Abstract
Sudden cardiac death continues to have a devastating impact on public health prompting the continued efforts to develop more effective therapies for cardiac arrhythmias. Among different approaches to normalize function of ion channels and prevent arrhythmogenic remodeling of tissue substrate, cardiac cell and gene therapies are emerging as promising strategies to restore and maintain normal heart rhythm. Specifically, the ability to genetically enhance electrical excitability of diseased hearts through voltage-gated sodium channel (VGSC) gene transfer could improve velocity of action potential conduction and act to stop reentrant circuits underlying sustained arrhythmias. For this purpose, prokaryotic VGSC genes are promising therapeutic candidates due to their small size (<1kb) and potential to be effectively packaged in adeno-associated viral (AAV) vectors and delivered to cardiomyocytes for stable, long-term expression. This article describes a versatile method to discover and characterize novel prokaryotic ion channels for use in gene and cell therapies for heart disease including cardiac arrhythmias. Detailed protocols are provided for: (1) identification of potential ion channel candidates from large genomic databases, (2) candidate screening and characterization using site-directed mutagenesis and engineered human excitable cell system and, (3) candidate validation using electrophysiological techniques and an in vitro model of impaired cardiac impulse conduction.
1. Introduction
Despite more than a century of research, congenital and acquired heart diseases continue to present tremendous health and socioeconomic burden. Underlying these diseases are deficiencies in CM numbers or function, as well as structural changes caused by fibrosis, hypertrophy, dilatation, or interventional/surgical treatments. Many of these pathologies are associated with impaired action potential (AP) conduction, making the heart vulnerable to reentrant arrhythmias and sudden cardiac death, which kill 1 in 7.4 people in the US (Virani et al., 2020). While the prevalence of cardiac arrhythmias is rising as the average population age is increased (Khurshid et al., 2018), current strategies for ventricular rate control and/or sinus rhythm restoration by drug or ablation therapies are suboptimal. Antiarrhythmic drugs carry arrhythmogenic risks and non-cardiovascular toxicities, while ablation therapies can have high recurrence rates requiring repeated procedures and can lead to serious complications (Markman & Nazarian, 2019).
The complexity of electrical disorders in the heart and the growing understanding of their molecular bases make gene therapies a viable treatment option for patients with difficult-to-manage acquired and inherited arrhythmias. In particular, the ability to directly augment velocity of AP conduction in diseased cardiac tissue could increase the width of propagating waves to prevent the induction and maintenance of reentry circuits that underlie lethal cardiac arrhythmias (Antzelevitch, 2001; Kleber & Rudy, 2004). Among other factors, cardiac conduction velocity depends on the strength of cardiomyocyte intercellular coupling mediated by gap junction protein Connexin-43 (Cx43). Overexpression of Cx43 in cardiomyocytes was shown to confer protection against atrial and ventricular arrhythmias (Bikou et al., 2011; Greener et al., 2012; Igarashi et al., 2012). Besides gap junctional coupling, speed of cardiac AP conduction is governed by current flow through voltage-gated sodium channels (VGSCs) which determines the slope of AP upstroke and electrical excitability of cardiomyocytes. Slow AP conduction induced by loss-of-function mutations in VGSCs or acquired conditions that reduce sodium current density or alter extracellular environment (e.g., fibrosis) (Carmeliet, 1999; Han, Tan, Sun, & Li, 2018; Janse & Wit, 1989; Liu, Yang, & Dudley, 2014; Meregalli et al., 2009; Rivaud et al., 2017) could benefit from gene therapies with cardiomyocyte overexpression of VGSCs. However, the size of mammalian VGSC genes (>6kb) is too large for effective packaging in therapeutic viruses (e.g., adeno-associated virus (AAV)), which precludes stable channel expression to enhance cardiac electrical excitability long term.
Previously, our group explored the superfamily of prokaryotic sodium channels (BacNav) as potential substitutes for mammalian VGSCs to stably generate de novo excitable cells (Nguyen, Kirkton, & Bursac, 2016). BacNav channels are encoded by genes only 0.7–0.9kb in size, approximately one-eighth to one-tenth the size of their mammalian counterparts, and thus are well-suited for gene therapy applications. We demonstrated the ability to engineer primary fibroblasts into electrically excitable and actively conducting cells by lentiviral co-delivery of the selected BacNav orthologs, a potassium channel (Kir2.1) to hyperpolarize membrane potential, and Cx43 to enhance intercellular electrical coupling (Nguyen et al., 2016). In addition, the engineered fibroblasts were shown to rescue cardiac conduction in an in vitro model of interstitial fibrosis (Nguyen et al., 2016).
Despite promising therapeutic potential, the biophysical properties of BacNav, especially the kinetics and voltage dependency of channel activation and inactivation, are different from those of the cardiac VGSCs (Nav1.5). Therefore, to engineer BacNav orthologs that have optimal biophysical properties for rescuing slow cardiac AP conduction, it will be critical to expand the pool of functional BacNavs beyond the few that have been characterized in previous studies (Charalambous & Wallace, 2011; Nguyen et al., 2016; Ren et al., 2001; Yue, Navarro, Ren, Ramos, & Clapham, 2002). In this chapter, we describe an ion channel engineering platform to screen, identify, and characterize novel prokaryotic ion channels for use in cell and gene therapies for cardiac arrhythmias. Such a platform is aimed to allow precise tailoring of engineered BacNav properties for patient-and disease-specific therapeutic applications (Fu, 2015; Gourraud, Barc, Thollet, Le Marec, & Probst, 2017; Huizar, Ellenbogen, Tan, & Kaszala, 2019; Kuriachan, Sumner, & Mitchell, 2015). Additionally, the discovery of novel prokaryotic ancestors of eukaryotic VGSCs is expected to promote basic understanding of the mammalian ion channel evolution and function.
2. Screening of novel prokaryotic ion channel orthologs
2.1. Target selection, DNA synthesis, and construct design
In order to effectively search for potential BacNav candidates, it is important to use a database that has comprehensive coverage of bacterial genomes and convenient built-in search tools. Two major genomic database collections are constructed by The National Center for Biotechnology Information (NCBI) at the National Institutes of Health in the United States and the European Molecular Biology Laboratory/European Bioinformatics Institute (EMBL-EBI). We recommend using the EMBL-EML database “The Universal Protein Resource (UniProt)” for its user-centered design, comprehensive and cross-linked databases, and easy-to-use search tools. The selected target genes will be codon optimized, synthesized, and incorporated into appropriate expression construct vectors.
2.1.1. Materials and equipment
Lentiviral mammalian expression plasmid with a constitutive and strong eukaryotic promoter, fluorescent reporter gene, and antibiotic resistance gene
cDNAs encoding target ion channels
Restriction Enzymes (New England Biolabs, NEB)
CutSmart Buffer (NEB)
Agarose
1 × Tris-acetate-EDTA (TAE) buffer
DNA Gel electrophoresis system (Fisher Scientifics)
Sybr Safe DNA Stain (Thermo Fisher)
1kb plus DNA Ladder (NEB)
Zymoclean Gel DNA Recovery Kit (Zymo Research)
UV illuminator
T4 DNA Ligase (NEB)
Chemically competent E. coli cells
Luria Bertani (LB) medium (broth)
Glycerol
Antibiotics
Bacterial Shaking Incubator
Spin Miniprep Kit (Qiagen)
NanoDrop™ 2000 (Thermo Fisher)
2.1.2. Protocols
Go the UniProt home page at http://www.uniprot.org/. For a detailed protocol on how to use UniProt database, please see reference paper (Pundir, Martin, O’Donovan, & UniProt, 2016).
