Abstract
We evaluated a recently described linear signal amplification method for sensitivity and specificity in detecting mutations associated with resistance to rifampin (RIF) and isoniazid (INH) in Mycobacterium tuberculosis. The assay utilizes the thermostable flap endonuclease Cleavase VIII, derived from Archaeoglobus fulgidus, which cleaves a structure formed by the hybridization of two overlapping oligonucleotide probes to a target nucleic acid strand. This method, termed the Invader assay, can discriminate single-base differences. Nine pairs of probes, encompassing five mutations in rpoB and katG that are associated with resistance to either RIF or INH, as well as the corresponding wild-type (drug-susceptible) alleles, were tested using amplified DNA. Fluorescent-labeled cleavage products, ranging from 4 to 13 nucleotides in length, depending on the genotype of the test sample, were separated by denaturing polyacrylamide (20 to 24%) gel electrophoresis and then detected by scanning. All nine alleles could be identified and differentiated on the basis of product size. Multiple mutations at a specific rpoB nucleotide in target PCR products could be identified, as could mutants that were present at ≥0.5% of the total population of target sequences. The Invader assay is a sensitive screen for some mutations associated with antituberculosis drug resistance in amplified gene regions.
The number of new cases of tuberculosis reported in the United States decreased from 26,673 (10.5 cases per 100,000 population) in 1992 to 18,361 (6.8 cases per 100,000 population) in 1998. From 1995 to 1997, resistance to isoniazid (INH) among Mycobacterium tuberculosis isolates from reported culture-positive cases decreased from 8.2 to 7.6%, and combined resistance to both INH and rifampin (RIF) dropped from 1.8 to 1.3% (2). These data underscore the success of tuberculosis control programs in the United States; however, the prevalence of this disease among foreign-born individuals continued to increase from 1992 until 1997 (2). Moreover, because the prevalence of resistance to primary antituberculosis drugs is much higher in some developing countries, timely diagnosis of drug resistance may be useful for the continued success of control programs in these countries as well as in the United States.
Genomic mutations frequently associated with resistance to each of the primary antituberculosis drugs have been identified (12, 13). For example, at least 95% of RIF-resistant M. tuberculosis isolates have mutations in the rpoB gene encoding the β subunit of RNA polymerase (10, 19). Approximately two-thirds of isolates resistant to INH harbor mutations in katG, which encodes a catalase-peroxidase required for INH activation (12, 22). Identification of the importance of rpoB and katG in antituberculosis drug resistance has led to the application of a variety of rapid methods to detect mutations in these genes (12, 13). These approaches typically rely on amplifying the affected genomic region by PCR, followed by DNA sequence-based analysis using direct sequencing or other assays such as oligotyping, e.g., the Line-Probe Assay (LiPA) (Innogenetics, Ghent, Belgium), heteroduplex analysis, dideoxy fingerprinting, RNA-RNA duplex analysis, single-strand conformation polymorphism (SSCP) analysis, and Cleavase fragment length polymorphism (CFLP) analysis (3–6, 11–13, 15, 17, 19, 21). Given the long turnaround times required to complete conventional antituberculosis drug susceptibility testing, these molecular methods could be of tremendous value in the rapid detection of drug resistance. However, since both wild-type (i.e., drug-susceptible) and mutant alleles are typically identified simultaneously by these methods, they have not proven effective in detecting clinically important drug-resistant subpopulations among predominantly susceptible bacilli. If these subpopulations exceed 1% of the total when tested against RIF or INH (10% for other primary antituberculosis drugs), therapeutic success for the patient may be compromised (1).
