Abstract
Objective:
The main objective of this study is to design a synthetic vaccine from the binder of sperm-1 (BSP1).
Materials and Methods:
This study was carried out using bioinformatics-related techniques. BSP-1 has been chosen as one of the biomarkers of a ruminant’s male fertility. We hypothesize that the BSP1 synthetic vaccines, which contain T-cell epitopes, can produce antibodies more effectively for the development of a sperm fertility detection kit. A sequence of BSP-1 peptides A0A0K1YXR5 from Bubalus bubalis (Domestic water buffalo) origin has been decided to be used to develop the peptide vaccine.
Results:
In this study, we succeeded in making synthetic vaccines from BSP-1 with a peptide sequence of LPEDSVPDEERVFPFTYRNRKHF. The three-dimensional theoretical prediction analysis of the peptide binding pattern to its ligand, as well as the molecular docking, has also been revealed.
Conclusions:
A synthetic vaccine from the BSP-1 has been developed in this study with the amino acid sequence LPEDSVPDEERVFPFTYRNRKHF, which is buffer-soluble, and the three-dimensional theoretical prediction analysis of the peptide binding pattern of BSP-1 to its ligand, as well as molecular docking, has also been revealed.
Keywords: BSP1, epitope, peptide, ruminant, synthetic vaccine, T-cell
Introduction
In a sustainable cattle industry, bull fertility plays a central role. Subfertile bulls might result in serious economic losses. As a result, efforts to develop a bull fertility detection device are continuously being carried out, including the development of peptide biomarker-based immunoassay methods. Since the early 2020s, the utilization of synthetic peptide antigens/immunogens to produce specific immunological reagents for specific purposes, including for studying animal fertility, has increased significantly [1–4]. One of the processes in designing synthetic peptides involves bioinformatics [3,5]. The purpose of this study was to determine how to utilize bioinformatics-related techniques to design peptide vaccines. In this study, we used the binder of sperm-1 (BSP-1) peptide as a model. BSP-1 is a family of proteins found in several species‘ seminal plasma (SP) [6]. BSP-1 can stabilize the sperm membrane and mediate the sperm‘s binding to the oviduct‘s epithelium [7,8]. Therefore, BSP-1 has been chosen as one of the biomarkers of male fertility. The research in an effort to find biomarkers for male fertility selection both proteomically and genomically has been carried out systematically for the last two decades [9,10]. These can be based on molecular compounds in sperm and SP [10–13]. Many promising biomarkers for bull fertility have been studied based on the proteomic landscape of SP [14,15]. A recent publication using matrix-assisted laser desorption ionization-time of flight/mass spectrometry (MALDI-TOF/MS) technology has identified 23 proteins associated with the fertility levels expressed in the spermatozoa of crossed bulls [16]. According to them, of the 23 protein biomarkers, there are two biomarkers with the most potential, namely the peptide Enolase-1 as a marker that a male is fertile, and BSP-1 as a marker that a male is infertile or sterile [16].
The MALDI-TOF/MS method is good for proteomic studies, including a study on bulls’ fertility or infertility, as has been done by Aslam et al. [16]. However, this method is relatively expensive and quite complicated. One of the ways that is quite straightforward is to use a peptide sequence that can be accessed by applying bioinformatics techniques. Then the peptide sequence obtained will be further analyzed to be used to make a vaccine. The antibodies obtained can then be used to explore the expression of these peptides immunologically [3]. Success in producing antibodies against synthetic peptide antigens is currently very important and needed not only for research purposes but also for the biotech industry‘s purposes. Several essential actions must be taken, including the T-cell epitope prediction of the target protein or peptide, in this study, the BSP-1. For this reason, a study using sequence-based and molecular docking methods was conducted.
