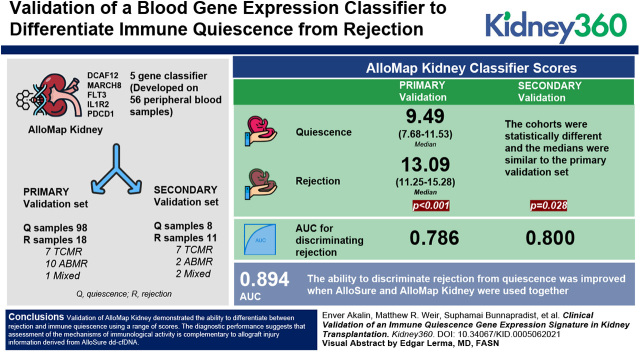
Key Points
AlloMap Kidney is a gene expression profile developed using candidate genes from the AlloMap assay broadly used in heart transplantation.
AlloMap Kidney was validated to differentiate quiescence from rejection in two independent sample sets using a quantitative scale.
Blood cell gene expression and donor-derived cell-free DNA contribute independent signals and inform on different aspects of allograft rejection.
Keywords: transplantation, allograft rejection, antibody-mediated rejection, donor-derived cell-free DNA, gene expression profiling, kidney transplantation, T cell-mediated rejection
Visual Abstract
Abstract
Background
Despite advances in immune suppression, kidney allograft rejection and other injuries remain a significant clinical concern, particularly with regards to long-term allograft survival. Evaluation of immune activity can provide information about rejection status and help guide interventions to extend allograft life. Here, we describe the validation of a blood gene expression classifier developed to differentiate immune quiescence from both T cell–mediated rejection (TCMR) and antibody-mediated rejection (ABMR).
Methods
A five-gene classifier (DCAF12, MARCH8, FLT3, IL1R2, and PDCD1) was developed on 56 peripheral blood samples and validated on two sample sets independent of the training cohort. The primary validation set comprised 98 quiescence samples and 18 rejection samples: seven TCMR, ten ABMR, and one mixed rejection. The second validation set included eight quiescence and 11 rejection samples: seven TCMR, two ABMR, and two mixed rejection. AlloSure donor-derived cell-free DNA (dd-cfDNA) was also evaluated.
Results
AlloMap Kidney classifier scores in the primary validation set differed significantly between quiescence (median, 9.49; IQR, 7.68–11.53) and rejection (median, 13.09; IQR, 11.25–15.28), with P<0.001. In the second validation set, the cohorts were statistically different (P=0.03) and the medians were similar to the primary validation set. The AUC for discriminating rejection from quiescence was 0.786 for the primary validation and 0.800 for the second validation. AlloMap Kidney results were not significantly correlated with AlloSure, although both were elevated in rejection. The ability to discriminate rejection from quiescence was improved when AlloSure and AlloMap Kidney were used together (AUC, 0.894).
Conclusion
Validation of AlloMap Kidney demonstrated the ability to differentiate between rejection and immune quiescence using a range of scores. The diagnostic performance suggests that assessment of the mechanisms of immunologic activity is complementary to allograft injury information derived from AlloSure dd-cfDNA. Together, these biomarkers offer a more comprehensive assessment of allograft health and immune quiescence.
Introduction
Despite modern immune suppression regimens, kidney allograft rejection continues to be both a common occurrence and the primary driver of unacceptably high graft failure rates (1–3). Ten percent of kidney transplant recipients experience allograft rejection in the first year after transplant (4). Although allograft biopsy is the current standard for diagnosis of rejection, optimizing the appropriateness of biopsies by noninvasive techniques is imperative due to the invasive nature of the procedure and the associated risk, and the sampling error and subjective nature of histopathologic interpretation. Analysis of a large series of renal transplant protocol biopsy specimens demonstrated a 2% major complication rate and a 5% risk of gross hematuria (5,6). Additionally, up to 15% of biopsies yield an inadequate specimen, exposing patients to procedural risk without diagnostic benefit (7). The ability to enhance the timing and diagnostic yield of biopsies could meaningfully improve post-transplant outcomes. In addition, methods for assessing response to rejection treatment and return to baseline allograft function frequently rely on additional follow-up biopsies, all associated with risks, expense, inconvenience, and diagnostic failure.