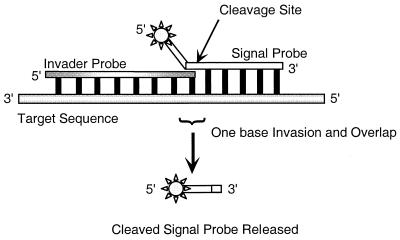
A potential solution to the problem of detecting drug-resistant subpopulations involves a novel linear signal amplification technology termed the Invader assay. This assay is based on cleavage of a unique secondary structure formed between two overlapping oligonucleotides that hybridize to the target sequence of interest to form a “flap” (Fig. 1) (9). The downstream signal probe oligonucleotide consists of a 5′ region that is noncomplementary to the target (i.e., an unpaired flap) and a 3′ portion that is complementary to the target. The upstream invasive oligonucleotide is complementary to the target and overlaps the 5′ end of the complementary portion of the signal probe oligonucleotide. This structure is a substrate for Cleavase VIII, a member of a class of naturally occurring and engineered enzymes, including a number of archaeal and eucaryotic flap endonucleases derived from Archaeoglobus fulgidus (8). Cleavage occurs 3′ to the last base in the overlap and releases the flap. By conducting reactions at high concentrations of signal probe and at temperatures near its melting temperature (Tm), this oligonucleotide can be made to cycle rapidly on and off the target. Successive rounds of cleavage then result in the generation of multiple products for each target molecule, i.e., linear signal amplification, without temperature cycling.
FIG. 1.
Schematic representation of the Invader linear signal amplification assay. The single-strand signal probe is cleaved by Cleavase VIII at the junction of the signal probe, invader probe, and target nucleic acid.
The specificity of the Invader method results from a combination of both the requirement for coordinate hybridization of two oligonucleotides spanning at least 35 to 40 bases and the structure-specific recognition properties of the enzymes. A nicked structure, i.e., one that possesses a flap but no overlap, fails to support cleavage (9). The 3′-terminal base of the Invader probe may be mismatched with the target; however, a mismatch between the signal probe and the target at the nucleotide immediately upstream of the intended cleavage site precludes the creation of the overlap (Fig. 1). This stringent requirement for complementarity at a defined position allows the Invader assay to be used as a sensitive indicator of base changes, including single point mutations (9, 14). Moreover, the linear amplification of the cleaved product makes possible the detection of low levels of the target, potentially enabling the discrimination of small subpopulations in mixed DNA samples.
In the present study, we evaluated the suitability of the Invader technology for identifying four point mutations in rpoB and one in katG, along with their corresponding wild-type alleles. Among the base changes we chose to evaluate are two that affect the same nucleotide, thereby challenging the method to be as precise as DNA sequencing in indicating the specific identity of a given base. We also investigated whether multiple alleles could be examined simultaneously in a single reaction. Finally, we tested the method's limits of detection on mixed target populations to mimic the situation in which an antimicrobial resistance allele exists in a predominantly wild-type population.
MATERIALS AND METHODS
Bacterial isolates.
Rifampin-resistant (Rifr) rpoB mutants of M. tuberculosis included strains 94-2850 (Ser531→Leu), 94-2856 (Asp516→Val), 94-3047 (His526→Tyr), and 94-3070 (His526→Asp) (4). An isoniazid-resistant (Inhr) katG mutant, strain 94-2856 (Ser315→Thr), and a strain that is wild type at both loci and susceptible to both drugs, H37Rv, were also tested. Methods used for culture, drug susceptibility testing, extraction of genomic DNA by agitation of cells in the presence of siliconized glass beads, and automated fluorescent DNA sequence analyses were as previously described (3, 4).
PCR amplification.
A 193-bp fragment of the rpoB gene was amplified by PCR. The sequence of the forward primer was 5′-CGTGGAGGCGATCACACCGCAGACGTT-3′ (rpoB nucleotides 2288 to 2314), and the sequence of the reverse primer was 5′-GACCTCCAGCCCGGCACGCTCACGT-3′ (nucleotides 2456 to 2480) (10). Amplification reactions were performed in a final volume of 100 μl, containing 2 μl of genomic DNA, 35 pmol of each primer, 50 μM each deoxyribonucleotide (Perkin-Elmer–Applied Biosystems Inc. [PE-ABI], Foster City, Calif.), 1× PCR buffer (20 mM Tris-HCl [pH 8.5], 50 mM KCl, 1.5 mM MgCl2, 0.05% Tween 20, 0.05% NP40), 1 M betaine, 5% dimethyl sulfoxide (DMSO), and 2.5 U of Taq polymerase (Roche Boehringer Mannheim, Indianapolis, Ind.). PCR cycling conditions consisted of an initial denaturation step at 95°C for 5 min; 30 cycles of denaturation at 95°C for 1 min, annealing at 68°C for 1 min, and extension at 72°C for 1 min; and a final extension at 72°C for 5 min. A 620-bp fragment of the katG gene was amplified using a forward primer with the sequence 5′-AGCTCGTATGGCACCGGAAC-3′ (nucleotides 904 to 923) and a reverse primer with the sequence 5′-TTGACCTCCCACCCGACTTG-3′ (nucleotides 1504 to 1523) (22). The PCR conditions were as above, with the exception that betaine and DMSO were excluded. PCR cycling conditions for katG amplifications consisted of an initial denaturation at 95°C for 3 min and 35 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min.