Materials and Methods
This study was mainly software-based experimental research; hence, the approval of the relevant ethics committee was not necessarily implied. The main material of this research is BSP-1, with amino acid sequences derived from tracking on protein banks such as ExPASy and UniProtKB (https://www.uniprot.org/), according to Depamede [17]. The retrieved sequences were aligned using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/; [18]. Prediction of the availability of B and T cell epitopes was conducted using the Immune Epitope Database (IEDB) (http://www.iedb.org/) and, when available, the prospective B and T cell epitopes were determined based on the conserved epitopes found [19]. In addition, to find peptides binding to major histocompatibility complex (MHC) class I and class II molecules, two servers, i.e., ProPred-I and ProPred-Server, were adopted [20,21]. In addition, the peptide sequences were also evaluated using NetMHCpan-4.1 (http://www.cbs.dtu.dk/services/NetMHCpan/index_4.1a.php), [22,23]. Finally, the theoretical prediction of the binding of BSP-1 with its ligands was carried out based on the I-TASSER software program [24].
Results
By typing BSP1, the tracking results on the Protein Bank data, namely, on ExPASy and UniProtKB, were found to have 170 entry identifier numbers. Furthermore, when retrieved based on species or organism, it found 11 entries (Table 1). BSP1 with entry identifier A0A0K1YXR5is belonging to Bubalus bubalis (Domestic water buffalo) consists of 38 residues (Fig. 1). We then conducted multiple alignments of the A0A0K1YXR5 hybrid cattle (Bos indicus × Bos taurus) with gene name binder of sperm protein homolog 1 (BSPH1), entry identifier A0A4W2IDX4, and B. indicus (Zebu) A0A6P5D798, as the cattle (Bos) representative, and the result is presented in Figure 2.
Table 1. Binder sperm protein in ruminants and its entry identifier retrieved from UnibProtKB.
| No. | Entry identifier | UniProtKB entry identifier | Protein names | Gene name | Organism | Sequence number |
|---|---|---|---|---|---|---|
| 1 | A0A6P3I827 | A0A6P3I827_BISBI | BSPH1 | BSPH1 | Bison bison bison | 115 |
| 2 | A0A4W2IDX4 | A0A4W2IDX4_BOBOX | BSPH1 | BSPH1 | B. indicus × B. taurus (Hybrid cattle) | 138 |
| 3 | A0A4W2CI70 | A0A4W2CI70_BOBOX | BSPH1 | B. indicus × B. taurus (Hybrid cattle) | 132 | |
| 4 | A0A4W2D2A3 | A0A4W2D2A3_BOBOX | BSPH1 | B. indicus × B. taurus (Hybrid cattle) | 138 | |
| 5 | A0A4W2GYS9 | A0A4W2GYS9_BOBOX | BSPH1 | BSPH1 | B. indicus × B. taurus (Hybrid cattle) | 132 |
| 6 | B3EWK6 | SFP1_BOSIN | SP protein PDC-109 | B. indicus (Zebu) | 32 | |
| 7 | A0A6P5D798 | A0A6P5D798_BOSIN | BSPH1 | BSPH1 | B. indicus (Zebu) | 102 |
| 8 | A0A0M4B6N2 | A0A0M4B6N2_BUBBU | BSP 3 | BSP3 | B. bubalis (Domestic water buffalo) | 140 |
| 9 | A0A0K1YXR5 | A0A0K1YXR5_BUBBU | BSP 1 | BSP1 | B. bubalis (Domestic water buffalo) | 138 |
| 10 | A0A097GTY0 | A0A097GTY0_BUBBU | BSP 1 | B. bubalis (Domestic water buffalo) | 138 | |
| 11 | A0A0M5I4D0 | A0A0M5I4D0_BUBBU | BSP 5 | BSP5 | B. bubalis (Domestic water buffalo) | 183 |
Figure 1. Amino acid sequence of BSP1 belongs to B. bubalis (Domestic water buffalo) retrieved from UniProtKB (https://www.uniprot.org/).

Figure 2. Multiple sequence alignment of Domestic water buffalo BSP1 (A0A0K1YXR5) against cattle binder sperm protein using Clustal Omega. Conserved residues are denoted by star (*).