Click “BLAST” on the left-side of the toolbar.
Follow the directions under the instruction and input protein sequence of desired reference candidate.
Choose the appropriate target database if proteins from a specific taxonomic domain (e.g., Archaea, Bacteria) is desired. Set E-Threshold to 0.001 and click the “Run BLAST” button.
On the BLAST result page, the protein hits are ranked based on percentage alignment with the reference protein.
- Click the entry identifier to show structural and sequence details of each protein. Several criteria are used to select potential BacNav candidates:
- presence of coiled-coil region in the C-terminal domain (CTD),
- presence of selectivity filter (e.g., TLESW), and (3) presence of conserved arginine residues in the S4 gating region.
-
Choose a commercial company for production of human codon-optimized DNA oligonucleotides (e.g., Integrated DNA Technologies (IDT)). Import the protein sequences of selected candidates and order the codon-optimized DNA oligonucleotides.
Note: It is important to choose a cloning strategy and design plasmid before ordering the fragments. Here we describe restriction cloning as it allows simple subcloning for different inserts using the same backbone plasmid; thus, adding appropriate restriction sites at 5’ and 3’ ends will facilitate subsequent cloning process. The choice of restriction sites depends on the particular plasmid backbone and insertion site. For primary screening as described below, it is important to choose a vector that contains a strong and ubiquitous promoter (e.g., human cytomegalovirus (CMV)) and fluorescence reporter gene (e.g., green fluorescence protein (GFP)).
Upon receipt of DNA oligonucleotides, digest the DNA fragments and lentivirus backbone plasmid with appropriate restriction enzymes as per NEB protocol.
Add DNA loading dye to the digested inserts and plasmid backbone and load the DNA and 1kb plus ladder onto a 1% agarose gel and perform gel electrophoresis.
-
After electrophoresis, remove the gel and image using UV transilluminator to visualize the DNA bands. The lentivirus plasmid digestion should result in two bands (backbone usually has larger size) while the digested inserts should contain only a single band.
Note: It is important to have correct size and number of bands, otherwise the sequencing of the plasmid and correct selection of restriction sites (single cutting sites in the backbone or insert) needs to be confirmed.
Cut out the appropriate bands that contain backbone and insert, perform gel purification to extract the DNA, and measure the concentration using NanoDrop.
Ligate the backbone and the insert at room temperature for 30min using T4 DNA Ligase as per NEB protocol.
Transform ligated DNA into chemically competent E. coli cells (e.g., Stbl3™) as per manufacturer’s protocol.
Plate the cells on agar plates containing the appropriate antibiotic corresponding to the resistance gene in the plasmid backbone and culture the plates overnight at 37 °C.
Pick individual colonies from the agar plates and inoculate them in culture tubes containing 5mL of LB broth with the appropriate antibiotic. Culture the tubes overnight in a bacterial shaking incubator at 37 °C and 250rpm.
Following overnight incubation, add 500μL 50% glycerol to 500μL of bacterial culture and store at −80°C as a glycerol stock. Perform DNA miniprep on the rest of the culture to purify plasmid DNA as per Qiagen’s miniprep protocol and measure the concentration.
2.2. Excitable HEK-293 expression system
For the initial screening of the candidate BacNav channels, we recommend using our previously described 293KC cell line, i.e., a human embryonic kidney 293 (HEK-293) monoclonal cell line stably expressing Kir2.1 and Cx43 (Kirkton & Bursac, 2011). The 293KC system has several advantages for BacNav screening including: (1) HEK-293 cells are a widely accepted heterologous expression system for electrophysiological studies with well-defined endogenous currents (Jiang, Sun, Cao, & Wang, 2002; Varghese, Tenbroek, Coles, & Sigg, 2006); (2) HEK-293 cells are highly proliferative allowing for quick, large-scale expansion to generate monoclonal lines with uniform morphology and properties; and (3) with already engineered hyperpolarized resting membrane potential and robust coupling via Cx43 junctions, 293KC cells can be readily converted into excitable, AP conducting cells by transient or stable expression of a functional sodium channel with appropriate biophysical properties. Recording AP conduction in 293KC monolayers expressing different BacNav candidates can then serve to identify functional channels with optimal properties. For generation of the 293KC cell line, please refer to our previous publication (Kirkton & Bursac, 2011).
2.2.1. Materials and equipment
Lentiviral mammalian expression plasmid with a constitutive and strong eukaryotic promoter driving the ion channel of interest, fluorescent reporter gene, and antibiotic resistance gene
Lentiviral VSV-G-expressing envelope plasmid pMD2.G (Addgene, plasmid no. 12259)
Lentiviral packaging plasmid psPAX2 (Addgene, plasmid no. 12260)
HEK-293 cells (ATCC)
150mm TC-treated Cell Culture Dish (Falcon)
Trypsin-EDTA (0.05%), phenol red (Thermo Fisher Scientific)
JetPRIME transfection reagent (Polyplus)
0.45μm cellulose acetate filter (Corning)
HEK-293 growth medium—Dulbecco’s Modified Eagle Medium (DMEM) (high glucose; Thermo Fisher Scientific), 10% (vol/vol) fetal bovine serum (FBS) (Hyclone), and 1% (vol/vol) penicillin-streptomycin (Thermo Fisher Scientific)
HEK-293 freezing medium—HEK-293 growth medium, 10% (vol/vol) dimethyl sulfoxide (DMSO)
Lenti-X concentrator (Clontech)
Polybrene (Sigma-Aldrich)
Inverted fluorescence microscope (Nikon)
Centrifuge (Eppendorf)
Tissue culture plate (6/12 wells; Thermo Fisher Scientific)
Phosphate Buffered Saline (PBS)
Tyrode’s solution—135mM NaCl, 5.4mM KCl, 1.8mM CaCl2, 1mM MgCl2, 0.33 mM NaH2PO4, 5mM HEPES, and 5mM glucose. pH adjusted to 7.4 with NaOH and osmolarity adjusted to 280mOsm
Voltage-sensitive dye (Di-4-ANEPPS, 10μM; Biotium)
Aclar coverslip (Electron Microscopy Sciences)
Solid-state excitation light source (Lumencor)
504-channel optical mapping photodiode array (Redshirt Imaging)
Optical filters (Chroma)
PSCRed photoresist (Brewer Science)
MATLAB software (Mathworks)
SD9 Grass stimulator (Grass Technologies)
Optical air table (Newport)
2.2.2. Protocol
2.2.2.1. Lentivirus production
The following protocol is intended for one 15-cm dish. We typically prepare virus from at least four dishes at a time. The protocol can be scaled up accordingly.
-
On the day before transfection, seed 12 million HEK-293 cells into a 15-cm tissue culture dish for transfection the subsequent day.
Note: For optimal lentivirus production, we recommend using newly thawed cells expanded for at least 3 but not more than 15 passages.
The following day, change medium prior to transfection and check cells every 1–2h until ~70–85% confluency is reached.
Combine 18μg of lentiviral vector, 9μg of psPAX2, and 3μg of pMD2. G into one Eppendorf tube and transfect HEK-293 cells using jetPRIME reagent as per manufacturer’s protocol.
Change to fresh HEK-293 media 6–18h after transfection.
Collect the culture medium containing lentiviral particles 72h after the start of transfection.
Spin down the media at 500 × g for 5min at room temperature to pellet cell debris, and pass the supernatant through 0.45μm filter.
Combine 1 volume of Lenti-X Concentrator with 3 volumes of supernatant. Gently invert the mixture 4–5 times and incubate at 4°C overnight.
Centrifuge the mixture at 1500 × g for 45min at 4°C in a pre-chilled centrifuge.
