, GNA11, somatic mutations, Chinese
Background
Uveal melanoma is the most common primary malignancy in the adult eye in Western countries.1 It is a cancer of the melanocytes in the choroid, ciliary body, and iris, which comprise the uvea. The most frequent (90%) site of uveal melanoma is in the choroid.2 Melanoma in the uvea comprises 5% of all melanomas,3 and has a particularly strong tendency to metastasize to the liver.4
The pathogenesis underlying uveal melanoma is unclear. To date, there are few known risk factors for uveal melanoma, one of which is nevus of Ota, which is an intradermal proliferation of melanocytes that manifests clinically as scleral and periorbital bluish-gray hyperpigmentation.5,6 Uveal melanoma is more commonly found in patients who are older.4 Phenotypic features such as light skin and high susceptibility to sunburn, as well as environmental exposure to ultraviolet radiation, are some of the risk factors that have been strongly implicated in the mutagenesis and carcinogenesis underlying cutaneous melanoma; however, the epidemiologic evidence to support the same connection in uveal melanoma remains inconsistent.7 Furthermore, a predisposition to uveal melanoma based on race remains unclear, despite the observation that uveal melanoma is much more common in people with light skin compared to those with darker skin.8 In Chinese, as in populations from the Western hemisphere, uveal melanoma is the most frequently occurring primary cancer in the adult eye, and was found to be more frequently encountered than previously suggested.9
Recent investigation by Van Raamsdonk et al. into the genetic predisposition to uveal melanoma has revealed that frequent somatic mutations (R183 or Q209) in GNAQ and its paralogous gene GNA11 are found in 83% of uveal melanomas, and this has established the oncogenic potential of these genes in uveal melanoma.6,10 In light of the finding of these mutational hotspots in GNAQ and GNA11 in Caucasians, we hypothesized that somatic mutations in GNAQ or GNA11 may also be frequently observed in uveal melanoma in the Chinese population. We sequenced and explored the most frequent mutations and novel missense somatic mutations in the entire coding regions of exons 4 and 5 in GNAQ and GNA11 in Chinese uveal melanoma samples.
Methods
Participants and clinical examinations
This study used de-identified tumor samples. Patient privacy sensitive information was removed and eye samples anonymized before study. The patients were diagnosed with uveal melanoma by clinical history, standard complete ophthalmic examination, and ancillary study. A total of 63 eyes enucleated from Chinese uveal melanoma patients were paraffin-embedded between 2005 and 2011.
PCR and DNA sequencing
Somatic DNA samples were extracted from paraffin–embedded uveal melanoma biopsies with a QIAamp DNA FFPE Tissue Kit (Qiagen Inc., Chatsworth, CA, USA) according to the manufacturer’s instructions. The exon 4 and exon 5 of GNAQ and GNA11 were amplified by nest PCR, which was performed in a Bio–Rad T100 thermal Cycler. Using outer set primers (Table S1) and 10ng extracted somatic DNA as template: 1st PCR procedure initial denaturation at 95 °C for 3 minutes, followed by 36 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, then extension at 72 °C for 5 min, and keep at 4 °C for 10 min. Using inner set primers (Table S1) and 1 µL 1st PCR product as template, 2nd PCR procedure is: initial denaturation at 95 °C for 3 minutes, followed by 33 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, then extension at 72 °C for 5 min, and keep at 4 °C for 10 min.
Direct nucleotide sequence analysis was completed for both GNAQ and GNA11. The exon 4 and exon 5 of GNAQ and GNA11 were amplified by PCR and sequenced on Genetic Analyzer 3130 (Applied Biosystems). Amplified exon 4 and exon 5 of GNAQ and GNA11 were sequenced on Genetic Analyzer 3130 (Applied Biosystems) using one of inner primers (Table S1).
Results
Demographic information
Of 63 Chinese samples with primary uveal melanoma, 55.6% were from males. The average age of patients was 51 ± 1.8 years old.