This study aimed to design a peptide vaccine to produce BSP1 antibodies. Work on finding epitopes associated with vaccine design is increasingly dependent on bioinformatics analytical tools and access to data stores pertinent to pathogens and specific immune reactions [24,25]. This requires a combination of techniques or methods. Based on these considerations, in this study, the IEDB program was also used based on Droppa-Almeida et al. [26]. This is shown in in Figure 3.
Figure 3. The results of the analysis of the BSP-1 Epitope prediction based on the IEDB. Predicted Emini surface accessibility of the proposed epitope, with a minimum propensity score of 0.060 and a maximum score of 5.195. Sequence positions and surface probabilities are directed by the X- and Y- axes, respectively. The areas above the threshold are antigenic, shown in yellow.

From Figure 3, it appears that there are seven variations of the peptide sequence related to the epitope that is possible to be developed as vaccine materials since they are predicted to be antigenic.
In addition to the IEDB program, we also used NetMHCpan-4.1 to evaluate peptide binding affinities between the BSP1 peptides and MHC class I and class II molecules. For this purpose, we searched for murine gene H-2Dq and found one sequence with a strong bind level and three weak bind levels (Table 2). The predicted affinity was classified according to the default NetMHCpan-4.1 program. Strong binding was ≤500 nM, while weak binding was >500 nM, with the rank threshold of strong and weak binding peptides being 0.5% and 2.0%, respectively.
Table 2. NetMHCpan prediction results of BSP1.
| Residue position | MHC | Peptide | Aff. (nM) | Bind level |
|---|---|---|---|---|
| 48 | H-2-Dq | VPDEERVFPF | 453.42 | Strong |
| 39 | H-2-Dq | RPAELPEDSV | 2,595.69 | Weak |
| 74 | H-2-Dq | FPWCSLDANY | 648.57 | Weak |
| 126 | H-2-Dq | SPNYDKDRAW | 4,302.52 | Weak |
We then combined the IEDB data in Figure 3 and NetMHCpan-4.1 (Table 2) and manually determined the vaccine candidate; in this study, i.e., LPEDSVPDEERVFPFTYRNRKHF (sequence of residues number 43–65). This peptide consists of strong and weak binding affinities, with most of them being noticed as antigenic. By using the meta-server approach, COACH, i.e., a combination of COFACTOR, TIM-SITE, and S-SITE programs on I-TASSER, the peptide was predicted in its 3D model along with its ligand-binding site [24]. The result is presented in Figure 4. Based on the PDB hit, the peptide target is close to GP1HB, classified as a DNA-binding protein/DNA. Meanwhile, the first rank for ligand binding site is 3h5bB, related to the crystal structure of wild-type HIV-1 protease, EC 3.4.23.16, known as HIV-1 retropepsin.
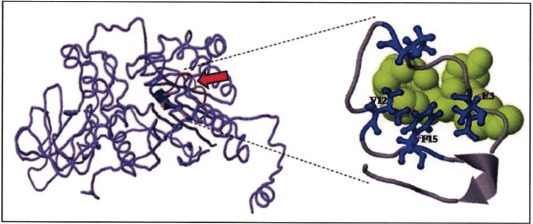
Figure 4. The results of the 3-dimensional theoretical prediction analysis of the peptide binding pattern of BSP1 to its ligand (red arrow), based on the I-TASSER Program. Enlarged exposure image shows the ligand bind site-residues (E3, P7, V12, and F15) of the peptide vaccine LPEDSVPDEERVFPFTYRNRKHF proposed in this study.
Discussion
The provision of antibodies for developing diagnostic tools for testing male livestock fertility is still limited, and in fact, this is urgently needed. There are several causative factors, including the availability of antigens to make these antibodies. In this study, we tried to develop a peptide-based vaccine from BSP1, which is known as a biomarker of male fertility.
