Abstract
For better understanding the genetic diversity and phylogeny of the cultivated Salvia miltiorrhiza populations, four intergenic spacer sequences, ETS, psbA-trnH, trnL-trnF, and ycf1-rps15 of the 40 populations collected from China were Polymerase Chain Reaction (PCR) amplified, analyzed both individually and in combination. Haplotype diversity analysis showed that the cultivated S. miltiorrhiza populations had a very rich genetic diversity and an excellent capacity to resist environmental pressure. The best-fit nucleotide substitution models for ETS, psbA-trnH, trnL-trnF, ycf1-rps15, and their combined sequences were HKY+I, T92, T92, T92+G, and T92+G, respectively; the nucleotide conversion frequency in the combined sequences was lower than the transversion, and the relatively high nucleotide substitution frequencies suggests its high genetic variability. Neutral tests showed that the spacer sequences of the populations conform with the neutral evolution model, and there has been no current expansion events occurred. Phylogeny analyses based on both the individual and the combined sequences showed that the 40 populations were clustered in two clades with a very similar topological structure. The discrimination rate of the combined sequence marker is significantly increased to 52.5% (21 populations) over the highest 35% (13 populations) by the single marker of ETS, though still inadequate but a big step forward. Further exploration of more DNA markers is needed. This study for the first time revealed the rich genetic diversity and phylogeny of the currently cultivated S. miltiorrhiza populations in China and provides novel alternative molecular markers for the genetic identification and resources evaluation of the cultivated S. miltiorrhiza populations.
Introduction
Salvia miltiorrhiza Bunge is a perennial medicinal plant of Salvia in the Labiatae family [1]. Its rhizomes have been traditionally and widely used as medicine in China, Japan, the United States and Europe to treat cardiovascular disorders, including atherosclerosis, high blood pressure, hyperlipidemia and stroke [2]. In the last decade, it has been also proved that S. miltiorrhiza has many other pharmacological effects, such as antioxidant [3], neuroprotective [4], anti-fibrosis [5], anti-inflammatory and antibacterial [6], and anti-tumor [7]; and its extensive application has been contributing greatly to patients care and human health. Although native to China, S. miltiorrhiza is also widely cultivated in other countries such as South Korea, Vietnam and Australia [8]. The pharmaceutically effective ingredients of S. miltiorrhiza mainly include fat-soluble tanshinones and water-soluble phenolic acids [9].
Genetic diversity, the genetic variability present within species that changes with time and space, is the product of recombination of genetic material by sexual reproduction, and by mutation of genes, genetic drift, and gene flow. It gives different physical attributes to the individual and capacity to adapt to stress, diseases and unfavorable environmental conditions. Low genetic diversity is an important factor affecting species survival, leading to endangerment or even to distinction. The genetic diversity at various levels provides the basis for the collection, preservation, utilization and evaluation. To ensure and maintain good levels of genetic diversity in crop populations is crucial. The genetic diversity at various levels provides the basis for the collection, preservation, utilization and evaluation of S. miltiorrhiza.
The external properties of S. miltiorrhiza cultivated in different producing areas are very similar and difficult to identify, and the yields and medicinal ingredient contents of different germplasm are also different, which affects the stability of the quality of S. miltiorrhiza medicinal materials and brings multiple difficulties to the standardized production of S. miltiorrhiza [10]. At present, traditional methods based on sources, morphological traits, physical and chemical and biological properties have been mainly used to identify the provenance of S. miltiorrhiza. They have played important roles in the identification and quality evaluation of Chinese medicinal materials, but also have their own shortcomings. Trait identification is difficult to identify materials with similar morphological traits, the microscopic identification requires the relevant professional knowledge, and the physical and chemical properties are easily affected by many environmental factors. In view of the limitations of traditional identification, new methods as supplements have been emerging to ensure more accurate and reliable identification. In recent years, molecular marker technology that reveals polymorphism at the DNA level has become a powerful tool for studying plant genetic diversity [11]. Both the chloroplast and nuclear gene markers are excellent for application for these purposes.
The nuclear ribosomal DNAs (nrDNAs) in higher plants are highly tandem repeating sequences consisted of the external transcribed spacer (ETS), the non-transcribed spacer (NTS), 18SrDNA, internal transcription spacer 1 (ITS1), 5.8SrDNA, internal transcription spacer 2 (ITS2), and 28SrDNA. The non-coding regions of nrDNA are hypervariable, due to the much less selective pressure they subjected. Sequence differences manifested in these regions have potentials for application in systematics, hybridization and population evolution researches among closely related groups [12]. Seven markers had been selected to barcode a wide range of medicinal plant species and their relatives, and found that ITS2 reached the highest discriminating rate of 92.7% at the species level among the 7 fragments [13]. The relatively less repetitive ETS evolves rapidly and has a high degree of polymorphism and variability, and is extensively used in the study of genetic variation, classification and phylogeny. For example, the phylogenetic study on the origin and spread of millet based on ETS detected both intra- and inter-species polymorphisms [14]; and analysis of the ETS of sycamore showed that all the Scandinavian populations composed of well-defined subgroups of the species’ genes and had a spontaneous origin closest with other spontaneous populations from neighboring areas of the Nordic continent [15]. The genetic diversity of 39 Chinese cultivated and wild pepper populations from 8 provinces has been assessed by two nrDNA non-coding markers (ETS and ITS2) and found that despite its long-term cultivation, the genetic variation was relatively high existing mainly within each province rather than between provinces [16].