Invader reactions.
Invader and signal probe oligonucleotide pairs were designed to detect each of the four rpoB mutant alleles and the corresponding wild-type sequences. Since two of the mutant alleles affect the same nucleotide position, only three wild-type oligonucleotide sets were required. One set of oligonucleotides was designed to detect the katG mutation and the corresponding wild-type sequence (Table 1). Signal probes contained a 3′ region that was complementary to the target sequence and a noncomplementary 5′ arm used for detection. Signal probes were labeled on the 5′ arm with either tetrachloro-6-carboxyfluorescein (TET), hexachloro-6-carboxyfluorescein (HEX), or 6-carboxyfluorescein (FAM) as indicated. The cleaved products were 13 nucleotides in length, respectively, for the wild-type rpoB and katG sequences, and 4 to 10 nucleotides for mutant products. Invader oligonucleotides were complementary to an 18- to 22-bp region immediately upstream of the signal probe, with an additional base at the 3′ end that “invades” the region hybridized to the signal probe by 1 base. Both invader and signal probes were complementary to either the sense or the antisense target DNA strand, depending on which resulted in the formation of the least number of predictable secondary structures (e.g., “hairpin” structures with negative free-energy values) that could negatively impact subsequent enzymatic cleavage. Probe sets for the rpoB516, rpoB526Y, rpoB526D, and katG315 loci were complementary to the antisense strand, and those for the rpoB531 locus were complementary to the sense strand (Table 1).
TABLE 1.
Nucleotide sequences of probes used in the Invader assay reactions
| Gene | Mutation | Invasive probe sequence (5′ to 3′) | 5′ signal probe label | Signal probe sequencea (5′ to 3′) | Product size (ntb) |
|---|---|---|---|---|---|
| rpoB | wt516 | GCCAGCTGAGCCAATTCATGGC | FAM | AACGAGGCGCACaCCAGAACAACCCGC | 13 |
| rpoB | Asp516→Val | GCCAGCTGAGCCAATTCATGGC | FAM | CAGGCGCACtCCAGAACAACCCGC | 10 |
| rpoB | wt526 | CCGCTGTCGGGGTTGACCA | HEX | AACGAGGCGCATcACAAGCGCCGACTGT | 13 |
| rpoB | His526→Tyr | CCGCTGTCGGGGTTGACCA | HEX | CGCTCTTtACAAGCGCCGACTGT | 8 |
| rpoB | His526→Asp | CCGCTGTCGGGGTTGACCA | HEX | CTCTTgACAAGCGCCGACTGT | 6 |
| rpoB | wt531 | CAGACCGCCGGGCCCCAGCGCCC | TET | AACGAGGCGCATgACAGTCGGCGCTTG | 13 |
| rpoB | Ser531→Leu | CAGACCGCCGGGCCCCAGCGCCC | TET | CTCaACAGTCGGCGCTTG | 4 |
| katG | wt315 | CGGTAAGGACGCGATCACCAT | TET | AACGAGGCGCACgCGGCATCGAGGTCG | 13 |
| katG | Ser315→Thr | CGGTAAGGACGCGATCACCAT | TET | GAGGCGCACcCGGCATCGAGGTCG | 10 |
In each sequence, the nucleotide where mismatch cleavage occurs is lowercased.
nt, nucleotides.