-
Carefully remove the supernatant and resuspend the pellet in 100μL PBS. Immediately aliquot the virus into Eppendorf tubes and store at −80°C. For each virus, also prepare a tube with 0.1 × dilution (100μL total, in PBS) for titering (see below).
Note: Everything in contact with lentiviral particles or supernatant during the collection process should be discarded into a waste container filled with bleach.
Note: Avoid multiple thawing-freezing cycles of the virus as this will reduce virus titer.
2.2.2.2. Small-scale, unconcentrated lentivirus production
For rapid extensive screening of multiple BacNav candidates, we recommend making small-scale, unconcentrated lentivirus.
On the day before transfection, seed 1 million HEK-293 cells into each well of a 6-well plate.
Next day, change media prior to transfection and keep checking cells every 1–2h until ~60–70% confluency is reached.
Combine 1.8μg of lentiviral vector, 0.9μg of psPAX2, and 0.3μg of pMD2.G into one Eppendorf tube and transfect HEK-293 cells using jetPRIME reagents as per protocol.
Change to 2mL fresh HEK-293 media 6–18h after transfection.
Collect the culture media with lentiviral particles 72h after the start of transfection.
Spin down the media at 500 × g for 5 min at room temperature and aliquot the virus-containing supernatant to appropriate volumes (20–100μL) and store at −80°C until use.
2.2.2.3. Determination of lentiviral titer
The following method to determine the viral titer is designed for constructs with a fluorescence reporter.
Seed cells into a 12-well plate to achieve roughly 30% confluency by the following day. For 293KC, this corresponds to about 80,000 cells/well.
The next day, transduce each well with 1, 5, and 20μL of the 0.1 × diluted BacNav virus by adding virus to the culture medium. Detach the cells from at least one well using 0.05% Trypsin-EDTA and count the number of cells at the time of infection.
72h later, determine the percentage of transduced cells at each dilution by imaging using a fluorescence microscope. Use a well that has between 1% and 20% of cells exhibiting fluorescence to determine titer using the following equation: Titer=% Positive cells × Total cell number at time of infection/Volume of virus used.
2.2.2.4. Lentivirus transduction
Thaw the virus aliquot on ice.
-
Determine appropriate volume of virus needed (Volume=Total number of cells/Titer). If the virus titer is low, adding polybrene (8μg/mL) will improve transduction efficiency.
Note: Some cell types are sensitive to polybrene. If toxicity is observed in the presence of polybrene, use a lower concentration or exclude entirely.
Add virus directly to the culture media and incubate overnight. Replace with fresh media 24h after transduction.
If using unconcentrated lentivirus for transduction, replace cell culture medium with virus-containing supernatant, add 1 volume of fresh media 24h after transduction, and change to fresh media 48h post-transduction.
Expression usually can be detected 48–72h post-transduction.
If the transduction efficiency is over 80%, freeze down the cells in HEK-293 freezing media and store in liquid nitrogen. A plate can also be maintained for further validation and characterization studies.
2.2.2.5. Optical mapping of action potential propagation
For primary screening, we recommend performing optical mapping of membrane potential to quantify the conduction properties of the engineered 293KC cells expressing candidate BacNav channels (293KCBacNav).
Place 21-mm Aclar® coverslips into a 50mL conical tube, fill the tube with 70% ethanol, and sonicate the coverslips for 1.5h.
Wash the coverslips with PBS three times.
Pipette fibronectin solution (30μg/mL) as single 150μL drops on a sterile non-treated 15-cm petri dish (12 drops per plate). Place coverslip on a fibronectin drop by starting at the edge of the coverslip so that the liquid coats one side of the coverslip evenly. The coverslip should be able to spin on top of the liquid droplet.
Place the petri dish with coverslips on a rocker for 1–2h at room temperature or 4 °C overnight for coating.
-
Seed 200,000 of 293KCBacNav cells onto a 21-mm fibronectin-coated Aclar coverslip in 12-well tissue culture plates with 2mL media and leave the cells in the incubator for 24–48h until they generate confluent monolayers.
Note: After adding seeding cell solutions to wells of a 12-well plate, gently shake the plate to disperse cells and make sure under a microscope that the cells are evenly distributed. Carefully move the plate to a 37 °C incubator and let the cells attach to the coverslips without disruption. A heterogeneously seeded monolayer could result in uneven AP propagation or formation of conduction obstacles and reentrant waves.
Note: The density and confluency of the monolayer at the time of mapping could significantly impact the optical mapping results. Exceedingly sparse cultures could exhibit propagation failure or low conduction velocity due to reduced cell coupling. Overly dense cultures could exhibit slower conduction due to increased spatial density of gap junctions.
On the day of mapping, warm Tyrode’s solution in a 37°C water bath.
-
Wash the coverslip with warmed Tyrode’s solution for 5min and stain with voltage-sensitive dye Di-4 ANEPPS (3nM) for 5min at room temperature.
Note: Perform steps 7–10 in the dark as Di-4-ANEPPS is readily photobleached. Use of a red color headlamp is recommended.
Wash the cells with Tyrode’s solution for 5min at room temperature to remove excess dye and transfer them to a pre-warmed optical mapping chamber mounted on an optical air table. After placing in the chamber, let coverslips equilibrate to 37 °C for 5min.
-
Apply stimulus pulses (10ms, 6V, 1Hz) at the edge of the coverslip with a bipolar point electrode connected to a Grass stimulator and illuminate the cell culture with green excitation light (520±20nm, Lumencore) for 2–5s. Light shutter control, data acquisition, and electrical stimulation are synchronized using Lab View. During illumination, the emitted fluorescence signal is recorded in the red spectrum (>590nm) through a PCSRed photoresist-coated glass coverslip filter and converted to voltage using an array of 504 photodiodes individually connected to 750-μm-diameter optical fibers arranged into a 19-mm-diameter hexagonal imaging bundle. The recorded voltage signals are amplified, sampled at 2.4kHz, and stored on a computer for MATLAB analysis. Increase the electrical stimulation rate with an increment of 1Hz and map fluorescence signals until maximum capture rate is reached.
Note: Make sure that the excitation light is angled relative to coverslip to avoid phototoxicity and potential signal saturation. Also ensure that all of the photodiode array, recording chamber, and light source are firmly mounted on the optical air table to avoid mechanical vibrations and noise.
Analyze the optical mapping results and calculate conduction velocity (CV) and action potential duration (APD) as a measure of conduction properties. We use a software that is custom-made for our optical mapping setup; please refer to our previous publications for a more detailed discussion (Badie & Bursac, 2009; Badie, Satterwhite, & Bursac, 2009; Scull, McSpadden, Himel, Badie, & Bursac, 2012).
3. Validation and characterization of selected prokaryotic ion channel orthologs
Even if a 293KC cell monolayer expressing a BacNav candidate fails to elicit action potential propagation during the primary optical mapping screen, the channel might be still altered to function in human excitable cells. Many of the wild-type BacNav channels have non-physiological (highly hyperpolarized inactivation or highly depolarized activation) voltage dependencies. For example, wild-type NavSheP, a bacterial channels previously studied by us (Nguyen et al., 2016), exhibits overly hyperpolarized, left-shifted inactivation. Use of site-directed mutagenesis to substitute one of the channel’s extracellular negative charge cluster (ENC) residues (aspartic acid) with a non-charged amino acid (alanine) at the position 60 (i.e., D60A) resulted in a right shift of activation and inactivation to enable current flow in mammalian excitable cells (Nguyen et al., 2016). Before and after site-directed mutagenesis voltage dependencies and kinetics of channel activation and inactivation are determined using whole-cell patch-clamp analysis.