Of these 170 entry identifiers related to BSP1 in the Protein Bank data on ExPASy, UniProtKB, 11 of these numbers are related to ruminants (Table 1). In this study, we used the BSP1 sequence of B. bubalis (Domestic water buffalo) as the main target (Fig. 1) and two representations of bovine, i.e., hybrid cattle (B. indicus × B. taurus) and B. indicus (Zebu). As might be expected, the multiple alignment results show that there are several conserved residues among the three (Fig. 2). The expected achievement target in vaccine development in this study is a strong epitope in the conserved part. However, the Emini surface accessibility results combined with NetMHCpanprediction analysis showed that the residue components with potential immunogenic or antigenic properties of buffalo BSP1 were not fully conserved (Fig. 3, Table 2).
It is generally based on B cell immunity in vaccine development, although the T cell epitope has also received attention, particularly for CD8+ T cells or MHC-I [27]. This study found that the dominant epitopes are primarily associated with MHC-I T cells or CD8 epitopes (Table 2), compared to B cells and MHC-II T cells (CD4 epitopes). Although the epitope found is mainly related to MHC-I (CD8 T Cells), the sequence of these residues can be appointed as a candidate for peptide vaccine. Several studies have shown that CD8 T cells are essential in inducing immune responses to certain diseases [28–30]. Therefore, we combined the prospective epitopes to make a candidate peptide vaccine consisting of 23 residues, i.e., LPEDSVPDEERVFPFTYRNRKHF.
The results of the three-dimensional structure modeling using I-TASSER showed a similar structure to the HIV-1 retropepsin enzyme (EC 3.4.24.45). When ligand analysis was performed, binding sites were observed at the residues E, P, V, and F, in sequences 3, 7, 12, and 15, respectively, of the peptide vaccine proposed in this study (Fig. 4a), or those of residues numbers 45, 49, 54, and 57 in Figures 1 and 2. Furthermore, When paired with the Emini surface accessibility data in Figure 3, these epitope residues fall into the region above the threshold, which is categorized as immunogenic. The data from I-TASSER was then used to perform molecular docking using MTiOpenScreen [31], and the results obtained (Fig. 4b) strengthened the analysis results of the ligand bonds of the I-TASSER.
Suppose we observed the position of the residues from the epitope of the proposed peptide vaccine. In that case, only F15 (phenylalanine No. 15) or F57 (Fig. 2) is conserved with amino acid sequences from hybrid cattle (B. indicus × B. taurus) and B. indicus (Zebu). This can be understood because B. bubalis is different from B. indicus and B. taurus. Whether in the future, the antibodies produced from the vaccine proposed in this study will produce antibodies specific to the sperm binder only for buffaloes or cross-react with the BSP protein from bulls still needs to be proven in vivo.
We sincerely hope that in the near future, we will be able to produce polyclonal antibodies against BSP-1. These antibodies are very important for developing sperm fertility kits or diagnostics for ruminants. The success of making a device to detect whether a male is fertile or infertile will play an important role in the livestock industry [32]. The review of Butler et al. [32] can also consider that male fertility traits can be inherited; hence they can be improved through male selection. Thus, the availability of biomarkers or kits for male fertility indeed plays a central role and is urgently needed.
One of the reasons for the lack of assessment of male quality based on the level of sperm fertility or infertility is the complexity of the male reproductive systems, starting from sperm production and ending with the success of the fertilization process by “selected” sperm in the female animals [33]. On the other hand, Foxcroft et al. [34] acknowledged that the reproductive capacity of males, including their fertility rate, greatly affects the reproductive efficiency of females, namely the impact of pregnancy rates and the number of non-productive days of females. In addition, it is also recognized that detecting bull fertility is extremely difficult, and there is no single test currently available to envisage or diagnose bull sub-fertility or fertility [9,35]. Therefore, the availability of kits to accurately measure bull fertility is very beneficial in managing and strengthening the impact of using superior fertile bulls on genetic advancement and the economic efficiency of artificial insemination programs [32,36]. The results of our study provide opportunities for the development of male fertility detection tools in the future. The results of bioinformatics observations led to the determination of the peptide sequence for the development of the BSP-1 vaccine, which was a peptide with the sequence “LPEDSVPDEERVFPFTYRNRKHF,” a modified part of the BSP-1 sequence in this study.