Mutations in Q209 in exons 5 of GNA11 and GNAQ
Of 63 Chinese samples with uveal melanoma, we found seven somatic Q209 mutations in GNAQ (11.1%) and 14 Q209 mutations in GNA11 (22.2%) (Table 1). This equates to a frequency of 33.3% of samples carrying a Q209 mutation in GNAQ or GNA11. Among the seven mutations affecting the glutamine codon 209 in GNAQ, there were five (71.4%) substitutions for proline (Q209P) and two (28.6%) substitutions for leucine (Q209L) (Fig. 1). Among the 14 mutations affecting the glutamine codon 209 in GNA11, there were 11 (78.6%) substitutions for leucine (Q209L), one (7.1%) substitution for histidine (Q209H), one (7.1%) substitution for proline (Q209P), and one (7.1%) substitution for lysine (Q209K) (Fig. 1).
Table 1 .
Frequencies of mutations in R183 (exon 4) and Q209 (exon 5) of GNAQ and GNA11.
| GNAQ | GNA11 | Study | ||||||
|---|---|---|---|---|---|---|---|---|
| R183 mutations | Q209 mutations | R183 mutations | Q209 mutations | |||||
| Number/total | Frequency, % | Number/total | Frequency, % | Number/total | Frequency, % | Number/total | Frequency, % | |
| 0/63 | 0 | 7/63 | 11.1 | 0 | 0 | 14/63 | 22.2 | Present study |
| 4/145 | 2.8 | 73/163 | 44.8 | 3/145 | 2.1 | 52/163 | 31.9 | Ref. [10] |
Figure 1 .
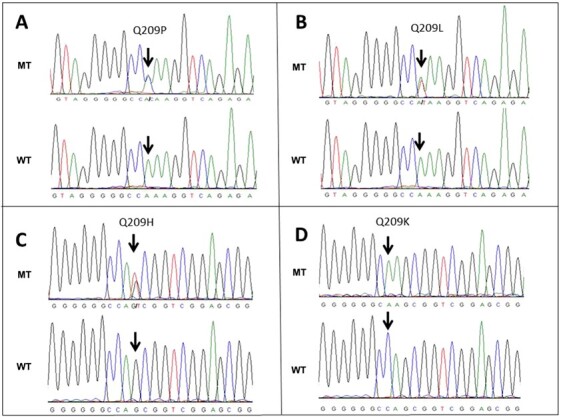
Mutations in Q209 in exons 5 of GNA11 and GNAQ. MT, Mutant; WT, Wild type.
Mutations in R183 in exons 4 of GNAQ and GNA11
No somatic mutations in R183 in exons 4 of GNAQ and GNA11 were found in 63 Chinese uveal melanoma samples (Table 1).
Novel mutations in GNAQ
We included the entire coding regions of exons 4 and 5 of GNAQ in our sequencing analysis (Fig. 2A). In exon 4 of GNAQ, we found three novel somatic mutations: Y192C, F194L and P170S (Table 2, Fig. 2B). Individual M6, in addition to carrying the novel mutation in codon 192, also had a compound heterozygous novel mutation in codon 166 in exon 4 of the paralogous gene GNA11. Individual M11 had a novel mutation in codon 194 (F194L) in addition to a compound heterozygous mutation Q209L. Individual M45 had two novel compound heterozygous mutations in GNAQ, including P170S in exon 4 and M227I in exon 5.
Figure 2 .