Serum creatinine is commonly used to assess kidney function as a screen for rejection. However, allograft damage sufficient to impair renal function is often irreversible (8), and serum creatinine has repeatedly been shown to be a poorly sensitive or specific indicator of rejection (9). Robust diagnostic and prognostic biomarkers that provide evidence of graft rejection ahead of pathologic findings are needed to help guide clinical management of transplant patients. Among the best studied advanced biomarkers in transplantation is the plasma level of donor-derived cell-free DNA (dd-cfDNA) used to assess allograft injury (9–12). dd-cfDNA has gained significant adoption since it became available as a clinically validated test (12), with numerous studies demonstrating clinical utility in a broad array of contexts (10,11,13–15).
Additional assays are those that use gene expression of circulating immune cells to evaluate immune activity. One example of a broadly integrated gene expression profile (GEP) assay is AlloMap, available as a surveillance tool for recipients of heart transplants since 2005 (16). The assay methodology has not changed since validation and subsequent publication of clinical utility (17); the high negative predictive value (NPV) has enabled avoidance of biopsies for >15 years by the heart transplant community (18). Gene expression profiling of the immune system in kidney transplantation, however, has been elusive as a consistently reliable and reproducible measure of rejection. Several plasma gene expression panels have been published (19–22), one of which is commercially available and designed for use specifically in place of the protocol biopsy (19). Another has faced concerns after independent studies were unable to replicate the validation (20,23). Other signatures with a strong association with fibrosis (21) or specific to antibody-mediated rejection (ABMR) (22) have not yet achieved routine clinical use.
A focused determination of the immune state as active compared with quiescent can help to assess the likelihood of allograft rejection. We describe validation of a blood GEP that can stratify samples according to likelihood of immune quiescence versus T cell–mediated rejection (TCMR) or ABMR. Building on the extensively demonstrated utility of the AlloMap gene set (17), we developed a novel classifier for kidney allograft rejection, condensing the 11 informative genes from AlloMap to a five-gene subset. The AlloMap Kidney classifier was validated using two sample sets independent of the training set, and the performance was evaluated in the primary validation both independently and in conjunction with AlloSure dd-cfDNA.
Materials and Methods
Study Design
Three sets of data were used in this study. The training set and the primary validation set consisted of randomly-assigned (per cohort), distinct sets of patients from the DART study (ClincalTrials.gov, NCT02424227), a multicenter, prospective, observational study to collect plasma in Streck Cell-Free DNA BCT for the purpose of dd-cfDNA measurement and whole blood RNA in PAXgene tubes for gene expression profiling (9). The institutional review board (IRB) at each site approved the study and all of the patients provided written informed consent. The study sponsor provided the statistical analysis, data management, and clinical operations coordination. A second validation set was used to further validate the performance of AlloMap Kidney. These samples, from Montefiore Medical Center (Bronx, NY), were from patients not included in the DART study. Samples were derived from an IRB-approved study of “immune monitoring of kidney transplant recipients” (IRB number 09-06-174). The clinical and research activities reported are consistent with the Principles of the Declaration of Istanbul, as outlined in the Declaration of Istanbul on Organ Trafficking and Transplant Tourism.
Patients and Samples
Patient samples were assigned to two general cohorts: Rejection or Quiescence. Each cohort was defined by established Banff criteria (24), and each contained subgroups as follows. The rejection samples comprised TCMR, ABMR, or mixed rejection (meeting criteria for both ABMR and TCMR). The quiescence samples were one of three types: healthy stable (HS), which had no clinical or laboratory indicators of concern for the graft (and, therefore, no clinically indicated biopsy) and a low level of dd-cfDNA, as measured by AlloSure (<0.5%); nonrejection (NR), which were determined to not have signs of rejection upon pathologist review after a clinically indicated biopsy; and protocol NR (pNR), which were determined to not have signs of rejection upon pathologist review after a protocol surveillance biopsy. Because it is standard to include the best-defined members of the two main cohorts when training a classifier, the Rejection sets for training and the primary validation set included TCMRIA, IB, IIA, and IIB along with ABMR. Borderline TCMR samples were a part of the rejection group in the second validation set. For ABMR, acute/active and chronic, active ABMR were included.