3.1. Characterization of channel function through whole-cell patch clamp
3.1.1. Materials and equipment
Patch-clamp glass pipette (OD 1.5mm, ID 1.16mm; Harvard Apparatus)
Aclar coverslip (Electron Microscopy Sciences)
Tyrode’s solution—135mM NaCl, 5.4mM KCl, 1.8mM CaCl2, 1mM MgCl2, 0.33mM NaH2PO4, 5mM HEPES, and 5mM glucose. pH adjusted to 7.4 with NaOH and osmolarity adjusted to 280mOsm
Patch-clamp pipette solution—140mM KCl, 10mM NaCl, 1mM CaCl2, 2mM MgCl2, 10mM EGTA, 10mM HEPES, and 5mM MgATP; pH adjusted to 7.3 with KOH and osmolarity adjusted to 270 mOsm
Silver wire (World Precision Instruments)
Patch-clamp setup—Multiclamp 700B amplifier (Axon Instruments), Patch clamp CV-7B headstage (Axon Instruments), BNC-2090 accessory (National Instruments), MPC-200 Micromanipulator (Sutter Instrument Company), Optical air table (Newport), NIDAQ-MX computer interface (National Instruments), WinWCP software (J. Dempster, University of Strathclyde), P-97 micropipette puller (Sutter Instrument)
3.1.2. Protocol
Culture 293KC cells expressing BacNav channel of interest (293KCBacNav) in a T-75 culture flask at 37 °C and 5% (vol/vol) CO2 until the cells grow to >90% confluency.
Detach the cells using 0.05% trypsin-EDTA and seed onto 21mm Aclar coverslips at a density of 5×103 cells per cm2 in 12-well tissue culture plates and incubate in 2mL of HEK growth media at 37°C and 5% CO2 for 5–8h to facilitate attachment. Make sure the cells are plated as single cells.
Before recording, place patch-clamp glass pipette in a micropipette puller and fabricate two recording glass pipettes via predefined cycles of heating and pulling. Optimize the protocol per the manufacturer’s instructions for desired tip size and shape.
Place the fabricated micropipette under a microscope and fire-polish the tip of the pipette using a heating filament. Optimize the heating temperature, duration, and filament position for desired tip diameter.
Turn on the nitrogen tank connected to the optical air table. Transfer the coverslip with the 293KCBacNav cells to a glass-bottom patch-clamp chamber perfused with Tyrode’s solution. Immerse the ground electrode in the bath solution. Heat the solution if needed.
-
Fill the pipette with patch-clamp pipette solution using a 1mL syringe with 0.22μm filter. Mount the pipette to the headstage with a pipette holder and ensure that the silver wire electrode is in contact with the pipette solution.
Note: If the silver wire electrode turns black, place it in bleach for few minutes and rinse with water multiple times.
Note: The patch-clamp pipette solution can be divided into aliquots and stored at −20°C for up to 6 months. Thaw a new aliquot before each patch-clamp session.
-
Under a fluorescence microscope, identify a single transduced cell based on its expression of a fluorescent reporter. Move the pipette toward the cell using the micromanipulator. As soon as the micropipette tip touches the batch solution, the pipette tip resistance will display on the software. For 293KC cells, we recommend using pipette tip resistance between 1 and 2MΩ. Once the pipette tip is close to the cell surface, change the micromanipulator settings to diagonal mode such that the pipette is moving along Z and X axis simultaneously. Approach the cell slowly and the pipette resistance will increase when the tip is in contact with the cell membrane. Slowly move the pipette forward until pipette resistance is increased ~1.5–2 times, then apply a brief negative pressure through suction to induce the giga seal formation between the pipette and cell membrane. Sometimes it is helpful to apply a negative holding voltage (−40mV) to induce the giga seal. Once the giga seal is formed, apply slight suction or a brief current pulse to rupture the membrane and achieve a whole-cell configuration. Filter the recordings with a 10-kHz low-pass Bessel filter and digitize at 40kHz.
Note: If the initial resistance is too high, this is often caused by presence of a bubble in the pipette solution. Apply slight negative pressure to remove the bubble.
Apply first voltage-clamp mode to record the ion currents when cell is subject to different step voltage pulses. To measure the activation properties of the channel, the membrane potential is held at −80mV and stepped up for 500ms to different holding potentials (−60 to 60mV) in increments of 10mV. If no current is observed, change to a more hyperpolarized holding potential at an increment of −10mV and repeat the protocol. We define candidate BacNav channel as non-functional when no observable currents can be recorded at holding voltages down to −180mV. To measure the inactivation properties of the channel, first apply a 10-s pre-pulse potential (−30 to −160mV), then step voltage for 1s to 0mV testing potential. In current-clamp mode, inject a 1-ms current pulse to trigger an action potential. The resting membrane potential of 293KC cell is usually between −75 and −80mV. Depolarized resting membrane potential is usually indicative of a leaky seal.
Export the peak currents (Ipeak) and testing potential (V) recorded during activation protocol to an Excel spreadsheet. Plot I-V curve and extract (sodium Nernst potential). Calculate (channel conductance) and m∞ (the steady-state activation gating variable) using Eqs. (1 and 2) and plot channel activation curve (m∞ vs V). Fit the current traces from the activation protocol with Eq. (3) using MATLAB to compute the time constant for inactivation kinetics (τh) and time constant for activation kinetics (τm). Fit the activation curve using the Boltzmann equation (Eq. 4) where V1/2 is the mid-point voltage of Boltzmann equation fit.
| (1) |
| (2) |
| (3) |
| (4) |
3.2. Site-directed mutagenesis
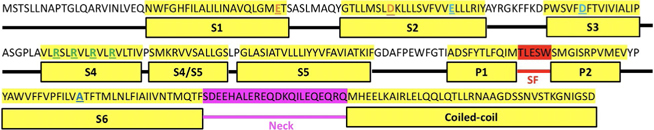
Mammalian VGSCs are formed by four structurally homologous transmembrane domains, each consisting of six transmembrane segments (S1–S6) comprising a voltage-sensing domain (VSD; S1–S4) and a pore-lining domain (PD; S5–S6). Although only containing one transmembrane domain, the six transmembrane segment architecture is well-conserved among BacNavs(Fig. 1 showing NavSheP channel). The S4 segment in VSD contains positively charged amino acid residues at every third position that act as gating charges, moving vertically across the membrane to trigger channel activation in response to membrane depolarization. Sodium ion flow through BacNav is achieved via a multi-ion mechanism, whereby four glutamic acid residues of the selectivity filter in each of S1–S4 (TLESW) serve as high-field-strength (HFS) sites that favor conduction of small cations whereas the adjacent backbone carbonyls (TLESW) form the central and inner coordination sites (Chakrabarti et al., 2013). As membrane potential is depolarized, change in electric field across the membrane induces an “outward” movement of the S4 segment, which then triggers a conformational change in the S4–S5 linker. This in turn shifts the positions of S5 and S6 helices, inducing a bend in the middle of S6 (hinge), which leads to opening of the intracellular gate (McCusker et al., 2012). Beneath this intracellular gate is the C-terminal domain (CTD), comprised of a neck region and a distal coiled-coil domain, which have been suggested to play various roles in channel assembly, stability, and gating (Arrigoni et al., 2016).
Fig. 1.
Structure and corresponding sequence of NavSheP. S1–S6, selectivity filter (SF), neck, and coiled-coil regions are indicated. Extracellular negative cluster (ENC) residues are highlighted orange. Intracellular negative cluster (INC) residues are highlighted in cyan. Positive gating charges are highlighted in green. Glycine hinge residue is highlighted in blue.