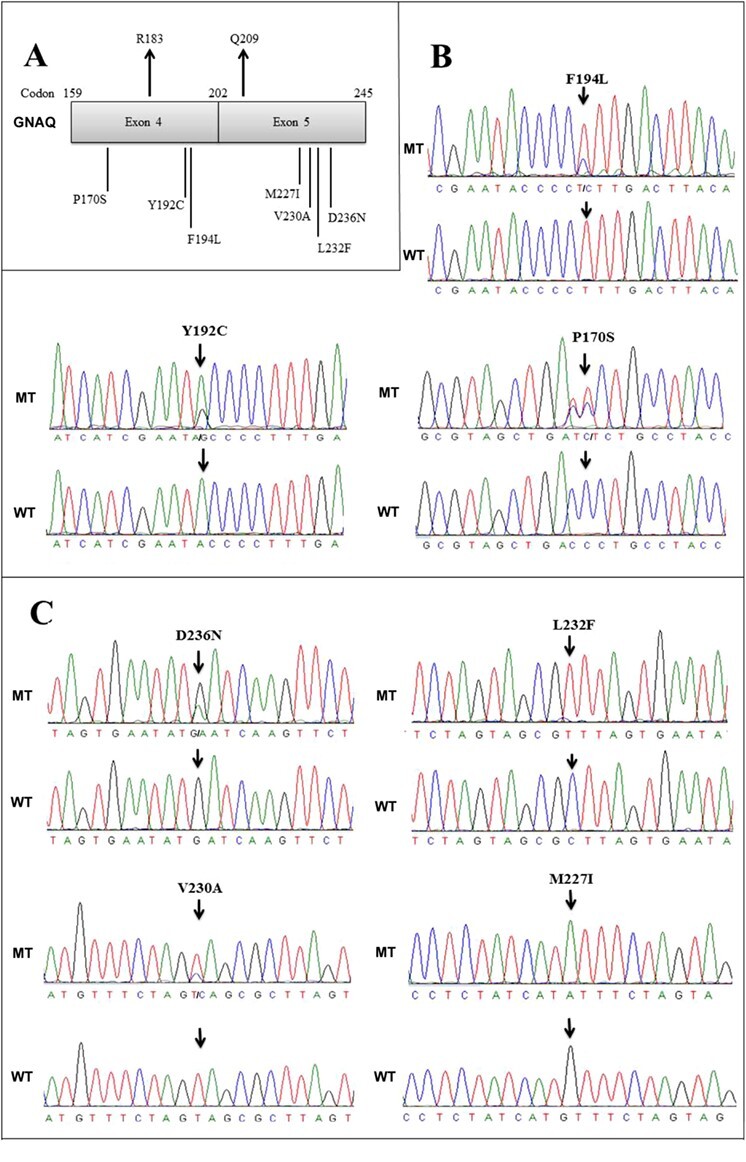
Novel mutations in exon 4 and exon 5 of GNAQ. (A) Relative position of mutations in GNAQ. (B) Novel mutations in exon 4. (C) Novel mutations in exon 5. MT, Mutant; WT, Wild type.
Table 2 .
Novel mutations in exons 4 and 5 of GNAQ and GNA11.
| Sample ID | DNA change | Codon change | Protein change | Comment |
|---|---|---|---|---|
| GNAQ exon 4 | ||||
| M6 | 575A > G | TAC > TGC | Y192C (Tyr192Cys) | Compound heterozygous for a second novel mutation in R166C in GNA11 |
| M11 | 580 T > C | TTT > CTT | F194L (Phe194Leu) | Compound heterozygous for a second mutation in Q209L of GNA11 |
| M45 | 508C > T | CCT > TCT | P170S (Pro170Ser) | Compound heterozygous for a second novel mutation in M227I in GNAQ |
| GNAQ exon 5 | ||||
| M5 | 706G > A | GAT>AAT | D236N (Asp236Asn) | Compound heterozygous for a second mutation in Q209P in GNAQ |
| M12 | 694C > T | CTT > TTT | L232F (Leu232Phe) | Homozygous for this mutation and homozygous for missense mutation Q209P in GNAQ |
| M36 | 689 T > C | GTA > GCA | V230A (Val230Ala) | |
| M45 | 681G > A | ATG > ATA | M227I (Met227Ile) | Homozygous for this mutation, and compound heterozygous with mutation P170S in GNAQ |
| GNA11 exon 4 | ||||
| M6 | 496C > T | CGC > TGC | R166C (Arg166Cys) | Compound heterozygous for a second novel mutation in Y192C in GNAQ |
| M17 | 599 T > C | ATC > ACC | I200T (Ile200Thr) | |
| GNA11 exon 5 | ||||
| M42 | 674C > T | TCC > TTC | S225F (Ser225Phe) | |
| M56 | 616G > A | GTG > ATG | V206M (Val206Met) |
In exon 5 of GNAQ, we found four novel mutations: D236N, L232F, V230A, and M227I (Table 2, Fig. 2C). Individual M5, in addition to carrying the mutation D236N, also had in the same exon a compound heterozygous mutation Q209P. Sample M12 carried homozygous missense mutations L232F in both alleles of GNAQ, and was also homozygous for mutation Q209P in the same exon. Sample M45 carried homozygous mutations V230A in GNAQ, in addition to carrying a novel compound heterozygous mutation in P170S in exon 4 of GNAQ.