RNA Purification
The DART PAXgene blood tubes were collected alongside Streck BCT plasma, shipped at ambient temperature, received within 3 days, and stored at −80°C until RNA extraction. After thawing, RNA was purified using the QIAsymphony PAXgene Blood RNA Kit (catalog number, 762635; Qiagen) on the QIAsymphony SP system, following the manufacturer’s instructions. The second validation set samples were extracted manually using PAXgene Blood RNA Kits (catalog number, 762164; Qiagen) according to the manufacturer’s instructions. The concentration and purity of the extracted RNA samples were determined by spectrophotometry. Samples were also analyzed for integrity by capillary electrophoresis (TapeStation; Agilent).
Quantitative RT-PCR Methodology
The purified RNA samples were subjected to quantitative RT-PCR (qRT-PCR) in the CareDx CLIA laboratory as described (16). The qPCR for each gene was run in triplicate, and the raw threshold cycle (CT) values (at which probe fluorescence reaches the measurement threshold) were used to calculate a smoothed mean CT. The mean CT was then used for the development of the AlloMap Kidney signature, as described below, without using additional data processing methods or procedures from AlloMap Heart.
Classifier Training
The mean CT for the candidate test genes was normalized against six reference genes (Supplemental Table 2), which were selected on the basis of their stability in this sample set using a scheme similar to what was described previously (16). The normalized results were assessed for statistical significance in a univariate model. Six genes identified as statistically significant were then crossvalidated via bootstrapping and leave-one-out validations. The five genes that passed these internal validations were grouped into three clusters on the basis of their normalized CT level across the full set of training samples. Each cluster has a pairwise correlation coefficient >0.6. A multivariate model that integrates the normalized expression of the five genes was built to optimize performance to differentiate rejection from quiescence in the training sample set.
RNA-Sequencing Methodology
RNA sequencing (RNA-seq) was chosen as a validation and testing platform to enable improved detection of low-expression genes, higher reproducibility, and accurate measurement of gene expression changes that can be readily expanded to additional gene sets and classifiers. A targeted RNA-seq panel (QIAseq; Qiagen), which includes the five informative genes, 15 reference genes (Supplemental Tables 1 and 2), genomic DNA contamination controls, and spike-in controls, was developed and optimized for PAXgene blood RNA samples on an RNA-seq platform using molecular tags (25). Single-read sequencing was performed on an Illumina NextSeq 550. Primary analysis of the sequencing data was performed using the Qiagen GeneGlobe QIAseq bioinformatics pipeline for adaptor trimming, read mapping, quality checks, and computing the molecular tag counts (MTs) for the targeted transcripts. Because the MTs are directly correlated with the initial copy number of the input RNA, a conversion could be defined and tested (Supplemental Appendix 1, Supplemental Figures 1 and 2) to convert MTs to a CT value that would match the CT generated on the same sample by qPCR. The corresponding CT number was derived using the equation X0=Eamp(b−CT), where Eamp is the exponential amplification value, b is the y intercept of log(copies) versus CT, using the average amplification efficiency of 98%, and b=39 for the AlloMap qRT-PCR tests. The CT values for the informative genes were normalized using the average CT of the reference genes. The normalized CT numbers were then used to compute the AlloMap Kidney score using the locked classifier algorithm trained on the qRT-PCR data. AlloSure measurement of dd-cfDNA was performed as previously described (26).
Statistical Analysis
The analysis of differences between groups was performed using an unpaired t test; performance metrics were calculated using standard methods in JMP version 13. Receiver operating characteristic (ROC) curves were generated using the pROC package in R (version 4.0.5). To generate ROC plots for combined AlloMap Kidney and AlloSure data, the AlloSure score was log transformed and then converted to the standard normal distribution. The AlloMap Kidney score was converted to the standard normal distribution similarly. The two converted scores were added arithmetically to create a single result for each sample to plot the ROC.