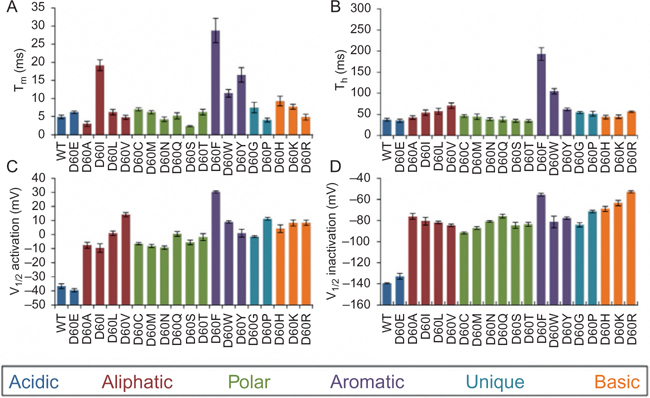
Based on previous studies of BacNav structure and biophysics, the voltage dependency properties of BacNav can be altered through site-directed mutagenesis in the highly conserved regions such as the activation gate, CTD, and intracellular negative charge cluster (INC) or extracellular negative charge cluster (ENC) of VSD (Blanchet, Pilote, & Chahine, 2007; Nguyen et al., 2016; Shaya et al., 2014; Shimomura, Irie, Nagura, Imai, & Fujiyoshi, 2011). Combined with our in vitro screening and characterization results, we have summarized potential mutations for BacNav candidates to alter the channel voltage dependency and kinetics in Table 1. Furthermore, as an example of a mutagenesis screen, Fig. 2 shows changes in electrophysiological properties of NavSheP when substituting different amino acids in D60 position.
Table 1.
Genetic manipulations for altering BacNav biophysical properties.
| Mutation | Intended effects |
|---|---|
| Altering membrane voltage dependencies of BacNav activation and inactivation | |
| D60X | Right-shifted inactivation (via substitution by neutral and positive amino acids) (Shimomura et al., 2011) |
| E43X | Right-shifted inactivation (via substitution by neutral and positive amino acids) (Shimomura et al., 2011) |
| E70X or D91X | Left-shifted activation (via substitution by neutral and positive amino acids) (Blanchet et al., 2007) |
| D237G, E238G, E239G | Left-shifted activation (Shaya et al., 2014) |
| M232A | Left-shifted activation (Shaya et al., 2014) |
| R110C, R113C, R116C, R119C | These are gating charges of the voltage-sensing domain. Right-shifted inactivation via substitution by neutral amino acids (Chahine, Pilote, Pouliot, Takami, & Sato, 2004) |
| Speeding up gating kinetics | |
| L64T | Faster activation and inactivation kinetics via substitution by hydrophilic amino acids (Lacroix, Campos, Frezza, & Bezanilla, 2013) |
| F218A | Faster activation and inactivation kinetics |
| A216G | Faster inactivation kinetics (Irie et al., 2010) |
Note: The amino acid residues shown in table are based on NavSheP. The actual position may vary in different BacNav orthologs.
Fig. 2.
Electrophysiological properties of NavSheP D60 mutants. (A and B) Time constants of activation (τm, A) and inactivation (τh, B) measured at −20mV for WT and D60E mutant. (C and D) Mid-points (V1/2) of activation (C) and inactivation (D) curves of NavSheP mutants exhibit different depolarization shifts relative to the V1/2 of the wild-type (WT) channel. Recordings were performed at 25°C (n=4–10).
3.2.1. Materials and equipment
Sequencing analysis software
Site-directed mutagenesis primers
Phusion® Hot Start Flex DNA Polymerase (NEB)
5 × Phusion HF Buffer (NEB)
10mM dNTPs (Thermo Fisher Scientific)
Thermal cycler (Bio-Rad)
Gel electrophoresis system (Bio-Rad)
T4 DNA ligase buffer (NEB)
T4 Polynucleotide Kinase (PNK) (NEB)
T4 DNA ligase (NEB)
DpnI (NEB)
Chemically competent E. coli cells
Agar plate
LB medium
Antibiotics
Sequencing primers
3.2.2. Protocol
-
1
Align the protein sequence of a candidate channel with previously characterized ion channel (NavSheP) and identify key residues as shown in Table 1.
-
2
Based on whole-cell patch-clamp results, choose potential mutations to alter BacNav voltage dependency or kinetics. If the channel has highly hyperpolarized activation (V1/2 < −25mV), choose the mutations that could yield a right shift in activation. If the channel has highly depolarized activation (V1/2 > 0mV), choose the mutations that would potentially yield a left shift in activation. If the channel is functional within physiological range, choose mutations in the “speeding up gating kinetics” section.
-
3
Design the site-directed mutagenesis primers using the NEBaseChanger tool (http://nebasechanger.neb.com/) as per instruction and order the primers from oligonucleotides synthesis company. Calculate the melting temperature using NEB Tm Calculator (https://tmcalculator.neb.com).
Note: When calculating Tm, use only the target-specific sequences.
-
4
Perform polymerase chain reaction (PCR) using Phusion Hot Start Flex DNA polymerase as per NEB protocol. For each reaction, prepare 4 PCR mixtures to run at 4 different annealing temperatures.
-
5
Run PCR product on 1% agarose gel. Cut out the bands with correct size and perform gel purification using commercial kit. If desired band size is observed under multiple conditions, combine all DNA together after elution.
-
6
Combine 1μL purified PCR product with 1μL T4 DNA ligase buffer, 1μL T4 PNK, 1μL T4 DNA ligase, 1μL DpnI and 5μL Nuclease-Free water and incubate at 37°C overnight.
-
7
Use ~2μL of reaction mixture to transform into chemically competent E. coli cells (e.g., Stbl3™) as per manufacturer’s protocol.
-
8
Plate the cells on agar plates containing the appropriate antibiotic in the plasmid backbone and culture the plates overnight at 37 °C.
-
9
Pick individual colonies from the agar plates and inoculate them in culture tubes containing 5mL of LB broth with the appropriate antibiotic. Culture the tubes overnight in a bacterial shaking incubator at 37 °C and 250rpm.
-
10
Following overnight incubation, add 500μL 50% glycerol to 500μL of bacteria and store at −80°C as glycerol stock. Perform DNA miniprep to purify plasmid DNA from the rest of the bacteria as per protocol (e.g., Qiagen miniprep) and measure the concentration.
-
11
Design and order sequencing primers flanking the mutation region and send plasmid for Sanger sequencing to commercial companies such as Genewiz.
-
12
Once the sequencing is confirmed, follow protocols described in Section 2.2 and 3.1 for characterization of channel function.
-
13
Select appropriate BacNav candidates based on electrophysiological and biophysical properties for subsequent in vitro assessment.