Novel mutations in GNA11
We included the entire coding regions of exons 4 and 5 of GNA11 in our sequencing analysis (Fig. 3A). In exon 4 of GNA11, we detected two novel somatic mutations, including R166C and I200T (Table 2, Fig. 3B). Of note, individual M6 carried a novel mutation in R166C as well as a compound heterozygous novel mutation Y192C in exon 4 of GNAQ. In exon 5 of GNA11, we detected two novel mutations, including S225F and V206M (Table 2, Fig. 3C).
Figure 3 .
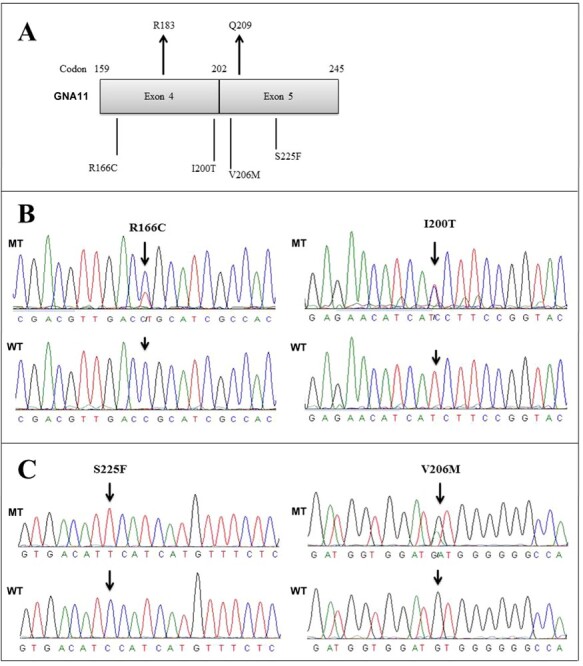
Novel mutations in exon 4 and exon 5 of GNA11. (A) Relative position of mutations in GNA11. (B) Novel mutations in exon 4. (C) Novel mutations in exon 5. MT, Mutant; WT, Wild type.
Conservation of the mutations
All novel missense mutations detected in exons 4 and 5 of GNAQ and GNA11 were located in revolutionarily conserved regions (Fig. 4).
Figure 4 .
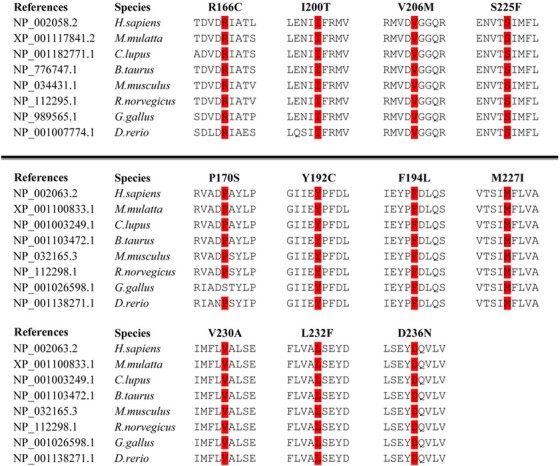
Conservation of the mutations of GNAQ and GNA11.
Discussion
Uveal melanoma is the most common primary cancer in the adult eye. Based on previous studies, 83% of uveal melanoma carries R183 or Q209 mutations in GNAQ or its paralogue GNA11. Exon 4 and exon 5 in GNAQ and GNA11 are mutational hotspots in uveal melanomas in Caucasians.10
In this study, one-third (33.3%) of 63 Chinese primary uveal melanoma samples carried mutations in Q209 of GNAQ or GNA11, while the frequency was 76.6% in the Caucasian population (Table 1). Compared to previous studies, we found seven novel missense somatic mutations in GNAQ (Y192C, F194L, P170S, D236N, L232F, V230A, and M227I) and four novel missense somatic mutations in GNA11 (R166C, I200T, S225F, and V206M). The high frequency of Q209 mutations and the large number of novel missense mutations found in this study suggest that somatic mutations in GNAQ and GNA11 are also frequent in uveal melanoma in Chinese people.