Results
The AlloMap Heart gene set was developed using PBMCs and comprises genes implicated in diverse immune pathways; therefore, this was chosen as a source of candidate genes for development of a classifier in kidney transplantation (Supplemental Table 1) (16). Due to the complexity of purifying PBMCs at the time of collection, whole blood samples were collected in PAXgene tubes from recipients of kidney transplants. Gene expression data were generated for the 11 AlloMap Heart genes from a subset of the patients from the DART study who were designated as the training sample set, with 38 samples from 22 patients classified as quiescence (HS with AlloSure <1%) and 18 samples from 16 patients classified as rejection (seven TCMR, eight ABMR, three mixed). The only demographic differences between the cohorts in the training sample set were that the Quiescence cohort were earlier post-transplant, and that the Rejection groups had higher serum creatinine and lower eGFR, as expected. No differences were observed in race, sex, type of transplant, recipient or donor cytomegalovirus serology, HLA mismatches, panel-reactive antibodies, induction therapy, or maintenance immunosuppression (Table 1).
Table 1.
Clinical characteristics of the DART analysis groups
| Clinical Characteristic | Training | Validation | ||||||
|---|---|---|---|---|---|---|---|---|
| Healthy Stable AlloSure <1% | Rejection | P Value | Nonrejection | Healthy Stable | Protocol Nonrejection | Rejection |
P Value (Nonrejection versus Rejection) |
|
| Patients, N | 22 | 16 | 44 | 20 | 19 | 16 | ||
| Samples, N | 38 | 18 | 47 | 22 | 29 | 18 | ||
| Race, n (%) | 0.35 | 0.63 | ||||||
| Black | 13 (34) | 8 (44) | 14 (30) | 0 | 8 (28) | 4 (22) | ||
| White | 21 (55) | 9 (50) | 25 (53) | 15 (68) | 19 (66) | 13 (72) | ||
| Native | 0 | 1 (6) | 0 | 0 | 1 (4) | 0 | ||
| Hispanic/Latino | 3 (8) | 0 | 5 (11) | 4 (18) | 1 (4) | 1 (6) | ||
| Asian | 0 | 0 | 1 (2) | 1 (5) | 0 | 0 | ||
| Other | 1 (3) | 0 | 2 (4) | 2 (9) | 0 | 0 | ||
| Sex, n (%) | 0.88 | 0.31 | ||||||
| Men | 24 (63) | 11 (61) | 30 (64) | 14 (64) | 21 (72) | 9 (50) | ||
| Women | 14 (37) | 7 (39) | 17 (36) | 8 (36) | 8 (28) | 9 (50) | ||
| Age at enrollment, yr, mean±SD | 48.8±13.7 | 47.9±17.8 | 0.85 | 50.9±14.5 | 54.9±11.8 | 53.8±12.7 | 39.4±10.9 | 0.001 |
| Post-transplant, d, mean±SD | 94.5±55.3 | 1091±1071 | <0.001 | 965±1360 | 244±200 | 242±192 | 1322±1213 | 0.32 |
| CMV serologic status, n (%) | 0.05 | 0.07 | ||||||
| D−/R+ | 10 (26) | 3 (17) | 13 (28) | 7 (32) | 12 (41) | 3 (17) | ||
| D+/R+ | 14 (37) | 3 (17) | 14 (30) | 7 (32) | 7 (24) | 3 (17) | ||
| D−/R− | 7 (18) | 2 (11) | 8 (17) | 3 (14) | 7 (24) | 4 (22) | ||
| D+/R− | 4 (10) | 3 (17) | 5 (11) | 2 (9) | 3 (10) | 0 | ||
| Unknown | 3 (8) | 7 (39) | 7 (15) | 3 (14) | 0 | 8 (44) | ||
| Donor type, n (%) | 0.47 | 0.12 | ||||||
| Deceased | 21 (55) | 13 (72) | 28 (60) | 13 (59) | 15 (52) | 11 (61) | ||
| Living, unrelated | 4 (10) | 1 (6) | 14 (30) | 3 (14) | 8 (28) | 2 (11) | ||