Conclusion
The synthetic vaccine from the BSP-1 has been successfully developed with the amino acid sequence LPEDSVPDEERVFPFTYRNRKHF, was buffer-soluble, and the three-dimensional theoretical prediction analysis of the peptide binding pattern of BSP-1 to its ligand, as well as molecular docking, has also been revealed.
Acknowledgment
This study was funded by the University of Mataram (DIPA BLU-PNBP), according to Research Contracts No. 2838I/UN18.L1/PP/2019 and No. 2874/UN18.L1/PP/2021.
List of abbreviations
BSP-1, Binders of Sperm-1; BSPH1, Binder of sperm protein homolog 1; IEDB, Immune Epitope Database; MALDI-TOF/MS, matrix-assisted laser desorption ionization-time of flight/mass spectrometry; MHC, major histocompatibility complex.
Conflict of interests
The authors declare that they have no conflict of interest.
Authors’ contribution
SND designed the study, interpreted the data, and drafted the manuscript. WW was involved in designing the study, collecting data and also contributed to manuscript preparation. MS, AR, and MA took part in preparing and critically checking this manuscript.
References
- [1].Vernekar VJ, Bandivdekar AH, Raghavan VP, Kamada M, Koide SS. Studies with synthetic peptides of 80 kDa human sperm antigen (80 kDa HSA) AJRI. 2004;51:106–11. doi: 10.1111/j.1600-0897.2004.00109.x. https://doi.org/10.1111/j.1600-0897.2004.00109.x. [DOI] [PubMed] [Google Scholar]
- [2].Bandivdekar AH. Development of anti fertility vaccine using sperm specific proteins. Indian J Med Res. 2014;140:73–7. [PMC free article] [PubMed] [Google Scholar]
- [3].Lee BS, Huang JS, Jayathilaka LP, Lee J, Gupta S. Antibody production with synthetic peptides. Methods Mol Biol. 2016;1474:25–47. doi: 10.1007/978-1-4939-6352-2_2. https://doi.org/10.1007/978-1-4939-6352-2_2. [DOI] [PubMed] [Google Scholar]
- [4].Sadat SM, Aghadadeghi MP, Yousefi M, Khodaei A, Larijani MS, Bahramali G. Bioinformatics analysis of SARS-CoV-2 to approach an effective vaccine candidate against COVID-19. Mol Biotechnol. 2021;63:389–409. doi: 10.1007/s12033-021-00303-0. https://doi.org/10.1007/s12033-021-00303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S. An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inf. 2015;53:405–14. doi: 10.1016/j.jbi.2014.11.003. https://doi.org/10.1016/j.jbi.2014.11.003. [DOI] [PubMed] [Google Scholar]
- [6].D’Amours O, Bordeleau LJ, Frenette G, Blondin P, Leclerc P, Sullivan R. Binder of sperm 1 and epididymal sperm binding protein 1 are associated with different bull sperm subpopulations. Reproduction. 2012;143:759–71. doi: 10.1530/REP-11-0392. https://doi.org/10.1530/REP-11-0392. [DOI] [PubMed] [Google Scholar]
- [7].Rodríguez-Villamil P, Hoyos-Marulanda V, Martins JAM, Oliveira AN, Aguiar LH, Moreno FB, et al. Purification of binder of sperm protein 1 (BSP1) and its effects on bovine in vitro embryo development after fertilization with ejaculated and epididymal sperm. Theriogenology. 2016;85:540–54. doi: 10.1016/j.theriogenology.2015.09.044. https://doi.org/10.1016/j.theriogenology.2015.09.044. [DOI] [PubMed] [Google Scholar]
- [8].Rodríguez-Villamil P, Mentz D, Ongaratto FL, Aguiar LH, Rodrigues JL, Bertolini M, et al. Assessment of binder of sperm protein 1 (BSP1) and heparin effects on in vitro capacitation and fertilization of bovine ejaculated and epididymal sperm. Zygote. 2020;28(6):489–94. doi: 10.1017/S0967199420000374. https://doi.org/10.1017/S0967199420000374. [DOI] [PubMed] [Google Scholar]