One microliter of each PCR product was added to 0.5 pmol of the appropriate Invader oligonucleotide, 10 ng of human genomic DNA (Promega Corp., Madison, Wisc.) as the carrier, and morpholinepropanesulfonic acid (MOPS) buffer (pH 8.0) at a final concentration of 10 mM in a volume of 7 μl. The mixtures were denatured for 5 min at 95°C and then cooled to a reaction temperature of 60°C. Invader reactions were initiated by the addition of a mixture containing 30 ng of Cleavase VIII (Third Wave Technologies, Inc., Madison, Wisc.), 25 mM MgCl2 (to yield a final concentration of 7.5 mM), and 10 pmol of the appropriate signal probe oligonucleotide in a volume of 3 μl. Reaction mixtures were incubated for 60 min. The reactions were terminated by the addition of 10 μl of 95% formamide–10 mM EDTA (pH 8.0)–0.05% crystal violet. Following termination, the reactions were diluted 1:10 in reagent-grade water (1:50 for the rpoB Asp516→Val mutant and its corresponding wild-type sequence). Samples of 2 μl were loaded and electrophoresed in a 24% denaturing polyacrylamide gel (18 cm by 25.5 cm by 2 mm) on an automated fluorescence sequencing apparatus (model 377; PE-ABI). The data were collected using filter set C and processed with GeneScan software (version 2.0; PE-ABI).
In addition, 5 μl of each sample was electrophoresed in 20% (acrylamide to bisacrylamide, 19:1) denaturing polyacrylamide gels at 20 W. Gel cassettes (20 cm by 20 cm by 0.5 mM) were scanned with a fluorescent scanner (FMBIO-100; Hitachi Corp., San Bruno, Calif.) by using a 585-nm filter for TET- and HEX-labeled probes and a 505-nm filter for FAM-labeled probes.
Molecular weight markers contained oligonucleotides that were 4, 6, 8, 10, 13, and 15 bases long and were labeled on their 5′ termini with 6-carboxy-x-rhodamine (ROX) during synthesis.
RESULTS
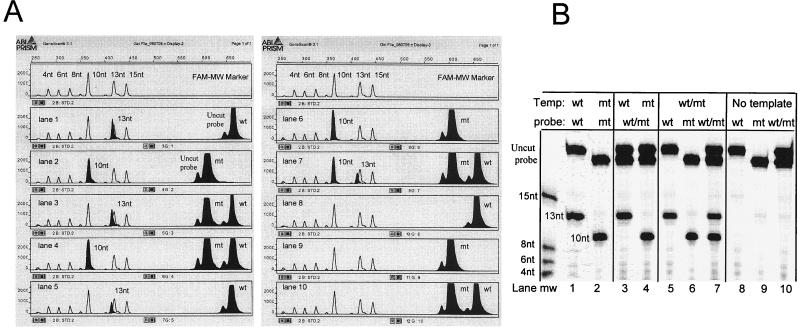
The nine sets of probes, four complementary to wild-type sequences and five complementary to mutant sequences, were each tested against both wild-type and mutant targets. Fluorescence peaks were specific for each of the target sequences when the samples were analyzed on either the automated sequencer or the fluorescence scanner (Fig. 2). Signal was generated only when the probe sets were complementary to the target sequence, i.e., probes added to targets that varied by as little as a single base failed to support detectable cleavage. Moreover, the products of each of the reactions could be readily distinguished by creating signal probes having noncomplementary 5′ flaps of different lengths (Table 1). The products of the reactions designed to detect rpoB Asp516→Val are shown in Fig. 2. The wt516 probe tested against the wild-type target resulted in the generation of a 13-nucleotide cleavage product (Fig. 2). Similarly, a 10-nucleotide product was observed when the rpoB Asp516→Val mutant probe was tested against this mutant. Equimolar mixtures of the Asp516 mutant and wild-type targets reacted with both mutant and wild-type probes to generate approximately equivalent amounts of both the 10- and 13-nucleotide products.
FIG. 2.
Identification of Asp516→Val mutation in the rpoB gene by the Invader linear signal amplification assay. A wild-type (lanes 1 and 3) or mutant (lanes 2 and 4) target or an equimolar mixture of PCR targets (lanes 5, 6, and 7) was tested against a wild-type (13-nt [lanes 1 and 5]) or mutant (10-nt [lanes 2 and 6]) FAM-labeled probe or against both (lanes 3, 4, and 7). Controls representing the signal probes in various combinations without target are shown in lanes 8, 9, and 10. (A) Samples were diluted 50-fold, and 2 μl was loaded onto a 24% 17-cm denaturing polyacrylamide gel and run on an ABI model 377 sequencing apparatus. Molecular weight markers of 4, 6, 8, 10, 13, and 15 nucleotides were FAM labeled. Data were collected using filter set C processed with GeneScan software. (B) samples (5 μl) were electrophoresed at 20 W on a 20% denaturing polyacrylamide gel and scanned at 505 nm using a Hitachi model FMBIO-100 fluorescence scanner. Molecular weight markers of 4, 6, 8, and 15 nucleotides were TET labeled. nt, nucleotide; wt, wild type; mt, mutant.