Of note, about 4.9% of uveal melanoma samples carried R183 mutations in Van Raamsdonk’s study.10 However, R183 mutations were absent in Chinese uveal melanoma samples in our study. The implication of the exclusive absence of this mutation in the Chinese population may need to be validated in a future study with a larger sample size.
GNAQ and GNA11 encode Gαq and Gα11, respectively, which make up the alpha subunits of heterotrimeric G proteins. The alpha subunit performs the essential GTPase function of hydrolyzing a GTP molecule and thereby turning off the signaling mechanism between membrane-bound G-protein-coupled receptors and their intracellular downstream effectors.11,12 Mutations that substitute amino acid residues at the GTP contact site, including glutamine at codon 209 and arginine at codon 183, were found to transform GNAQ and GNA11 into oncogenes and induce the constitutive activation of the mitogen-activated protein (MAP) kinase pathway.13–15 Recently, it was predicted that constitutive activation of Gq or G11 in uveal melanomas results in abnormal Yes-associated protein (YAP) activation, which contributes to uveal melanoma development.16,17
There is abundant evidence that uveal melanoma and cutaneous melanoma are distinct tumors.18,19 Uveal melanoma and cutaneous melanoma differ significantly in their epidemiological, clinical, immunophenotypical, and cytogenetic characteristics.20BRAF and NRAS are common mutation targets in cutaneous melanoma.21,22 However, it has been reported that mutations in BRAF and NRAS are absent in uveal melanoma.23,24
Clinically, uveal melanoma constitutes a grave diagnosis because of the risk of vision loss and high potential for metastatic disease, which is almost always fatal. Current therapy includes plaque brachytherapy, proton beam radiotherapy, transscleral local resection, transpupillary thermotherapy, and enucleation. Recently, the use of inhibitors of MAPK pathway and its downstream effectors has shown promise in treating GNAQ mutant uveal melanoma.6,25,26
In conclusion, this study found that somatic mutations in GNAQ and GNA11 occur frequently in uveal melanoma in Chinese people. The recognition of novel genetic aberrants, especially those in mutational hotspots of uveal melanoma, is an ongoing effort toward a more complete genetic understanding of this disease and may aid discovery of effective clinical therapies.
Supplementary Material
Acknowledgements
This work was supported by State Key Laboratory of Ophthalmology (Zhongshan Ophthalmic Center, Sun Yat-Sen University), “100 talents plan” from Sun Yat-sen University, the Open Research Funds of the State Key Laboratory of Ophthalmology (2017KF05), Funds of the Guangdong Provincial Key Laboratory of Ophthalmology and Visual Science (2017B030314025), Medical Research Fund of Guangdong Province (No. A2018337), Guangzhou Institute of Pediatrics/Guangzhou Women and Children’s Medical Center (No. IP-2018-002).
Conflict of interest
None declared.