The chloroplasts of plants have higher evolving rates than mitochondria; its uniparental inheritance enables the researchers to target DNA markers in the genome [17]. Initially, functional genes for rubisco large subunit (rbcL), maturase K (matK), NAD(P)H dehydrogenase F (ndhF), the β-subunit of ATP synthase (atpB), the RNA polymerase B and C subunit (rpoB and rpoC) genes, and intergenic spacers between tRNALeu and tRNAPhe genes (trnL-trnF), between D1 protein of PSII and tRNAHis genes (psbA-trnH), and between CP47 protein and small subunit I genes of PSII (psbK-psbI) in the chloroplast genome were used individually for plant species identification. Later, it was proposed to use different combinations of these fragments. For example, the relatively conserved psbA [18] has been studied as markers in lake and marine ecosystems [19]; the second longest hypothetical chloroplast open reading frame1(ycf1) in the chloroplast genome [20] which has been proved to be essential for plant survival [21] has only recently begun to be explored in phylogenetic studies; the chloroplast rps15 gene encoding a ribosomal protein S15 (rps15), mediating the interaction between the subunits and interacting with mRNA and tRNA [22] plays certain role in the production of ribosomes [23]. It has been also used for phylogenetic studies [13].
Current researches on plant genetic geography have been under transition from individual sequences in the past to the joint analysis of the sequences from the chloroplast genome or nuclear ribosomal genes. For example, the trnH-psbA spacer and partial coding region of rbcL gene has been combined as two global terrestrial plant molecular genetic markers and provided the necessary versatility and species differentiation [24]. A higher species resolution has been obtained with the combination of matK + atpF-atpH + psbK-psbI [25]; the monophyly and placement of Lepechina in the Lamiaceae plants have been tested based on separate and combined datasets of cpDNA (ycf1, ycf1-rps15, and trnL-trnF) and the combined nrDNA (ITS and ETS) datasets [26]. The phylogeny and biogeographic history of Lamiaceae plants based on a combined sequence of 1144bp containing 519 mutation sites and 339 parsimony information sites from five plastid sequences (rbcL, rps16, rpl32-trnH, psbA-trnH and trnL-trnF) and two nuclear sequences (ITS and ETS) has been explored to interpret the disjunct distribution of Northern Hemisphere herbaceous plants [27]. The phylogeny of East Asian sage of the most comprehensive geographic location, taxonomy, and genetic sampling data has been reconstructed by two separate matrixes, one combined the two nrDNA (ITS, ETS), and the other combined the four cpDNA (psbA-trnH, ycf1-rps15, trnL-trnF, and rbcL) [28].
The chloroplast genome of S. miltiorrhiza has been sequenced and its detailed structure provides an excellent basis for systematic analyses [29]. Its more variable non-coding regions could be readily used for intra- and inter-specific phylogenetic studies.
As one of China’s traditional bulk medicinal materials, S. miltiorrhiza has been playing increasingly important roles in clinical and health-care utilization. However, the random introduction of unidentified source varieties in production and nonstandard management have resulted in a chaotic variety sources and unstable quality; and studies concerning the genetic diversity and molecular identification of the germplasm resources have been also inadequate. So far as we known, no reports on the genetic diversity of the cultivated S. miltiorrhiza populations have been seen. In this study, the nuclear ETS and three chloroplast intergenic spacers of the 40 cultivated S. miltiorrhiza populations in China were for the first time analyzed in order to identify the SNP fingerprints, understand the genetic diversity and phylogeny both individually and in combination. A basis for the protection, introduction, domestication, novel variety selection, and breeding of the cultivated S. miltiorrhiza germplasm resources, was provided.
Materials and methods
Sampling
Seeds of 40 cultivated S. miltiorrhiza populations representing more than 30 regions of China were collected from three major seed industries (Table 1). Uniform seeds preliminarily selected were used for sowing in Southwest University Agricultural Station, Chongqing; and leaves of morphologically representative single plants of each population were used for extraction of genomic DNAs.
Table 1. Cultivated S. miltiorrhiza populations used in this study.
| No. | Population/Voucher | Production Region | Source | No. | Population/Voucher | Production Region | Source |
|---|---|---|---|---|---|---|---|
| 01 | V-HBAG-V-2 | Anguo, Hebei Hebei | Fenghong Seed Industry | 21 | B-SCZJ-V-2 | Zhongjiang, Sichuan | Hengda Seed Industry |
| 02 | V-CQ-V-2 | Chongqing | Hengda Seed Industry | 22 | B-SD-V-1 | Shandong | Tongda Seed Industry |
| 03 | V-JXJA-V-2 | Ji-an, Jiangxi Jiangxi | Hengda Seed Industry | 23 | R-HBAG-V-2 | Anguo, Hebei | Fenghong Seed industry |
| 04 | V-JLCC-V-2 | Changchun, Jilin | Hengda Seed Industry | 24 | R-HNFC-V-2 | Fangcheng, Henan | Hengda Seed Industry |
| 05 | V-JSSY-V-2 | Shuyang, Jiangsu | Hengda Seed Industry | 25 | R-SDJX-V-2 | Juxian, Shandong | Hengda Seed Industry |
| 06 | V-GZ-V-1 | Guizhou | Tongda Seed Industry | 26 | R-GX-V-2 | Guangxi | Hengda Seed Industry |
| 07 | V-GD-V-1 | Guangdong | Tongda Seed Industry | 27 | R-NM-V-2 | Nemeng | Hengda Seed Industry |
| 08 | V-GD-V-2 | Guangdong | Hengda Seed Industry | 28 | R-HNCS-V-2 | Changsha, Hunan Hunan | Hengda Seed Industry |
| 09 | V-GSLX-V-2 | Longxi, Gansu | Fenghong Seed industry | 29 | R-GSJQ-V-2 | Jiuquan, Gansu | Hengda Seed Industry |
| 10 | V-BJ-V-2 | Beijing | Fenghong Seed industry | 30 | W-SCHY-W-1 | Hongyuan, Sichuan | Self-collected |
| 11 | V-YNLJ-V-2 | Lijiang, Yunnan | Hengda Seed Industry | 31 | W-SXXA-bV-2 | Xi’an, Shaanxi | Hengda Seed Industry |
| 12 | V-GZZY-V-2 | Zunyi, Guizhou | Hengda Seed Industry | 32 | W-LNSY-V-2 | Shenyang, Liaoning | Hengda Seed Industry |
| 13 | V-SC-V-1 | Sichuan | Tongda Seed Industry | 33 | W-FJLY-V-2 | Luoyuan, Fujian | Hengda Seed Industry |
| 14 | V-SD-V-1 | Shandong | Tongda Seed Industry | 34 | W-GZ-V-1 | Guizhou | Tongda Seed Industry |
| 15 | V-JS-V-1 | Jiangsu | Tongda Seed Industry | 35 | W-SD-V-1 | Shandong | Tongda Seed Industry |
| 16 | V-HNYZ-bV-2 | Yongzhou, Hunan | Hengda Seed Industry | 36 | W-JS-V-1 | Jiangsu | Tongda Seed Industry |
| 17 | B-SC-V-1 | Sichuan | Tongda Seed Industry | 37 | W-SC-V-1 | Sichuan | Tongda Seed Industry |
| 18 | B-AHQJ-V-2 | Quanjiao, Anhui | Hengda Seed Industry | 38 | W-GD-V-1 | Guangdong | Tongda Seed Industry |
| 19 | B-GD-V-1 | Guangdong | Tongda Seed Industry | 39 | W-HBJM-V-2 | Jingmen Hubei | Hengda Seed Industry |
| 20 | B-JS-V-1 | Jiangsu | Tongda Seed Industry | 40 | W-YNLJ-V-2 | Lijiang, Yunnan | Hengda Seed Industry |
Methods
Genomic DNA extraction
The total genomic DNA of S. miltiorrhiza was extracted by the CTAB method [30], the purity was confirmed by 1% agarose gel electrophoresis, and was stored at -20°C.