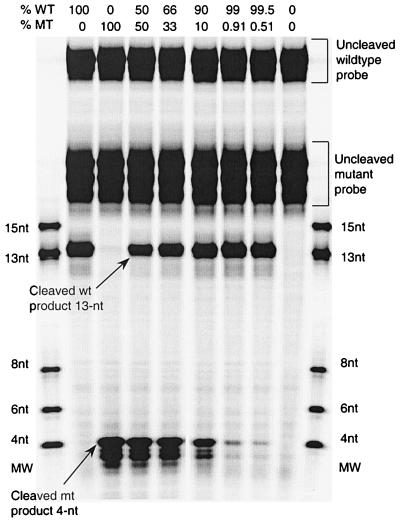
In addition to examining the specificity of the cleavage reactions for each allele at this position, we tested the ability of the Invader assay to detect low levels of the mutant sequence in a background of the wild-type sequence (Fig. 3). We mixed decreasing amounts of DNA amplified from the Ser531→Leu rpoB mutant with DNA amplified from the corresponding wild-type (RifS) allele, keeping the total amount of DNA in the reaction constant. Invader and signal probes to detect the mutant alleles were added to each mixed template sample; the signal probe was designed to yield a 4-nucleotide product (Fig. 3). The signal from the mutant allele was readily distinguishable over background at all levels tested, including when it comprised as little as 0.5% of the total DNA in the sample (Fig. 3).
FIG. 3.
Invader linear signal amplification identification of heterogeneous populations of rpoB mutant (Ser531→Leu) and wild-type PCR products. Mixtures of mutant and wild-type targets, containing 0, 100, 50, 33, 10, 0.9, or 0.5% mutant, were examined with TET-labeled wild-type and mutant signal probes. Cleavage products were electrophoresed on vertical denaturing polyacrylamide gels (20%) and scanned at 585 nm using a Hitachi model FMBIO-100 fluorescence scanner (data shown here).
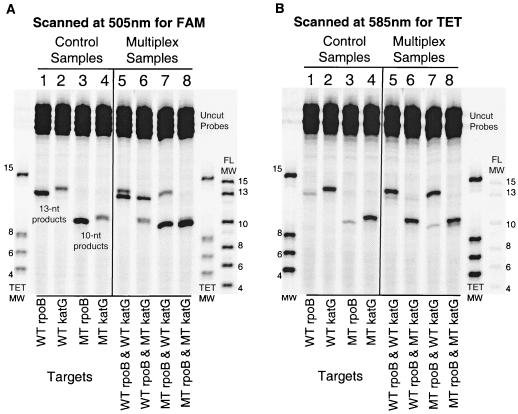
Finally, we investigated the degree to which the Invader assay could be multiplexed to detect multiple alleles in a single reaction. Cleavage products of 6, 8, and 13 nucleotides were observed when mixtures of targets containing multiple alleles at a single nucleotide position (wild-type, His526→Tyr, and His526→Asp) were examined by Invader reactions containing the three respective sets of invasive and HEX-labeled signal probes (data not shown). Similarly, mixtures of targets containing both the rpoB mutant (Asp516→Val) and the katG mutant (Ser315→Thr) and their corresponding wild-type sequences were simultaneously examined with their respective FAM- or TET-labeled probes (Table 1; Fig. 4). Cleavage products could be distinguished by size or emission wavelength in ABI GeneScan chromatograms (data not shown) or by size alone in gels examined by fluorescence scanning (Fig. 4). The latter method required scanning at two wavelengths (505 and 585 nm) to efficiently detect both the FAM and TET labels. Although the cleavage products for the wild-type rpoB and katG alleles were the same size (13 nucleotides), as were the corresponding rpoB and katG mutants (10 nucleotides), all of the four fragments displayed differences in their electrophoretic migration. These variations were most likely related to the charge-to-mass ratio as well as to secondary structural differences conferred by the different nucleotide sequences and fluorescent labels of the cleaved signal probes. The effects of dye charge and size on electrophoretic migration can be clearly seen by examining the two molecular weight markers presented in Fig. 4. Even though the nucleotide sequences for the individual size markers are identical between the FAM and TET standards, the FAM-labeled fragments migrate slightly faster than the TET-labeled fragments (20).