| Living, related | 13 (34) | 4 (22) | 5 (11) | 6 (27) | 6 (20) | 5 (28) | ||
| Creatinine, mg/dl, mean±SD | 1.46±0.53 | 2.47±1.15 | <0.001 | 2.27±1.49 | 1.36±0.53 | 1.61±0.57 | 2.83±0.91 | 0.08 |
| eGFR, ml/min per 1.73 m2, mean±SD | 55.0±14.9 | 33.6±13.1 | <0.001 | 38.9±19.8 | 56.3±19.6 | 48.2±13.2 | 25.9±11.3 | 0.002 |
| HLA class 1, n mismatches (A, B), mean±SD | 2.7±1.1 | 3.1±1.3 | 0.30 | 2.5±1.3 | 2.1±1.1 | 2.9±1.2 | 1.7±1.1 | 0.02 |
| HLA class 2, n mismatches (DR), mean±SD | 1.2±0.6 | 1.2±0.5 | 0.95 | 1.1±0.8 | 1.1±0.8 | 1.2±0.7 | 0.9±0.8 | 0.32 |
| PRA class I (%) | ||||||||
| Mean PRA | 1.72 | 1.73 | >0.99 | 24.1 | 10.8 | 9.03 | 13.4 | 0.17 |
| Samples | 36 | 15 | 39 | 16 | 29 | 15 | ||
| PRA class II (%) | ||||||||
| Mean PRA | 4.67 | 18.1 | 0.12 | 14.8 | 10.9 | 2.9 | 20.9 | 0.52 |
| Samples | 36 | 14 | 39 | 16 | 29 | 15 | ||
| Induction patients, n (%) | 0.05 | 0.46 | ||||||
| ATG | 2 (9) | 8 (50) | 9 (21) | 2 (10) | 0 | 5 (31) | ||
| Alemtuzumab | 3 (14) | 3 (19) | 9 (21) | 1 (5) | 0 | 2 (13) | ||
| Basiliximab | 2 (9) | 0 | 0 | 1 (5) | 0 | 1 (6) | ||
| Other | 3 (14) | 3 (19) | 8 (18) | 2 (10) | 0 | 3 (19) | ||
| None | 14 (65) | 5 (31) | 23 (52) | 15 (75) | 19 (100) | 8 (50) | ||
| Immunosuppression samples, n (%) | 0.26 | 0.52 | ||||||
| Cyclosporin | 2 (5) | 2 (11) | 5 (11) | 1 (4) | 0 | 2 (11) | ||
| Tacrolimus | 35 (92) | 16 (89) | 38 (81) | 20 (20) | 27 (93) | 15 (83) | ||
| Mycophenolate | 34 (90) | 14 (78) | 40 (85) | 20 (91) | 28 (97) | 15 (83) | ||
| Prednisone | 17 (45) | 14 (78) | 28 (60) | 16 (73) | 10 (34) | 13 (72) | ||
| Rapamycin | 2 (5) | 1 (6) | 3 (6) | 0 | 0 | 2 (11) | ||
| Azathioprine | 2 (5) | 1 (6) | 3 (6) | 0 | 1 (3) | 1 (6) | ||
| Belatacept | 1 (3) | 0 | 2 (4) | 0 | 2 (7) | 0 | ||
| Other | 0 | 3 (17) | 0 | 0 | 0 | 2 (11) | ||
CMV, cytomegalovirus; D−/R+, donor negative/recipient positive; PRA, panel-reactive antibody; ATG, antithymocyte globulin.
Given the differences in sample type and the transplanted organ, a new gene expression classifier was developed starting from the AlloMap Heart gene set. Of the 11 informative genes included in AlloMap Heart (16), six were significantly different between the Quiescence and Rejection cohorts in the training set in a univariate model (P<0.02, Supplementary Table 1). Bootstrap and leave-one-out testing within the training set indicated that five of the six genes were used in >75% of instances; subsequent stepwise selection yielded three important clusters with five genes. These genes represent biologic functions related to immune response pathways: DCAF12 and MARCH8 are involved in modulating immune reactivity, FLT3 and IL1R2 are steroid-responsive genes, and PDCD1 is expressed on activated T lymphocytes (Supplemental Table 1) (27). In the training set, the AlloMap Kidney classifier readily distinguished rejection from quiescence, with P<0.001 and with an area under the ROC curve (AUC) of 0.939 (95% CI, 0.89 to 0.99).