3.3. Assessing therapeutic potential of engineered prokaryotic ion channels using an arrhythmogenic cell culture model
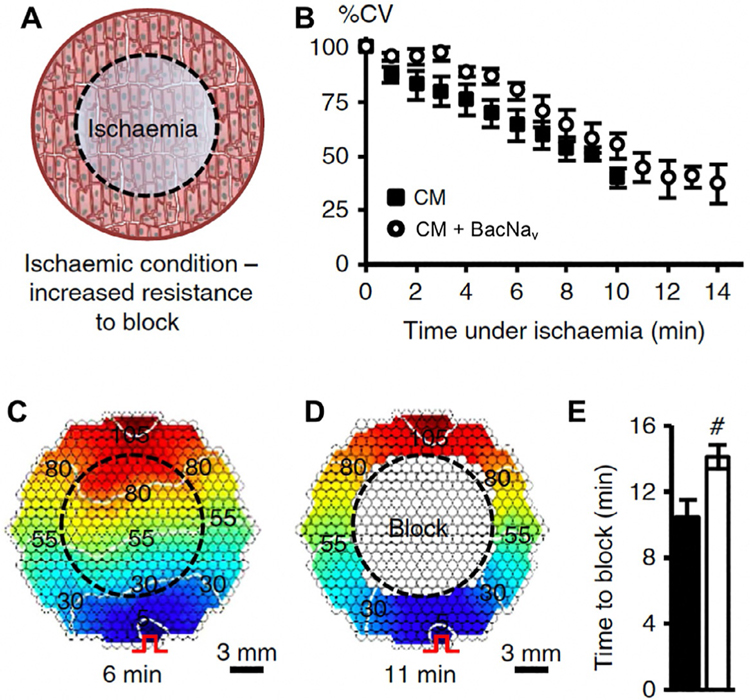
The described platform is intended to enable discovery, characterization, and optimization of BacNav channels for use in cell and gene therapies for improving action potential propagation and preventing conduction block and cardiac arrhythmias. Previously we have shown that lentiviral BacNav expression can augment cardiac tissue excitability and rescue AP conduction under simulated pathological conditions in vitro (Fig. 3). To be able to additionally assess the therapeutic potential of top BacNav candidates in vitro, we have established arrhythmogenic neonatal rat ventricular myocyte (NRVM) monolayers with high incidence of reentry induction by programmed electrical stimulation. Similar to Fig. 3, this NRVM model (described below) allows direct assessment of how expression of BacNav candidates in cardiomyocytes affects action potential conduction and incidence of reentry.
Fig. 3.
Improvement of mammalian AP conduction by BacNav in modeled pathological conditions. (A) Schematic depicting exogenous expression of BacNav in neonatal rat cardiomyocytes (CMs) to increase resistance to conduction block in ischemic conditions. Black dashed circle denotes position of glass coverslip used to induce regional ischemia in CM monolayer. (B) Progressive conduction slowing until block with time of ischemia in control (CM) and NavSheP D60A transduced monolayers. (C and D) Representative isochrone maps showing conduction slowing after 6min (C) and complete block after 11 min of ischemia (D). Pulse signs indicate location of stimulating electrode. (E) Under ischemic condition, CMs transduced with NavSheP D60A lentivirus (white) resisted conduction block longer than control CMs (black). Error bars indicate s.e.m.; statistical significance was determined by an unpaired Student’s t-test.
3.3.1. Materials and equipment
-
14
BacNav and control lentivirus
-
15
Neonatal rat ventricular myocyte
-
16
Aclar coverslip (Electron Microscopy Sciences)
-
17
Inverted fluorescence microscope (Nikon)
-
18
Centrifuge (Eppendorf)
-
19
Tissue culture plate (12 wells; Thermo Fisher Scientific)
-
20
Fibronectin (Sigma)
-
21
Mitomycin-C (Sigma)
-
22
NRVM seeding media: DMEM/F-12 medium (Gibco) supplemented with 10% FBS, 0.2% penicillin, and 0.2% B12
-
23
NRVM serum-free maintenance media: DMEM/F12+0.2% penicillin+ 0.2% B12+2.5μg/mL L-ascorbic acid+5nM Triiodo-L-Thyronine+1 × Insulin-Transferrin-Selenium supplement
-
24
NRVM maintenance media: DMEM/F12, 0.2% penicillin, 0.2% B12, and 2% horse serum
-
25
Tyrode’s solution—135mM NaCl, 5.4mM KCl, 1.8mM CaCl2, 1mM MgCl2, 0.33mM NaH2PO4, 5mM HEPES, and 5mM glucose; pH adjusted to 7.4 with NaOH and osmolarity adjusted to 280mOsm
-
26
Voltage-sensitive dye (Di-4-ANEPPS, 10μM; Biotium)
-
27
Aclar coverslip (Electron Microscopy Sciences)
-
28
Solid-state excitation light source (Lumencor)
-
29
504-channel optical mapping photodiode array (Redshirt Imaging)
-
30
Optical filters (Chroma)
-
31
PSCRed photoresist (Brewer Science)
-
32
MATLAB software (Mathworks)
-
33
SD9 Grass stimulator (Grass Technologies)
-
34
NanoDrop™ One/OneC Microvolume UV–Vis Spectrophotometer (Thermo Scientific)
-
35
Immunostaining blocking solution: 5% (vol/vol) donkey serum and 0.1% (vol/vol) Triton X-100 (diluted in PBS). The solution can be stored at 4 °C for up to 2 weeks
3.3.2. Protocol
3.3.2.1. Neonatal rat ventricular myocte (NRVM) arrhythmogenic model
-
Make lentivirus and perform virus titration as described in Section 2.2.
Note: For these studies, we recommend using promoters such as MHCK7 or minimum chicken cardiac troponin T promoter (cTnT) (Prasad, Xu, Yang, Acton, & French, 2011; Salva et al., 2007) to confer cardiomyocyte-specific BacNav expression.
For NRVM isolation protocol, please refer to our previous publications (Jackman, Carlson, & Bursac, 2016; Nguyen et al., 2016).
On Day 0, seed isolated NRVMs (8 × 104 cells/cm2) on 21 mm fibronectin-coated (30μg/mL) Aclar coverslips in NRVM seeding media.
Next day (Day 1), treat cells with 10μg/mL mitomycin-C and change to fresh seeding media 2–3h after treatment.
On Day 2, change media to NRVM serum-free maintenance media and transduce the cells with lentivirus. On Day 3, change culture media to NRVM maintenance media and perform media change every 2 days. Check fluorescence after 48h and make sure the transduction efficiency is over 90%.
On Day 6, perform optical mapping as described in Section 2.2.
Once the cells are placed in the mapping chamber and equilibrated for 5min, turn on the excitation light for 2s without stimulation and turn off for 20s, then repeat this twice to photobleach the excess dye before applying long light exposure.
First record 5s of spontaneous activity without any stimulation.
If reentry is observed, check the cardiomyocyte activation rate and to terminate the reentry, stimulate the coverslip with field electrode at a rate above reentry rate for 2s.
Repeat the 5s recording without stimulation to check if the reentry is stopped.
If reentry is stopped, start pacing at 1Hz for 2s and record from 0.5s before to 1.5s after pacing. Continue to increase pacing rate with increment of 1Hz until maximum capture rate is reached. The conduction velocity can be calculated at different pacing rates.
If reentry is not induced at maximum capture rate, increase the pacing rate by 0.5Hz in the next induction attempt. If 1:1 capture during pacing is lost, decrease the pacing rate by 0.25Hz and stimulate the monolayer again. The resulting success or loss of 1:1 capture is then followed by an increase or decrease of pacing rate by 0.125Hz, respectively, as the last attempt at induction. In the case of successful reentry induction, record without stimulation for 3s at 1, 2, and 5min after reentry induction to assess if reentry is sustained long term.
After mapping, save the coverslips for RNA extraction and immunostaining (see below).
3.3.2.2. RNA extraction and quantitative polymerase chain reaction (qPCR)
Assessment of gene expression levels of different BacNav candidates in NRVMs is important for being able to compare their functional effects. Moreover, due to the bacteria origin of BacNav channels, their effects on regulation and expression of other electrophysiology-related genes have not been studied in mammalian cells. The following qPCR protocol can be used to assess the level of BacNav gene expression or any potential changes in expression of important ion channel genes and other genes of interest resulting from BacNav transduction.
Go to NCBI website (https://www.ncbi.nlm.nih.gov/) and choose “Gene” from the database pool down menu.
Enter the gene of interest and click on the species of interest from the results (e.g., rat).