- [9].Özbek M, Hitit M, Kaya A, Jousan FD, Memili E. Sperm functional genome associated with bull fertility. Front Vet Sci. 2021;8:610888. doi: 10.3389/fvets.2021.610888. https://doi.org/10.3389/fvets.2021.610888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Westfalewicz B, Dietrich MA, Mostek A, Partyka A, Bielas W, Nizanski W, et al. Analysis of bull (Bos taurus) seminal vesicle fluid proteome in relation to seminal plasma proteome. J Dairy Sci. 2017;100(3):2282–98. doi: 10.3168/jds.2016-11866. https://doi.org/10.3168/jds.2016-11866. [DOI] [PubMed] [Google Scholar]
- [11].Feugang JM, Rodriguez-Osorio N, Kaya A, Wang H, Page G, Ostermeier GC, et al. Transcriptome analysis of bull spermatozoa: implications for male fertility. Reprod Biomed Online. 2010;21(3):312–24. doi: 10.1016/j.rbmo.2010.06.022. https://doi.org/10.1016/j.rbmo.2010.06.022. [DOI] [PubMed] [Google Scholar]
- [12].De Oliveira RV, Dogan S, Belser LE, Kaya A, Topper E, Moura A, et al. Molecular morphology and function of bull spermatozoa linked to histones and associated with fertility. Reproduction. 2013;146:263–72. doi: 10.1530/REP-12-0399. https://doi.org/10.1530/REP-12-0399. [DOI] [PubMed] [Google Scholar]
- [13].Bromfield JJ. A role for seminal plasma in modulating pregnancy outcomes in domestic species. Reproduction. 2016;152(6):R223–32. doi: 10.1530/REP-16-0313. https://doi.org/10.1530/REP-16-0313. [DOI] [PubMed] [Google Scholar]
- [14].Kumar P, Kumar D, Singh I, Yadav PS. Seminal plasma proteome: promising biomarkers for bull fertility. Agric Res. 2012;1:78–86. https://doi.org/10.1007/s40003-011-0006-2. [Google Scholar]
- [15].Viana AGA, Martins AMA, Pontes AH, Fontes W, Castro MS, Ricart CAO, et al. Proteomic landscape of seminal plasma associated with dairy bull fertility. Sci Rep. 2018;8:16323. doi: 10.1038/s41598-018-34152-w. https://doi.org/10.1038/s41598-018-34152-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Aslam MKM, Sharma VK, Pandey S, Kumaresan A, Srinivasan A, Datta TK, et al. Identification of biomarker candidates for fertility in spermatozoa of crossbred bulls through comparative proteomics. Theriogenology. 2018;119:43–51. doi: 10.1016/j.theriogenology.2018.06.021. https://doi.org/10.1016/j.theriogenology.2018.06.021. [DOI] [PubMed] [Google Scholar]
- [17].Depamede SN. Proteomic analysis of a 14.2 kDa protein isolated from bali cattle (Bos sondaicus/javanicus) saliva using 1-D SDS-PAGE gel and MALDITOF-TOF mass spectrometer. Italian J Anim Sci. 2013;12:371–4. https://doi.org/10.4081/ijas.2013.e59. [Google Scholar]
- [18].Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–41. doi: 10.1093/nar/gkz268. https://doi.org/10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky1006. https://doi.org/10.1093/nar/gky1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Singh H, Raghava GPS. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17(12):1236–37. doi: 10.1093/bioinformatics/17.12.1236. https://doi.org/10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- [21].Singh H, Raghava GPS. ProPred1: prediction of promiscuous MHC class-I binding sites. Bioinformatics. 2003;19(8):1009–14. doi: 10.1093/bioinformatics/btg108. https://doi.org/10.1093/bioinformatics/btg108. [DOI] [PubMed] [Google Scholar]