References
- 1. Singh AD, Topham A. Survival rates with uveal melanoma in the United States: 1973-1997. Ophthalmology 2003;110:962–5. doi: 10.1016/S0161-6420(03)00077-0. [DOI] [PubMed] [Google Scholar]
- 2. Damato B. Developments in the management of uveal melanoma. Clin Experiment Ophthalmol 2004;32:639–47. doi: 10.1111/j.1442-9071.2004.00917.x. [DOI] [PubMed] [Google Scholar]
- 3. Chang AE, Karnell LH, Menck HR. The National Cancer Data Base report on cutaneous and noncutaneous melanoma: a summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer 1998;83:1664–78. doi: 10.1002/(sici)1097-0142(19981015)83:8<1664: :aid-cncr23>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 4. Singh AD, Bergman L, Seregard S. Uveal melanoma: epidemiologic aspects. Ophthalmol Clin North Am 2005;18:75–84viii. doi: 10.1016/j.ohc.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 5. Singh AD, De Potter P, Fijal BA, et al. Lifetime prevalence of uveal melanoma in white patients with oculo (dermal) melanocytosis. Ophthalmology 1998;105:195–8. doi: 10.1016/s0161-6420(98)92205-9. [DOI] [PubMed] [Google Scholar]
- 6. Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shah CP, Weis E, Lajous M, et al. Intermittent and chronic ultraviolet light exposure and uveal melanoma: a meta-analysis. Ophthalmology 2005;112:1599–607. doi: 10.1016/j.ophtha.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 8. Singh AD, Rennie IG, Seregard S, et al. Sunlight exposure and pathogenesis of uveal melanoma. Surv Ophthalmol 2004;49:419–28. doi: 10.1016/j.survophthal.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 9. Kuo PK, Puliafito CA, Wang KM, et al. Uveal melanoma in China. Int Ophthalmol Clin 1982;22:57–71. doi: 10.1097/00004397-198202230-00007. [DOI] [PubMed] [Google Scholar]
- 10. Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med 2010;363:2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neves SR, Ram PT, Iyengar RG. G protein pathways. Science 2002;296:1636–9. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 12. Markby DW, Onrust R, Bourne HR. Separate GTP binding and GTPase activating domains of a G alpha subunit. Science 1993;262:1895–901. doi: 10.1126/science.8266082. [DOI] [PubMed] [Google Scholar]
- 13. Landis CA, Masters SB, Spada A, et al. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature 1989;340:692–6. doi: 10.1038/340692a0. [DOI] [PubMed] [Google Scholar]
- 14. Sondek J, Lambright DG, Noel JP, et al. GTPase mechanism of Gproteins from the 1.7-A crystal structure of transducin alpha-GDP-AIF-4. Nature 1994;372:276–9. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 15. Kalinec G, Nazarali AJ, Hermouet S, et al. Mutated alpha subunit of the Gq protein induces malignant transformation in NIH 3T3 cells. Mol Cell Biol 1992;12:4687–93. doi: 10.1128/mcb.12.10.4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu FX, Zhao B, Panupinthu N, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012;150:780–91. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu FX, Luo J, Mo JS, et al. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating yap. Cancer Cell 2014;25:822–30. doi: 10.1016/j.ccr.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shors AR, Iwamoto S, Doody DR, et al. Relationship of uveal and cutaneous malignant melanoma in persons with multiple primary tumors. Int J Cancer 2002;102:266–8. doi: 10.1002/ijc.10703. [DOI] [PubMed] [Google Scholar]
- 19. Eskelin S, Pyrhonen S, Hahka-Kemppinen M, et al. A prognostic model and staging for metastatic uveal melanoma. Cancer 2003;97:465–75. doi: 10.1002/cncr.11113. [DOI] [PubMed] [Google Scholar]
- 20. Houghton AN, Polsky D. Focus on melanoma. Cancer Cell 2002;2:275–8. doi: 10.1016/s1535-6108(02)00161-7. [DOI] [PubMed] [Google Scholar]
- 21. Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 22. Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 23. Cruz F 3rd, Rubin BP, Wilson D, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res 2003;63:5761–6. PMID: 14522897. [PubMed] [Google Scholar]
- 24. Rimoldi D, Salvi S, Lienard D, et al. Lack of BRAF mutations in uveal melanoma. Cancer Res 2003;63:5712–5. PMID: 14522889. [PubMed] [Google Scholar]
- 25. Ambrosini G, Musi E, Ho AL, et al. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol Cancer Ther 2013;12:768–76. doi: 10.1158/1535-7163.MCT-12-1020. [DOI] [PubMed] [Google Scholar]
- 26. Khalili JS, Yu X, Wang J, et al. Combination small molecule MEK and PI3K inhibition enhances uveal melanoma cell death in a mutant GNAQ- and GNA11-dependent manner. Clin Cancer Res 2012;18:4345–55. doi: 10.1158/1078-0432.CCR-11-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