Go the section titled “mRNA and protein(s)” and click on the accession number of the mRNA sequence.
Under the “Analyze this sequence” section, click on “Pick Primers.”
Change the parameters to meet the requirement that PCR product size is less than 250bp and set “Exon junction span” to “Primer must span an exon-exon junction.” Check the “Intron inclusion” box.
Click “Get Primers” and click Check on the results window. Find the primer sets that have 0 score of self 3’ complementarity and do not pick primer sets that detect more than one target.
Order the primer sets from a commercial company (e.g., IDT).
-
Extract the RNA from NRVM monolayers immediately after mapping using a commercial kit (e.g., RNeasy Mini) per manufacturer’s protocol. Measure the concentration and purity of RNA using NanoDrop.
Note: To ensure high RNA quality, we recommend cleaning the working bench with surface decontaminant and using filter-containing pipette tips and RNase-free reagents. For NanoDrop, the ratio of absorbance at 260 and 280nm (260/280) >2.0 is considered as pure RNA, otherwise the sample may contain protein. The 260/230 ratio should be >2.0 (usually 2.0–2.2), otherwise samples might have phenol, Trizol, or other contaminants.
-
Synthesize cDNA using a commercial kit (e.g., Bio-Rad) per manufacturer’s protocol. It is recommended that you calculate and use the same amount of RNA in each sample. Include a sample without adding reverse transcriptase as nRT control.
Note: 5–20ng of starting RNA is sufficient for each qPCR reaction.
Dilute the cDNA to 3.75ng/μL and prepare 10μM primer mixture (forward + reverse). Set up a qPCR plate and prepare technical triplicates for each group. For each well, add 4μL of cDNA, 1μL of primer mixture and 5μL of 2 × SYBR Green master mix. For each primer set, include an nRT control and no template control (water) wells.
Spin down the qPCR plate at 2000 × g for 2min.
Run qPCR cycles using a real-time PCR system (e.g., CFX96, Bio-Rad).
Ensure that the melting curves for each primer have one single peak and the Ct (cycle threshold) difference between technical replicates is less than 1.
For each group, average the Ct values from all three technical replicates and calculate gene expression using following equations:
Expression fold change (EFC) = 2^(−ΔΔCt); calculate average EFC for biological replicates.
3.3.2.3. Immunostaining
Assessment of BacNav expression in cardiomyocytes and non-myocytes in NRVM monolayers permits better understanding of their functional effects. The following immunostaining protocol can be used to assess the distribution of cardiomyocytes and non-myocytes (e.g., fibroblasts) in NRVM cultures as well as BacNav expression level in different cell types based on expression of fluorescent reporters (e.g., GFP) built in the BacNav lentiviral vectors.
Fix the NRVM monolayer after optical mapping with 4% PFA in PBS for 10min at room temperature. Wash three times in DPBS for 5min.
Block and permeabilize the monolayer with immunostaining blocking solution for 30min at room temperature.
Incubate the cells with primary antibodies diluted in blocking solution overnight at 4 °C. Wash three times in DPBS. Common antibodies we use are listed in Table 2.
Incubate the cells with secondary antibodies diluted in blocking solution for 2h at room temperature. Wash three times in DPBS.
Move coverslips to microscope slide and add appropriate amount of antifade mounting medium. Let the medium harden overnight at room temperature before imaging.
Table 2.
List of common antibodies.
| Antigen | Host | Dilution | Source | Catalog number |
| Primary antibody | ||||
| GFP | Mouse | 1:100 | Thermo Fisher Scientific | A-11120 |
| Vimentin | Mouse | 1:500 | Santa. Cruz Biotechnology | sc6260 |
| Cardiac troponin T (cTnT) | Rabbit | 1:100 | Abcam | ab45932 |
| Sarcomeric α-actinin (SAA) | Mouse | 1:200 | Sigma-Aldrich | A7811 |
| Secondary antibody | ||||
| Antibody | Dilution | Source | Catalog number | |
| Donkey anti-Mouse, Alexa Fluor Plus 647 | 1:200 | Invitrogen | A32787 | |
| Donkey anti-Rabbit, Alexa Fluor Plus 594 | 1:200 | Invitrogen | A32754 | |
4. Summary
Prokaryotic voltage-gated sodium channels-based gene therapies hold promise as an effective and translatable strategy to eliminate cardiac arrhythmias, yielding a viable solution for one of the leading causes of death and disability. As different antiarrhythmogenic applications (e.g., ischemic ventricular tachycardias, congenital arrhythmogenic disorders, chronic atrial fibrillation, right and left ventricular bundle brunch blocks) might benefit from different biophysical properties of the engineered ion channels, it is important to expand the pool of functional BacNav beyond the few that have previously been characterized in mammalian heterologous expression systems. This protocol describes experimental framework for discovery and engineering of prokaryotic ion channels and their in vitro validation and characterization. Following identification and characterization on this platform, top BacNav candidates will need to be tested by AAV delivery in small and large animal models of cardiac arrhythmias. Versatile screening and mutagenesis strategies described in this protocol provide foundation for eventual translation of BacNav gene therapies to clinical practice.
Acknowledgment
We thank S. DeLuca for critically reading the manuscript. This work was supported by National Institutes of Health grants HL134764, HL132389, and HL126524.
References
- Antzelevitch C (2001). Basic mechanisms of reentrant arrhythmias. Current Opinion in Cardiology, 16(1), 1–7. [DOI] [PubMed] [Google Scholar]
- Arrigoni C, Rohaim A, Shaya D, Findeisen F, Stein RA, Nurva SR, et al. (2016). Unfolding of a temperature-sensitive domain controls voltage-gated channel activation. Cell, 164(5), 922–936. 10.1016/j.cell.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badie N, & Bursac N (2009). Novel micropatterned cardiac cell cultures with realistic ventricular microstructure. Biophysical Journal, 96(9), 3873–3885. S0006–3495(09)00572–4 [pii] 10.1016/j.bpj.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badie N, Satterwhite L, & Bursac N (2009). A method to replicate the microstructure of heart tissue in vitro using DTMRI-based cell micropatterning. Annals of Biomedical Engineering, 37(12), 2510–2521. 10.1007/s10439-009-9815-x. [DOI] [PubMed] [Google Scholar]
- Bikou O, Thomas D, Trappe K, Lugenbiel P, Kelemen K, Koch M, et al. (2011). Connexin 43 gene therapy prevents persistent atrial fibrillation in a porcine model. Cardiovascular Research, 92(2), 218–225. 10.1093/cvr/cvr209. [DOI] [PubMed] [Google Scholar]
- Blanchet J, Pilote S, & Chahine M (2007). Acidic residues on the voltage-sensor domain determine the activation of the NaChBac sodium channel. Biophysical Journal, 92(10), 3513–3523. 10.1529/biophysj.106.090464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet E (1999). Cardiac ionic currents and acute ischemia: From channels to arrhythmias. Physiological Reviews, 79(3), 917–1017. 10.1152/physrev.1999.79.3.917. [DOI] [PubMed] [Google Scholar]
- Chahine M, Pilote S, Pouliot V, Takami H, & Sato C (2004). Role of arginine residues on the S4 segment of the Bacillus halodurans Na+ channel in voltage-sensing. The Journal of Membrane Biology, 201(1), 9–24. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/15635808. [DOI] [PubMed] [Google Scholar]