- [22].Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved peptide MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–8. doi: 10.4049/jimmunol.1700893. https://doi.org/10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020 doi: 10.1093/nar/gkaa379. https://doi.org/10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2014 doi: 10.1093/nar/gkv342. https://doi.org/10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fleri W, Paul S, Dhanda SK, Mahajan S, Xu X, Peters B, et al. The immune epitope database and analysis resource in epitope discovery and synthetic vaccine design. Front Immunol. 2017;8:278. doi: 10.3389/fimmu.2017.00278. https://doi.org/10.3389/fimmu.2017.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Droppa-Almeida D, Franceschi E, Padilha FF. Immune-informatic analysis and design of peptide vaccine from multi-epitopes against Corynebacterium pseudotuberculosis. Bioinf Biol Insights. 2018;12:1–9. doi: 10.1177/1177932218755337. https://doi.org/10.1177/1177932218755337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hart J, MacHugh ND, Sheldrake T, Nielsen M, Morrison WI. Identification of immediate early gene products of bovine herpes virus 1 (BHV-1) as dominant antigens recognized by CD8 T cells in immune cattle. J Gen Virol. 2017;98(7):1843–54. doi: 10.1099/jgv.0.000823. https://doi.org/10.1099/jgv.0.000823. [DOI] [PubMed] [Google Scholar]
- [28].Rodrigues MM, Boscardin SB, Vasconcelos JR, Hiyane MI, Salay G, Irene Soares S. Importance of CD8 T cell-mediated immune response during intracellular parasitic infections and its implications for the development of effective vaccines. An Acad Bras Cienc. 2003;75(4):443–68. doi: 10.1590/s0001-37652003000400005. https://doi.org/10.1590/S0001-37652003000400005. [DOI] [PubMed] [Google Scholar]
- [29].Oany AR, Sharmin T, Chowdhury AS, Jyoti TP, Hasan A. Highly conserved regions in Ebola virus RNA dependent RNA polymerase may be act as a universal novel peptide vaccine target: a computational approach. In Silico Pharmacol. 2015;3:7. doi: 10.1186/s40203-015-0011-4. https://doi.org/10.1186/s40203-015-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cosma G, Eisenlohr L. CD8 T-cell responses in vaccination: reconsidering targets and function in the context of chronic antigen stimulation. F1000Res. 2018;7:508. doi: 10.12688/f1000research.14115.1. https://doi.org/10.12688/f1000research.14115.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Labbé CM, Rey J, Lagorce D, Vavrusa M, Becot J, Sperandio O, et al. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res. 2015;43:W448–54. doi: 10.1093/nar/gkv306. https://doi.org/10.1093/nar/gkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Butler ML, Bormann JM, Weaber RL, Grieger DM, Rolf MM. Selection for bull fertility: a review. Transl Anim Sci. 2019;4(1):423–41. doi: 10.1093/tas/txz174. https://doi.org/10.1093/tas/txz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Diether N, Dyck MK. Male fertility evaluation using biomarkers in livestock. JSM Biomarkers. 2017;3(1):1011. [Google Scholar]
- [34].Foxcroft GR, Dyck MK, Ruiz-Sanchez A, Novak S, Dixon WT. Identifying useable semen. Theriogenology. 2008;70:1324–36. doi: 10.1016/j.theriogenology.2008.07.015. https://doi.org/10.1016/j.theriogenology.2008.07.015. [DOI] [PubMed] [Google Scholar]
- [35].Castilla JA, Zamora S, Gonzalvo MC, Castillo JDLD, Roldan-Nofuentes JA, Clavero A, et al. Sperm chromatin structure assay and classical semen parameters: systematic review. Reprod Biomed Online. 2010;20:114–24. doi: 10.1016/j.rbmo.2009.10.024. https://doi.org/10.1016/j.rbmo.2009.10.024. [DOI] [PubMed] [Google Scholar]
- [36].Nagata MPB, Egashira J, Katafuchi N, Endo K, Ogata K, Yamanaka K, et al. Bovine sperm selection procedure prior to cryopreservation for improvement of post thawed semen quality and fertility. J Anim Sci Biotechnol. 2019;10:91. doi: 10.1186/s40104-019-0395-9. https://doi.org/10.1186/s40104-019-0395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]