Primers and PCR amplification
Well-documented 4 primer pairs respectively for the 4 intergenic spacers (Table 2) were adopted. The spacers were PCR amplified in a 25μl reaction system consisted of 2×SanTaq PCR Mix (Sangon, Shanghai, China) 11μl, primers (10μmol/L) each 1μl, DNA template 1μl, ddH2O 11μl, with the program of pre-denaturation at 94°C for 5min, denaturation at 94°C for 30s, annealing for 30s, extension at 72°C for 30s, 35 cycles, and a final extension at 72°C for 10min. Products were electrophoresed by agarose gel, purified and then bidirectionally sequenced by dideoxy chain termination (Sangon, Chengdu, China).
Table 2. Primers used in this study.
| Locus | Primer code | Sequences(5’→3’) | Ta (°C) | Length (nt) | Reference |
|---|---|---|---|---|---|
| ETS | ETS-bdf1 (F) | ATAGAGCGCGTGAGTGGTG | 55 | 19 | [31] |
| 18S-IGS (R) | GACAAGCATATGACTGGATCAA | 22 | [32] | ||
| psbA-trnH | psbAF | GTTATGCATGAACGTAATGCTC | 55 | 22 | [33] |
| trnHR | CGCGCATGGTGGATTCACAAATC | 23 | [33] | ||
| trnL-trnF | trn-c (F) | CGAAATCGGTAGACGCTACG | 57 | 20 | [34] |
| trn-f (R) | ATTTGAACTGGTGACACGAG | 20 | [34] | ||
| ycf1–rps15 | ycf15711f | CTTGTATGRATCGTTATTGKTTTG | 53 | 24 | [26] |
| ycf1rps15r | CAATTYCAAATGTGAAGTAAGTCTCC | 26 | [26] |
Notes: F(f) and R(r): Forward and reverse primers respectively; Ta: Annealing temperature; nt: Nucleotides.
Data processing
The manually checked and confirmed sequence data were subject to BLAST analysis and a corresponding reference sequence with the highest identity percentage for each locus was determined and used for manual finishing and multiple alignment with BioEdit 7.09 and Vector NTI Advance 11.5.3, respectively. The best nucleotide substitution models were identified via the Find Best DNA/Protein Models in MEGA 7.0 [35], and the displayed substitution frequencies were used to make histograms with Excel. MEGA 7.0 was used for genetic evolution analysis and phylogenetic tree construction; unrooted phylogenetic trees with a cut-off value of 50% for consensus were constructed with MEGA 7.0 based on Neighbor-Joining (NJ) with a bootstrap value of 1000. DnaSP v5 was used for analyses of polymorphic sites, genetic diversity index, and neutral detection [36].
Results
General features of the intergenic spacers
The four intergenic spacer sequences of ETS, psbA-trnH, trnL-trnF and ycf1-rps15 of the 40 cultivated S. miltiorrhiza populations were assorted and submitted to GenBank, and the corresponding accession numbers were obtained (S1 Table). Comparison analyses with the highest identity reference sequences, MG824361.1, KJ025055.1, KC414292.1 and MG824106.1 respectively for ETS, psbA-trnH, trnL-trnF and ycf1-rps15, showed that the ETS sequences of the S. miltiorrhiza populations was 421–433 bp in length, with a GC content of 56.8~62.8%, a variable rate as high as 54.8%, and a total of 241 SNP variable sites consisting of 205 parsimony informative sites, 36 singleton variable sites; the psbA-trnH was 335~348bp with a GC content of 22.4~26.2%, a variable rate of 24.7%, and a total of 90 SNP variable sites consisting of 71 parsimony informative site and 19 singleton variable sites; the trnL-trnF was 298~306bp with a GC content of 35.0~36.9%, a variable rate of 7.7%, and a total of 25 SNP variable sites consisting of 24 parsimony informative sites and 1 singleton information site; the ycf1-rps15 was 421-433bp with a GC content of 25.9–30.5%, a variable rate of 23.1%, and a total of 105 SNP variable sites consisting of 88 parsimony informative sites and 17 singleton variable sites. The combined sequence length (in the order of ETS, psbA-trnH, trnL-trnF, and ycf1-rps15), was 1488-1517bp, with a GC content of 35.9–38.8%, a variable rate of 29.4%, and a total of 466 SNP variable sites consisting of 392 parsimony informative sites and 74 singlet variable sites (Table 3).