- Chakrabarti N, Ing C, Payandeh J, Zheng N, Catterall WA, & Pomes R (2013). Catalysis of Na+ permeation in the bacterial sodium channel Na(V)Ab. Proceedings of the National Academy of Sciences of the United States of America, 110(28), 11331–11336. 10.1073/pnas.1309452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous K, & Wallace BA (2011). NaChBac: The long lost sodium channel ancestor. Biochemistry, 50(32), 6742–6752. 10.1021/bi200942y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu DG (2015). Cardiac arrhythmias: Diagnosis, symptoms, and treatments. Cell Biochemistry and Biophysics, 73(2), 291–296. 10.1007/s12013-015-0626-4. [DOI] [PubMed] [Google Scholar]
- Gourraud JB, Barc J, Thollet A, Le Marec H, & Probst V (2017). Brugada syndrome: Diagnosis, risk stratification and management. Archives of Cardiovascular Diseases, 110(3), 188–195. 10.1016/j.acvd.2016.09.009. [DOI] [PubMed] [Google Scholar]
- Greener ID, Sasano T, Wan X, Igarashi T, Strom M, Rosenbaum DS, et al. (2012). Connexin43 gene transfer reduces ventricular tachycardia susceptibility after myocardial infarction. Journal of the American College of Cardiology, 60(12), 1103–1110. https://doi. org/10.1016/j.jacc.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Tan H, Sun C, & Li G (2018). Dysfunctional Nav1.5 channels due to SCN5A mutations. Experimental Biology and Medicine (Maywood, N.J.), 243(10), 852–863. 10.1177/1535370218777972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizar JF, Ellenbogen KA, Tan AY, & Kaszala K (2019). Arrhythmia-induced cardiomyopathy: JACC state-of-the-art review. Journal of the American College of Cardiology, 73(18), 2328–2344. 10.1016/j.jacc.2019.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Finet JE, Takeuchi A, Fujino Y, Strom M, Greener ID, et al. (2012). Connexin gene transfer preserves conduction velocity and prevents atrial fibrillation. Circulation, 125(2), 216–225. 10.1161/CIRCULATIONAHA.111.053272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K, Kitagawa K, Nagura H, Imai T, Shimomura T, & Fujiyoshi Y (2010). Comparative study of the gating motif and C-type inactivation in prokaryotic voltage-gated sodium channels. The Journal of Biological Chemistry, 285(6), 3685–3694. 10.1074/jbc.M109.057455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman CP, Carlson AL, & Bursac N (2016). Dynamic culture yields engineered myocardium with near-adult functional output. Biomaterials, 111, 66–79. https://doi. org/10.1016/j.biomaterials.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janse MJ, & Wit AL (1989). Electrophysiological mechanisms of ventricular arrhythmias resulting from myocardial ischemia and infarction. Physiological Reviews, 69(4), 1049–1169. [DOI] [PubMed] [Google Scholar]
- Jiang B, Sun X, Cao K, & Wang R (2002). Endogenous Kv channels in human embryonic kidney (HEK-293) cells. Molecular and Cellular Biochemistry, 238(1–2), 69–79. 10.1023/a:1019907104763. [DOI] [PubMed] [Google Scholar]
- Khurshid S, Choi SH, Weng LC, Wang EY, Trinquart L, Benjamin EJ, et al. (2018). Frequency of cardiac rhythm abnormalities in a half million adults. Circulation. Arrhythmia and Electrophysiology, 11(7), e006273. 10.1161/CIRCEP.118.006273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkton RD, & Bursac N (2011). Engineering biosynthetic excitable tissues from unexcitable cells for electrophysiological and cell therapy studies. Nature Communications, 2, 300. 10.1038/ncomms1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleber AG, & Rudy Y (2004). Basic mechanisms of cardiac impulse propagation and associated arrhythmias. Physiological Reviews, 84(2), 431–488. 10.1152/physrev.00025.2003. 84/2/431 [pii]. [DOI] [PubMed] [Google Scholar]
- Kuriachan VP, Sumner GL, & Mitchell LB (2015). Sudden cardiac death. Current Problems in Cardiology, 40(4), 133–200. 10.1016/j.cpcardiol.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Lacroix JJ, Campos FV, Frezza L, & Bezanilla F (2013). Molecular bases for the asynchronous activation of sodium and potassium channels required for nerve impulse generation. Neuron, 79(4), 651–657. 10.1016/j.neuron.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yang KC, & Dudley SC Jr. (2014). Cardiac sodium channel mutations: Why so many phenotypes? Nature Reviews. Cardiology, 11(10), 607–615. 10.1038/nrcardio.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markman TM, & Nazarian S (2019). Treatment of ventricular arrhythmias: What’s new? Trends in Cardiovascular Medicine, 29(5), 249–261. 10.1016/j.tcm.2018.09.014. [DOI] [PubMed] [Google Scholar]
- McCusker EC, Bagneris C, Naylor CE, Cole AR, D’Avanzo N, Nichols CG, et al. (2012). Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nature Communications, 3, 1102. 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meregalli PG, Tan HL, Probst V, Koopmann TT, Tanck MW, Bhuiyan ZA, et al. (2009). Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm, 6(3), 341–348. 10.1016/j.hrthm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Nguyen HX, Kirkton RD, & Bursac N (2016). Engineering prokaryotic channels for control of mammalian tissue excitability. Nature Communications, 7, 13132. 10.1038/ncomms13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KM, Xu Y, Yang Z, Acton ST, & French BA (2011). Robust cardiomyocyte-specific gene expression following systemic injection of AAV: In vivo gene delivery follows a Poisson distribution. Gene Therapy, 18(1), 43–52. 10.1038/gt.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundir S, Martin MJ, O’Donovan C, & UniProt C (2016). UniProt tools. Current Protocols in Bioinformatics, 53, 1.29.21–21.29.15. 10.1002/0471250953.bi0129s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, & Clapham DE (2001). A prokaryotic voltage-gated sodium channel. Science, 294(5550), 2372–2375. 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Rivaud MR, Agullo-Pascual E, Lin X, Leo-Macias A, Zhang M, Rothenberg E, et al. (2017). Sodium channel remodeling in subcellular microdomains of murine failing cardiomyocytes. Journal of the American Heart Association, 6(12), e007622. 10.1161/JAHA.117.007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salva MZ, Himeda CL, Tai PW, Nishiuchi E, Gregorevic P, Allen JM, et al. (2007). Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Molecular Therapy, 15(2), 320–329. 10.1038/sj.mt.6300027. [DOI] [PubMed] [Google Scholar]
- Scull JA, McSpadden LC, Himel HD, Badie N, & Bursac N (2012). Single-detector simultaneous optical mapping of V(m) and [Ca(2+)](i) in cardiac monolayers. Annals of Biomedical Engineering, 40(5), 1006–1017. 10.1007/s10439-011-0478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, et al. (2014). Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. Journal of Molecular Biology, 426(2), 467–483. 10.1016/j.jmb.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura T, Irie K, Nagura H, Imai T, & Fujiyoshi Y (2011). Arrangement and mobility of the voltage sensor domain in prokaryotic voltage-gated sodium channels. The Journal of Biological Chemistry, 286(9), 7409–7417. 10.1074/jbc.M110.186510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese A, Tenbroek EM, Coles J Jr., & Sigg DC (2006). Endogenous channels in HEK cells and potential roles in HCN ionic current measurements. Progress in Biophysics and Molecular Biology, 90(1–3), 26–37. 10.1016/j.pbiomolbio.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, et al. (2020). Heart disease and stroke statistics—2020 update: A report from the American Heart Association. Circulation, 143(8). 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- Yue L, Navarro B, Ren D, Ramos A, & Clapham DE (2002). The cation selectivity filter of the bacterial sodium channel, NaChBac. The Journal of General Physiology, 120(6), 845–853. 10.1085/jgp.20028699. [DOI] [PMC free article] [PubMed] [Google Scholar]