FIG. 4.
Identification of rpoB (Asp516→Val) and katG (Ser315→Thr) mutants using the Invader linear signal amplification assay. Invader assays with FAM-labeled rpoB probes and TET-labeled katG probes were analyzed at two wavelengths to examine mutant (MT) and wild-type (WT) target DNAs in four combinations. (A) The gel was scanned at 505 nm for FAM fluorescence. (B) The gel was scanned at 585 nm for TET fluorescence. Each of the eight lanes contains all four probes. In each panel, lanes 1 to 4 contain control DNA targets and show the cleavage products of 13 nucleotides (WT) and 10 nucleotides (MT) for identification of each of the rpoB and katG genotypes. Lanes 5 through 8 contain combinations of rpoB and katG targets, as indicated below each lane. The cleavage products in these lanes correctly identify the target combinations when referenced to the control samples. Due to the wide absorption spectrum of the TET dye, considerable crossover emission on the 505-nm scan can be seen. Similarly, the FAM-labeled products are visible at 585 nm. The sizes of the TET-labeled molecular weight markers are 15, 8, 6, and 4 nucleotides. The sizes of the FAM-labeled molecular weight markers are 15, 13, 10, 8, 6, and 4 nucleotides.
DISCUSSION
Genotypic drug resistance screening of M. tuberculosis isolates is an attractive alternative to conventional phenotypic susceptibility testing because of its ability to provide results rapidly and because highly prevalent mutations associated with resistance to five primary antituberculosis drugs have now been described (12, 13). These mutations occur in the genes rpoB (RIF), katG (INH), embB (ethambutol), rpsL (streptomycin), and pncA (pyrazinamide) (12, 13). Although the diversity of mutations associated with phenotypic drug resistance varies for each of these genes, some mutations have been shown to be more prevalent than others. For example, two mutations within an 81-bp “hot spot” region of rpoB (affecting codons 526 and 531) may be responsible for 70% or more of Rifr M. tuberculosis cases, depending on the collection of isolates under study (12, 13). The Invader assay detected mutations at these two loci, including two alleles affecting the same nucleotide in codon 526, as well as one at codon 516. The ability to distinguish multiple rpoB mutations at a common locus is particularly important because as many as 11 mutations resulting in amino acid substitutions at one codon within the 81-bp hot spot in rpoB have been described among Rifr M. tuberculosis strains (12).
The Invader assay could also simultaneously identify mutations in different genes. The katG mutation included in this study (Ser315→Thr) could be detected either alone or in combination with the rpoB Asp516→Val mutation. The katG Ser315→Thr mutation was chosen for study because it has been reported in 58 to 96% of Inhr strains in various studies, making it the most common mutation associated with INH resistance (12). Multiplex genetic screens for RIF and INH resistance may be particularly useful, since resistance to these drugs may be preliminary predictors of resistance to other antituberculosis drugs. Furthermore, these first-line antimicrobial agents are often the only drugs used in developing countries. It may be possible to combine rpoB and katG probes with additional Invader probes to identify mutations associated with resistance to other primary drugs. Likely candidates for this multiplex screen are probes for embB and rpsL mutations, which have been found to be highly prevalent among ethambutol-resistant and streptomycin-resistant M. tuberculosis isolates, respectively (3, 18). Identification of mutations associated with pyrazinamide resistance (Pzar), however, may pose a greater challenge for genotypic methods, in general, because of the diversity of these mutations and the overall absence of prevalence data. Although most Pzar M. tuberculosis isolates have mutations in pncA, which encodes pyrazinamidase, there appears to be virtually no limit on their nature and location, which makes the design of probe sets difficult (16). Limits on the extent of multiple mycobacterial gene analyses by Invader reactions, including the number of genes or the number of mutations within each gene capable of simultaneous testing, have not been determined. For example, it is not likely that all 47 mutations within the rpoB gene that have been reported (12) could be included in a single multiplex assay. Perhaps the most plausible scenario would be a multiplex screen for 10 or fewer of the most common mutations (and corresponding wild-type alleles) in each of three genes. The degree of multiplexing is limited by the number of fluorescent reporter molecules excited at 488 nm that are currently available as cleavage product labels for simultaneous automatic analysis using the PE-ABI sequencing instrument. Each of these reporter molecules should be reserved for analysis of a single gene, especially since electrophoretic migration of cleavage products is affected not only by their sequences but also by the label.