Table 3. General features of the intergenic spacers of the cultivated S. miltiorrhiza populations.
| Intergenic spacer | ETS | psbA-trnH | trnL-trnF | ycf1–rps15 | Combined seq. |
|---|---|---|---|---|---|
| Length range (bp) | 421~433 | 335~348 | 298~306 | 431~434 | 1488–1517 |
| Matrix length | 440 | 364 | 325 | 455 | 1584 |
| Average GC (%) | 56.8~62.8 | 22.4~26.2 | 35.0~36.9 | 25.9~30.5 | 35.9–38.8 |
| Sites with alignment gaps or missing data | 36 | 46 | 43 | 55 | 179 |
| Variable sites (%) | 241 (54.8) | 90 (24.7) | 25 (7.7) | 105 (23.1) | 466 (29.4) |
| Singleton variable sites | 36 | 19 | 1 | 17 | 74 |
| Parsimony informative sites (%) | 205 (46.6) | 71 (19.5) | 24 (7.4) | 88 (19.3) | 392 (24.7) |
| Best fit model | HKY+I | T92 | T92 | T92+G | T92+G |
| Relative identification (%) | 14 (35.0) | 8 (20.0) | 4 (10.0) | 11 (27.5) | 21 (52.5) |
Based on the matrix length of ETS, psbA-trnH, trnL-trnF, ycf1-rps15, or the combined sequence (Table 3), the parsimony informative sites accounted for 46.6, 19.5, 7.4, 19.3, and 24.7% respectively, indicating that the ETS of the cultivated S. miltiorrhiza populations had the highest in parsimony information site rate, the combined sequence, second; the chloroplast genome psbA-trnH and ycf1-rps15, lower and comparable, and trnL-trnF was the least.
In the studying of the processes of gene evolution, researchers have proposed different nucleotide substitution models to demonstrate the DNA substitution process. By MAGA 7.0 using the maximum likelihood method, the best nucleotide substitution models for the four DNA molecular markers under this study, ETS, psbA-trnH, trnL-trnF, ycf1-rps15, and the combined sequence were revealed to be HKY+I, T92, T92 and T92+G, and T92+G, respectively (Table 3).
The specific SNP fingerprints based on the ETS can discriminate 14 S. miltiorrhiza populations (V-JXJA-V-2, V-GD-V-1, V-GSLX-V-2, V-SC-V-1, V -SD-V-1, V-JS-V-1, B-GD-V-1, B-SD-V-1, W-SCHY-W-1, W-GZ-V-1, W-SD-V-1, W-GD-V-1, W-HBJM-V-2 and W-YNLJ-V-2) with a relative identification rate of 35%, the specific SNP fingerprints of the psbA-trnH can distinguish 8 S. miltiorrhiza populations (V- GD-V-2, V-BJ-V-2, V-HNYZ-bV-2, B-JS-V-1, B-SD-V-1, W-JS-V-1, W-SC- V-1 and W-GD-V-1) with a relative identification of 20%, the specific SNP fingerprints of trnL-trnF can distinguish 4 S. miltiorrhiza populations (V-HBAG-V-2, V-SC-V-1, R-GSJQ -V-2 and W-SC-V-1) with a relative identification of 10%, the specific SNP fingerprints of ycf1-rps15 can distinguish 11 S. miltiorrhiza populations (V-SC-V-1, V-SD-V-1, V- JS-V-1, B-GD-V-1, B-JS-V-1, B-SD-V-1, W-GZ-V-1, W-JS-V-1, W-SC-V-1, W-GD-V-1 and W-YNLJ-V-2) with a relative identification of 27.5%. Specific SNP fingerprints based on the composite DNA marker could discriminate 21 S. miltiorrhiza populations (V-HBAG-V-2, V-JXJA-V-2, V-GD-V-1, V-GSLX-V-2, V-SC-V-1, V-SD-V-1, V-JS-V-1, V- GD-V-2, V-BJ-V-2, V-HNYZ-bV-2, B-GD-V-1, B-SD-V-1, B-JS-V-1, R-GSJQ-V-2, W-SCHY-W-1, W-SD-V-1, W-GD-V-1, W-HBJM-V-2, W-YNLJ-V-2, W-JS-V-1, W-SC-V-1) with a discrimination rate of 52.5%.
The results demonstrated that highest relative identification rate was by the combined sequence marker. For single markers, the relative identification rate by ETS sequence was the highest (Table 3).
Nucleotide variation frequency analysis of the intergenic spacers
Statistics on the nucleotide substitution frequencies (Fig 1) showed that for the nuclear ribosomal ETS sequences, the conversion frequency (40.1%) is lower than the transversion (59.9%), while for the other three intergenic spacers of the chloroplast genome, conversion frequencies were higher than those of transversion. The conversion and transversion frequencies of the combined sequences were 43.4% and 56.8% respectively, in good accordance with the those of the three non-coding regions of the chloroplast genome.
Fig 1. Nucleotide substitution frequencies of the individual and combined intergenic spacers of the cultivated S. miltiorrhiza populations.
Genetic diversity analysis of the intergenic spacers
Dna SP 5.10 was used to analyze the ploidy polymorphism and neutrality detection, and the haplotype, haplotype diversity (Hd), nucleotide diversity (π), variance of haplotype diversity (Vh) and standard deviation of haplotype diversity (Sh) of the four intergenic regions and the combined sequence. The results of neutral analysis showed that the Fu and Li’s D* and F* test statistics for ETS were 0.22815 and -0.37550 respectively, and the Tajima’s D was -1.27644, not significant at the level of P> 0.10, indicating its conformity with the neutral evolution model; the Fu and Li’s D* and F* test statistics for the psbA-trnH were 0.19802 and 0.53273 respectively, and the Tajima’s D was 0.90040, not significant at the level of P> 0.10, conforming also with the neutral evolution model; the Fu and Li’s D* test statistic for the trnL-trnF was 1.44651, with significant difference at the level of P <0.05, Fu and Li’s F* test statistic was 1.89841, showing a significant difference at the level of P <0.02, and the Tajima’s D was 1.90978, not significant at the level of 0.10> P> 0.05, suggesting its inconformity with the neutral evolution model; the Fu and Li’s D* and F* test statistics for the ycf1-rps15 were -0.02679 and -0.19784 respectively, and the Tajima’s D was -0.41630, not significant at the level of P> 0.10, in line with the neutral evolution model; the Fu and Li’s D* and F* test statistics of the combined sequence were 0.34297 and 0.08141respectively, and the Tajima’s D was -0.41550, insignificant at the level of P> 0.10, and conforms with the neutral evolution model (Table 4).
Table 4. Nucleotide diversity of the intergenic spacers of the cultivated S. miltiorrhiza populations.
| Marker | Hap | Hd | π | Vh | Sh | Fu and Li’s D* | Fu and Li’s F* | Tajima’s D |
|---|---|---|---|---|---|---|---|---|
| ETS | 16 | 0.64 | 0.11217 | 0.00795 | 0.089 | 0.22815 | -0.37550 | -1.27644 |
| psbA-trnH | 11 | 0.601 | 0.08478 | 0.00763 | 0.087 | 0.19802 | 0.53273 | 0.90040 |
| trnL-trnF | 4 | 0.619 | 0.03260 | 0.00326 | 0.057 | 1.44651 | 1.89841 | 1.90978 |
| ycf1-rps15 | 13 | 0.795 | 0.06830 | 0.00205 | 0.045 | -0.02679 | -0.19784 | -0.41630 |
| Combined seq. | 26 | 0.950 | 0.07870 | 0.0005 | 0.023 | 0.34297 | 0.08141 | -0.41550 |
The results suggested that the three intergenic spacers, ETS, psbA-trnH, ycf1-rps15, and the combined sequence of the cultivated S. miltiorrhiza populations conform with the neutral evolution at the species level, while the trnL-trnF does not.
Evolutionary relationship of the intergenic spacers
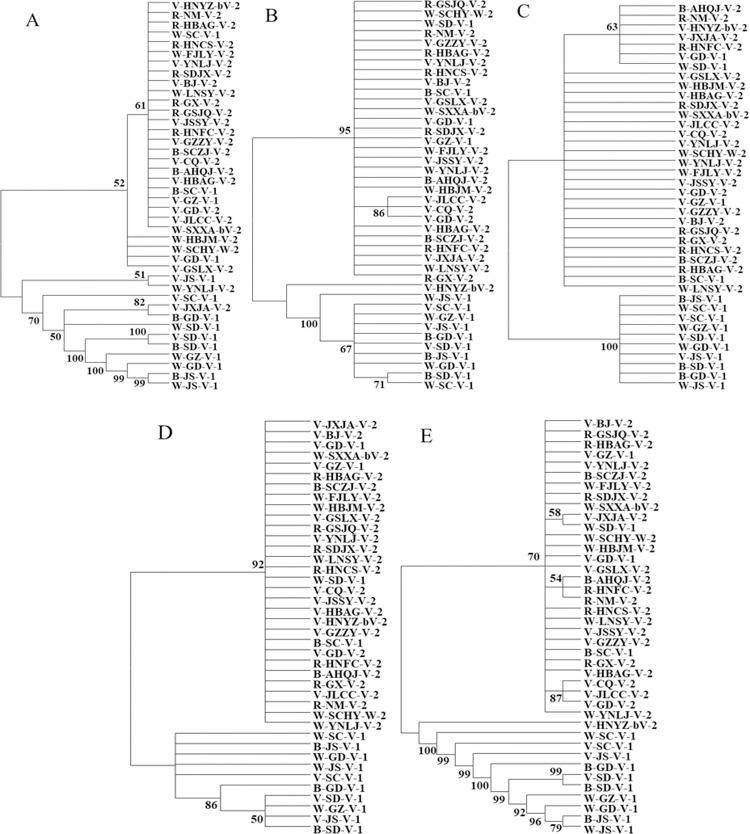
Unrooted phylogenetic trees with a cut-off value of 50% for consensus based on the four intergenic spacers (ETS, psbA-trnH, trnL-trnF, and ycf1-rps15) of the 40 cultivated Danshen populations were constructed. The phylogenetic trees based on ETS and psbA-trnH of S. miltiorrhiza were similarly shown a two-clade structure: for ETS, 28 populations clustered as one, and the remaining 12 populations clustered in the other clade (Fig 2A), and for psbA-trnH, 29 populations clustered in one clade, and the remaining 11 in the other (Fig 2B). The phylogenetic trees based on the trnL-trnF and ycf1-rps15 of S. miltiorrhiza showed a very similar structure in that the same 30 populations clustered in one clade, and the remaining 10 clustered in the other (Fig 2C and 2D). The phylogenetic tree based on the combined sequence of S. miltiorrhiza showed that 40 populations clustered in 2 clades, among which 29 populations are clustered in one clade, and the remaining 11 populations clustered in the other (Fig 2E), very similar to psbA-trnH (Fig 2B). The four intergenic spacers and the combined sequence of the 40 cultivated S. miltiorrhiza populations all showed a similar two-clade topological structure in the phylogenetic trees with only slight differences.
Fig 2. Phylogenetic trees based on the individual and combined intergenic spacers of the cultivated S. miltiorrhiza populations.
A: ETS; B: psbA-trnH; C: trnL-trnF; D: ycf1-rps15; E: Combined sequence.