The Invader assay also detected mutant subpopulations in heterogeneous samples. An rpoB mutation (Ser531→Leu) was identified when present at a level of ≥0.5% of the predominant wild-type allele. This level of sensitivity supports the use of this assay as a genotypic screen for identifying clinically important resistant subpopulations of bacilli. Preliminary experiments with other genetic systems have demonstrated that the Invader assay may be capable of detecting subpopulations representing as little as 0.1% or less of the total DNA in a mixed sample (7). Among other methods applied to the task of identifying mutations associated with drug resistance in M. tuberculosis, only SSCP has been shown capable of identifying mutant subpopulations, and only when they are present at relatively high levels, i.e., at least 10% of the total DNA (5).
Most nucleic acid-based methods for identifying drug resistance in M. tuberculosis have at least four limitations regarding their usefulness: (i) these genes are chromosomal, which may necessitate their amplification prior to applying detection methods; (ii) an amplification and primer set will be required to examine each of the loci implicated in resistance to each of the drugs; (iii) high degrees of sequence conservation with other microbial genera may create difficulties in interpreting results using templates derived from polymicrobic samples; and (iv) some mutations may be silent and not associated with phenotypic drug resistance.
A recently described modification of the Invader assay which has been shown to detect single-base changes in low-copy-number genes from as little as 40 ng of human DNA without the need for target amplification steps (9) has potential application for genotypic antituberculosis drug susceptibility screening. This modified Invader assay does not depend on electrophoretic separation of cleavage products but rather upon on analysis of fluorescence emissions in microdilution plate wells mediated by fluorescence resonance energy transfer (FRET) (14). Since Invader assays can be designed to identify specific nucleotides, only substitutions known to be associated with phenotypic resistance may be included, eliminating the identification of previously undocumented, perhaps silent, mutations. Identification of specific base substitutions means, however, that a potentially important novel mutation could remain undetected. To date, the FRET Invader assay has been designed and successfully tested on approximately 400 individual single-nucleotide polymorphisms (unpublished data). Thus, the Invader assay offers the potential to become a high-throughput screen for identifying mutations proven to be associated with antituberculosis drug resistance.
ACKNOWLEDGMENTS
We acknowledge Laura Heisler for her contributions to the manuscript and Marianne Siebert for technical assistance.
REFERENCES
- 1.Canetti G, Fox W, Khomenko A, Mahler H T, Menon N K, Mitchison D A, Rist N, Smelev N A. Advances in techniques of testing mycobacterial drug sensitivity, and the use of sensitivity tests in tuberculosis control programmes. Bull W Hl O. 1969;41:21–43. [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Tuberculosis morbidity—United States, 1997. Morbid Mortal Weekly Rep. 1998;47:253–257. [PubMed] [Google Scholar]
- 3.Cooksey R C, Morlock G P, McQueen A, Glickman S E, Crawford J T. Characterization of streptomycin resistance mechanisms among Mycobacterium tuberculosis isolates from patients in New York City. Antimicrob Agents Chemother. 1996;40:1150–1152. doi: 10.1128/aac.40.5.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooksey R C, Morlock G P, Glickman S, Crawford J T. Evaluation of a line probe assay kit for characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from New York City. J Clin Microbiol. 1997;35:1281–1283. doi: 10.1128/jcm.35.5.1281-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooksey R C, Morlock G P, Holloway B P, Mazurek G H, Abaddi S, Jackson L K, Buzard G S, Crawford J T. Comparison of two nonradioactive, single-strand conformation polymorphism electrophoretic methods for identification of rpoB mutations in rifampin-resistant isolates of Mycobacterium tuberculosis. Mol Diagn. 1998;3:73–80. doi: 10.154/MODI00300073. [DOI] [PubMed] [Google Scholar]
- 6.De Beenhouwer H, Lhiang Z, Jannes G, Mijs W, Machtelinckz L, Rossau R, Traore H, Portaels F. Rapid detection of rifampin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tubercle Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 7.Hall J, Breitzman R, Wood N, Kwiatkowski R, Hennesy K, Vavra S, Rosenblate H, Silver A, Fagan E A. Detection of HBV variants associated with resistance to nucleoside analogues using the Invader assay. Hepatology. 1998;98:583A. [Google Scholar]
- 8.Kaiser M W, Lyamicheva N, Ma W, Miller C, Neri B, Fors L, Lyamichev V I. A comparison of eubacterial and archaeal structure-specific 5′-exonucleases. J Biol Chem. 1999;274:21387–21394. doi: 10.1074/jbc.274.30.21387. [DOI] [PubMed] [Google Scholar]
- 9.Lyamichev V, Mast A, Hall J, Prudent J, Kaiser M, Takova T, Kwiatkowski R, Sander T, de Arruda M, Arco D, Neri B, Brow M A. Polymorphism identification and quantitative detection of genomic DNA by invasive cleavage of oligonucleotide probes. Nat Biotechnol. 1999;17:292–296. doi: 10.1038/7044. [DOI] [PubMed] [Google Scholar]
- 10.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash K A, Gaytan A, Inderlied C B. Detection of rifampin resistance in Mycobacterium tuberculosis by use of a rapid, simple, and specific RNA/RNA mismatch assay. J Infect Dis. 1997;176:533–536. doi: 10.1086/517283. [DOI] [PubMed] [Google Scholar]
- 12.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tubercle Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 13.Rattan A, Kalia A, Ahmad N. Multidrug-resistant Mycobacterium tuberculosis: molecular perspectives. Emerg Infect Dis. 1998;4:195–209. doi: 10.3201/eid0402.980207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan D, Nuccie B, Arvan D. Non-PCR-dependent detection of the Factor V Leiden mutation from genomic DNA using a homogeneous Invader microtiter plate assay. Mol Diagn. 1999;4:135–144. doi: 10.1016/s1084-8592(99)80037-x. [DOI] [PubMed] [Google Scholar]
- 15.Sander T, Olson S, Hall J, Siebert M, Grooms K, Heisler L, de Arruda M, Neri B. Comparison of detection platforms and post-polymerase chain reaction DNA purification methods for use in conjunction with Cleavase fragment length polymorphism analysis. Electrophoresis. 1999;20:1131–1140. doi: 10.1002/(SICI)1522-2683(19990101)20:6<1131::AID-ELPS1131>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 16.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreevatsan S, Bookout J, Ringpis F, Moghazeh S, Kreiswirth B, Pottathil R, Barathur R. Comparative evaluation of Cleavase fragment length polymorphism with PCR-SSCP and PCR-RFLP to detect antimicrobial agent resistance in Mycobacterium tuberculosis. Mol Diagn. 1998;3:81–91. doi: 10.154/MODI00300081. [DOI] [PubMed] [Google Scholar]
- 18.Sreevatsan S, Stockbauer K E, Pan X, Kreiswirth B N, Moghazeh S L, Jacobs W R, Jr, Telenti A, Musser J M. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob Agents Chemother. 1997;41:1677–1681. doi: 10.1128/aac.41.8.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti A, Imboden P, Marchesi P, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 20.Tu O, Knott T, Marsh M, Bechtol K, Harris D, Barker D, Bashkin J. The influence of fluorescent dye structure on the electrophoretic mobility of end-labeled DNA. Nucleic Acids Res. 1998;26:2797–2802. doi: 10.1093/nar/26.11.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams D L, Spring L, Gillis T P, Salfinger M, Persing D H. Evaluation of a polymerase chain reaction-based universal heteroduplex generator assay for direct detection of rifampin susceptibility of Mycobacterium tuberculosis from sputum specimens. Clin Infect Dis. 1998;26:446–450. doi: 10.1086/516313. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Heym B, Allen B, Young D, Cole S T. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358:591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]