Discussion and conclusion
For phylogenetic and genetic diversity studies, the key is the selection of suitable genetic markers [37]. It has been suggested that DNA barcodes are used for species identification by sequencing the standard regions of DNA [38]. Although the ribosomal DNA contains very rich genetic information and undergoes a high rate of evolution, it is often prone to recombination and heterozygous due to the biparental inheritance. It is difficult to find single- or low-copy nuclear genes with sufficient variation [39]. Thus it is often confronted with the problem of orthology or paralogy in pedigree geography studies. In contrast, chloroplast DNA is generally maternally inherited in most angiosperms [40] and its non-coding regions are subject to low selective pressures with no gene recombination, which can unambiguously reflect the pedigree history, and has gradually been widely used in studies of plant genetic structure evaluation, population genetic diversity, phylogeny and pedigree geography [41]. The nuclear ribosomal ETS evolve rapidly and have high polymorphism and variability, and play important roles in the studies of genetic variation, classification and phylogeny. However, it is difficult to find suitable universal primers to amplify the entire region of ETS. So there are relatively few related reports in systematic studies [42].
In this study, the four intergenic spacers of ETS, psbA-trnH, trnL-trnF and ycf1-rps15 of the 40 cultivated S. miltiorrhiza populations in China were successfully amplified by PCR. Based on the matrix length, the number of parsimony informative sites accounted for 205, 71, 24, and 88, respectively (Table 3). The results indicate that, among single markers, the ETS of the cultivated S. miltiorrhiza populations were the highest number of parsimony information sites. The PIS rates (Table 3) were generally consistent with the study of the phylogenetic relationship and haplosystem of Lamiaceae plants [26], which showed parsimony information rates of 39.8, 23.95, and16.67%, respectively, for the ETS, ycf1-rps15, and trnL-trnF, markers. The lower PIS of the chloroplast IGS than that of ETS demonstrated both in our research and in haplosystem of Lamiaceae plants [26] indicate that ETS within the nuclear genome are more varied, more informative, and more discriminating than those IGS in the chloroplast genome (psbA-trnH, trnL-trnF, and ycf1-rps15). However, the number of parsimony informative sites of the combined sequence was even higher (392), reaching a discrimination rate of 52.5%.
It is generally accepted that for most of the common species, the more widely distributed, the higher its genetic diversity, while the genetic diversity of endemic, rare or narrowly distributed species is low, and the low genetic diversity is an important factor leading to species endangerment or even extinction [43]. In this study, the genetic diversity analysis of 40 cultivated populations of S. miltiorrhiza showed that the Hd of ETS, psbA-trnH, trnL-trnF and ycf1-rps15 sequences were all greater than 0.5, indicating that S. miltiorrhiza has very rich genetic diversity at the species level, which is consistent with the result based on EST-SSR markers for S. miltiorrhiza [44]. The Hd of the combined sequences is 0.950, much higher than that of the four individual intergenic spacers, suggesting the better reflection of genetic diversity of the cultivated S. miltiorrhiza populations by multi-sequence combination, and could be important in evaluating its genetic variation and adaptability. The best-fit nucleotide substitution models for ETS, psbA-trnH, trnL-trnF, ycf1-rps15 and the combined sequence are HKY+I, T92, T92, T92+G and T92+G respectively. The lower conversion versus transversion frequency of the joint sequences is consistent with all the three IGSs from the chloroplast genome. The relative high nucleotide substitution rates found in this research might be the reason for the high genetic variation among the cultivated S. miltiorrhiza populations, as suggested [45]. Results from Fu and Li’s D* and F* test statistics, together with the Tajima’s D neutral test show that the ETS, psbA-trnH and ycf1-rps15 all conform with the neutral evolution model, while trnL-trnF does not. The combined sequence is not significant at the level of P>0.10, and conforms with the neutral evolution model. It is suggested that the cultivated S. miltiorrhiza populations has not recently experienced expansion events.
It was also found, in this research, that the phylogenetic trees based on both the individual and the combined sequences of the four intergenic spacers of the 40 cultivated populations of S. miltiorrhiza exhibited topologically very similar two-clade structures. For single sequences, ETS has the highest identification rate of 35.0%, and the trnL-trnF sequence has the lowest identification rate of 10.0%. The combined DNA marker has an identification rate as high as 52.5%. But still, they could not discriminate all the tested populations. Further endeavor to explore more and effective DNA markers for the identification of more reliable composite molecular markers is needed so that convenient, fast, efficient and standard identification at the gene level could be established.
Supporting information
(DOCX)
Acknowledgments
Our thanks go to Dr. Chongmo Yuan in our lab for technical assistance in bioinformatics analyses, and to Ms. Dandan Li in Pass College of Chongqing Technology & Business University for graphics.
Data Availability
The accession numbers are provided in S1 Table: GenBank accessions of the four intergenic spacers of the 40 cultivated S. miltiorrhiza populations (in Doc.), by which the readers can access the sequence data thru the URL: National Center for Biotechnology Information (nih.gov).
Funding Statement
This work was part of the Study on the Identification of Rare Animal and Plant Germplasm Activity and the Traceability Technology, the National Key R & D Program of China “Research and Application of National Quality Infrastructure” (Project/Subject SN 2017YFF0205305 and 2017YFF0210303) funded by the Ministry of Science and Technology of the People’s Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tsung OC. Danshen: a popular Chinese cardiac herbal drug. Journal of the American College of Cardiology. 2006; 47 (7): 1498. doi: 10.1016/j.jacc.2006.01.001 . [DOI] [PubMed] [Google Scholar]
- 2.Chan K, Chui SH, Wong DYL, Ha WY, Chan CL, Wong RNS. Protective effects of Danshensu from the aqueous extract of Salvia miltiorrhiza (Danshen) against homocysteine-induced endothelial dysfunction. Life Sciences. 2004; 75(26). doi: 10.1016/j.lfs.2004.06.010 . [DOI] [PubMed] [Google Scholar]
- 3.Fei AH, Cao Q, Chen SY, Wang HR, Wang FL, Pan SM, et al. Salvianolate inhibits reactive oxygen species production in H2O2-treated mouse cardiomyocytes in vitro via the TGFβ pathway. Acta Pharmacologica Sinica. 2013; 34 (4): 496–500. doi: 10.1038/aps.2012.209 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang HJ, Song P, Li J, Zhao DS. Effect of Salvia miltiorrhiza on acetylcholinesterase: enzyme kinetics and interaction mechanism merging with molecular docking analysis. International Journal of Biological Macromolecules. 2019; 135 (1): 303–313. doi: 10.1016/j.ijbiomac.2019.05.132 . [DOI] [PubMed] [Google Scholar]
- 5.Yang L, Zou XJ, Gao X, Chen H, Luo JL, Wang ZH, et al. Sodium tanshinone IIA sulfonate attenuates angiotensin II-induced collagen type I expression in cardiac fibroblasts in vitro. Experimental & Molecular Medicine. 2009; 41 (7): 508. doi: 10.3858/emm.2009.41.7.056 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren XQ, Wang C, Xie BB, Hu LF, Chai H, Ding L, et al. Tanshinone IIA induced cell death via miR30b-p53-PTPN11/SHP2 signaling pathway in human hepatocellular carcinoma cells. European Journal of Pharmacology. 2017; 796: 233–241. doi: 10.1016/j.ejphar.2016.11.046 . [DOI] [PubMed] [Google Scholar]
- 7.Lin CY, Chang TW, Hsieh WH, Hung MC, Lin IH, Lai SC, et al. Simultaneous induction of apoptosis and necroptosis by Tanshinone IIA in human hepatocellular carcinoma HepG2 cells. Cell Death Discovery. 2016; 2, 16065. doi: 10.1038/cddiscovery.2016.65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheng SJ, Pang ECK, Xue CCL, Li CG. Seasonal variations in bioactive marker contents in australian-grown Salvia miltiorrhiza roots. Chemistry & Biodiversity, 2009, 6 (4): 551–560. doi: 10.1002/cbdv.200800090 . [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Morris-Natschke SL, Lee KH. New developments in the chemistry and biology of the bioactive constituents of tanshen. Medicinal Research Reviews. 2007; 27 (1): 133–148. doi: 10.1002/med.20077 . [DOI] [PubMed] [Google Scholar]
- 10.Tang YJ, Zhao MY, Zhang J, Liu JX, Wang HL, Zhang XY. Analysis of genetic difference of different Salvia miltiorrhiza resources and comparison and evaluation of biological characteristics. Lishizhen Medicine and Materia Medica Research. 2017; 28 (4): 975–977. 10.3969/j.issn.1008-0805.2017.04.078. [DOI] [Google Scholar]
- 11.Keneni G, Bekele E, Imtiaz M, Dagne K, Getu E, Assefa F. Genetic diversity and population structure of Ethiopian chickpea (Cicer arietinum L.) germplasm accessions from different geographical origins as revealed by microsatellite markers. Plant Molecular Biology Reporter. 2012; 30 (3): 654–665. 10.1007/s11105-011-0374-6. [DOI] [Google Scholar]
- 12.David M, Michael T. Ribosomal DNA: molecular evolution and phylogenetic inference. The Quarterly Review of Biology. 1991; 66 (4): 411–453. doi: 10.1086/417338 . [DOI] [PubMed] [Google Scholar]
- 13.Chen SL, Yao H, Han JP, Liu C, Song JY, Shi LC, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010; 5: e8613. doi: 10.1371/journal.pone.0008613 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukunaga K, Ichitani K, Kawase M. Phylogenetic analysis of the rDNA intergenic spacer subrepeats and its implication for the domestication history of foxtail millet, Setaria italica. Theoretical and Applied Genetics. 2006; 113 (2): 261–269. doi: 10.1007/s00122-006-0291-5 . [DOI] [PubMed] [Google Scholar]
- 15.Thell A, Hansson M, Persson PE, Seaward M, Veste M, Hedrén M. ETS and plastid sequence data indicate a spontaneous origin of Scandinavian betony, Betonica officinalis L. Biologia Futura: A Quarterly of the Hungarian Academy of Sciences. 2019; 70 (4): 218–239. 10.1556/019.70.2019.27. [DOI] [PubMed] [Google Scholar]
- 16.Feng SJ, Niu JS, Liu ZS, Tian L, Wang XY, Wei AZ. Genetic diversity and evolutionary relationships of Chinese pepper based on nrDNA markers. Forests. 2020; 11 (5): 543. 10.3390/f11050543. [DOI] [Google Scholar]
- 17.Fu T, Wang ZL, Qian PX, Li W, Yuan DM, Yan CF. The latest research progress and application of the DNA barcode in higher plants. Journal of Nuclear Agricultural Sciences. 2016; 30 (5): 0887–0896. 10.11869/j.issn.100-8551.2016.05.0887 [DOI] [Google Scholar]
- 18.Mulo P, Sakurai I, Aro EM. Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta. 2012; 1817 (1): 247–257. doi: 10.1016/j.bbabio.2011.04.011 . [DOI] [PubMed] [Google Scholar]
- 19.Zhong X, Jacquet S. Prevalence of viral photosynthetic and capsid protein genes from cyanophages in two large and deep perialpine lakes. Applied and Environmental Microbiology. 2013; 79 (23): 7169–7178. doi: 10.1128/AEM.01914-13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry RJ. Plant diversity and evolution: genotypic and phenotypic variation in higher plants. Cambridge: CABI Publishing. 2005; 45–68. 10.1079/9780851999043.0000. [DOI] [Google Scholar]
- 21.Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R. The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. The Plant Journal. 2000; 22 (2): 97–104. doi: 10.1046/j.1365-313x.2000.00722.x . [DOI] [PubMed] [Google Scholar]
- 22.Khairulina J, Graifer D, Bulygin K, Ven’yaminova A, Frolova L, Karpova G. Eukaryote-specific motif of ribosomal protein S15 neighbors a site codon during elongation and termination of translation. Biochimie. 2010; 92 (7):820–825. doi: 10.1016/j.biochi.2010.02.031 . [DOI] [PubMed] [Google Scholar]
- 23.Ferreira-Cerca S, Pöll G, Kühn H, Neueder A, Jakob S, Tschochner H, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Molecular Cell. 2007; 28 (3): 446–457. doi: 10.1016/j.molcel.2007.09.029 . [DOI] [PubMed] [Google Scholar]
- 24.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007; 2 (6): e508. doi: 10.1371/journal.pone.0000508 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aron JF, Kevin SB, Prasad RK, Sean WG, Steven GN, Brian CH, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE, 2008, 3 (7): e2802. doi: 10.1371/journal.pone.0002802 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drew BT, Sytsma KJ. Testing the monophyly and placement of Lepechinia in the Tribe mentheae (Lamiaceae). Systematic Botany. 2011; 36 (4):1038–1049. 10.1600/036364411X605047. [DOI] [Google Scholar]
- 27.Deng T, Nie ZL, Drew BT, Volis S, Kim C, Xiang CL, et al. Does the Arcto-Tertiary biogeographic hypothesis explain the disjunct distribution of Northern Hemisphere herbaceous Plants? The case of Meehania (Lamiaceae). PloS ONE. 2015; 10 (2): e0117171. doi: 10.1371/journal.pone.0117171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu GX, Takano A, Drew BT, Liu ED, Soltis DE, Soltis PS, et al. Phylogeny and staminal evolution of Salvia (Lamiaceae, Nepetoideae) in East Asia. Annals of Botany. 2018; 122 (4):649–668. doi: 10.1093/aob/mcy104 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian J, Song JY, Gao HH, Zhu YJ, Xu J, Pang XH, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLOS ONE. 2013; 8 (2): e57607. doi: 10.1371/journal.pone.0057607 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amani J, Kazemi R, Abbasi AR, Salmanian AH. A simple and rapid leaf genomic DNA extraction method for polymerase chain reaction analysis. Iranian Journal of Biotechnology. 2011; 9 (1): 69–71. [Google Scholar]
- 31.Beardsley PM, Olmstead RG. Redefining Phrymaceae: the placement of Mimulus, tribe Mimuleae, and Phryma. American Journal of Botany. 2002; 89 (7): 1093–1102. doi: 10.3732/ajb.89.7.1093 . [DOI] [PubMed] [Google Scholar]
- 32.Baldwin BG, Markos S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S-26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae). Molecular Phylogenetics and Evolution. 1998; 10 (3): 449–463. doi: 10.1006/mpev.1998.0545 . [DOI] [PubMed] [Google Scholar]
- 33.Sang T, Crawford D, Stuessy T. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany. 1997; 84 (8):1120. 10.2307/2446155 . [DOI] [PubMed] [Google Scholar]
- 34.Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991; 17 (5): 1105–1109. doi: 10.1007/BF00037152 . [DOI] [PubMed] [Google Scholar]
- 35.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2016; 33: 1870–1874. doi: 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25 (11): 1451–1452. doi: 10.1093/bioinformatics/btp187 . [DOI] [PubMed] [Google Scholar]
- 37.Shaw J, Lickey EB, Beck JT, Farmer SB, Liu WS, Miller J, et al. The trotoise and the hareⅡ: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany. 2005; 92 (1): 142–166. doi: 10.3732/ajb.92.1.142 . [DOI] [PubMed] [Google Scholar]
- 38.CBOL Plant Working Group1. A DNA barcode for land plants. Proceedings of the National Academy of Sciences. 2009; 106 (31): 12794–12797. doi: 10.1073/pnas.0905845106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences of the United States of America. 1987; 84 (24): 9054–9058. doi: 10.1073/pnas.84.24.9054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biological Journal of the Linnean Society. 1996; 58 (3): 247–276. 10.1111/j.1095-8312.1996.tb01434.x. [DOI] [Google Scholar]
- 41.King RA, Ferris C. Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Molecular Ecology. 1998; 7 (9): 1151–1162. 10.1046/j.1365-294x.1998.00432.x. [DOI] [Google Scholar]
- 42.Roalson EH, Friar EA. Phylogenetic relationships and biogeographic patterns in North American members of Carex section Acrocystis (Cyperaceae) using nrDNA ITS and ETS sequence data. Plant Systematics and Evolution. 2004; 243 (3–4): 175–187. 10.1007/s00606-003-0089-8. [DOI] [Google Scholar]
- 43.Morris CA, Hickey SM, Clarke JN. Genetic and environmental factors affecting lamb survival at birth and through to weaning. New Zealand Journal of Agricultural Research. 2000; 43 (4): 515–524. 10.1080/00288233.2000.9513448. [DOI] [Google Scholar]
- 44.Song ZQ, Wang JH, Chen WX, Zhu HY. Genetic diversity analysis of Salvia miltiorrhiza Bge from different habitats by EST-SSR markers. Journal of Nuclear Agricultural Sciences. 2014; 8 (02): 193–199. 10.11869/j.issn.100-8551. 2014.02.0193. [DOI] [Google Scholar]
- 45.Newton AC, Allnot TR, Gilliesa ACM, Lowe AJ, Ennos RA. Molecular phylogeography, intraspecific variation and conservation of tree species. Trendsin Ecology and Evolution. 1991; 14 (4): 140–145. doi: 10.1016/s0169-5347(98)01555-9 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
The accession numbers are provided in S1 Table: GenBank accessions of the four intergenic spacers of the 40 cultivated S. miltiorrhiza populations (in Doc.), by which the readers can access the sequence data thru the URL: National Center for Biotechnology Information (nih.gov).