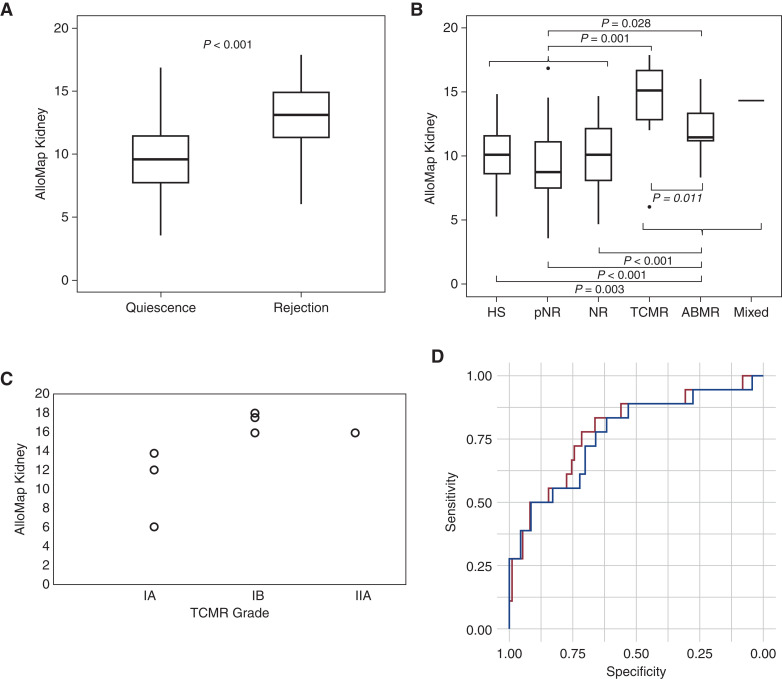
A second set of DART samples from patients not contributing to the training sample set was used as the primary validation set. These 99 unique patients contributed 98 quiescence samples (22 HS, 29 pNR, and 47 NR) and 18 rejection samples (seven TCMR, ten ABMR, and one mixed rejection) (Table 1). The only demographic differences between the cohorts in the primary validation set were that the rejection cohort was younger and had fewer HLA class 1 mismatches. There is not a statistical difference in time post-transplant between the Rejection and Quiescence cohorts. The Rejection cohort had higher serum creatinine levels and lower eGFR than the Quiescence cohort, as expected. No differences were observed in race, sex, type of transplant, time post-transplant, HLA class II mismatches, panel-reactive antibodies, induction therapy, or maintenance immunosuppression. This set of independent samples validated that the AlloMap Kidney classifier distinguished quiescence (median, 9.49; interquartile range [IQR], 7.68–11.53) from rejection (median, 13.09; IQR, 11.25–15.28; P<0.001; Figure 1A). The medians (IQRs) for each of the sample groups were as follows: HS, 10.04 (8.38–11.85); pNR, 8.73 (7.45–11.13); NR, 10.07 (8.05–12.14); TCMR, 15.09 (11.99–17.42); ABMR, 11.48 (10.95–13.68), and mixed rejection, 14.33. Each of the three defined Quiescence groups was significantly different from the Rejection cohort (for rejection versus HS, P=0.003; for rejection versus pNR, P<0.001; for rejection versus NR, P<0.001; Figure 1B). Each of the defined types of rejection was different from the Quiescence cohort (Figure 1B), with P=0.03 and P=0.001 for ABMR and TCMR versus Quiescence, respectively. Although insufficient numbers were available for a robust analysis of the association of AlloMap Kidney scores with TCMR grade, the data suggest that higher grades of TCMR may have higher scores (Figure 1C). The AUC for quiescence versus rejection in the primary validation set was 0.786 (95% CI, 0.66 to 0.91), demonstrating the excellent performance of the GEP across the score range (Figure 1D).
Figure 1.
AlloMap Kidney classifier differentiates quiescence from rejection in the primary independent validation set. (A) Box-and-whisker plots show that biopsy specimen–defined rejections (n=18) were significantly different from quiescence (n=98). (B) No difference among quiescence subgroups, including nonrejection (NR), protocol NR (pNR), and healthy stable (HS) subgroups; all three were significantly lower than T cell–mediated rejection (TCMR) and antibody mediated rejection (ABMR). HS, n=22; pNR, n=29; NR, n=47; TCMR, n=7; ABMR, n=10; mixed rejection, n=1. (C) TCMR results stratified by grade suggest a trend for AlloMap Kidney and TCMR grade. (D) Receiver operating characteristic plot for the primary validation set with rejection compared with quiescence (red line) or NR (blue line). Unpaired t test.
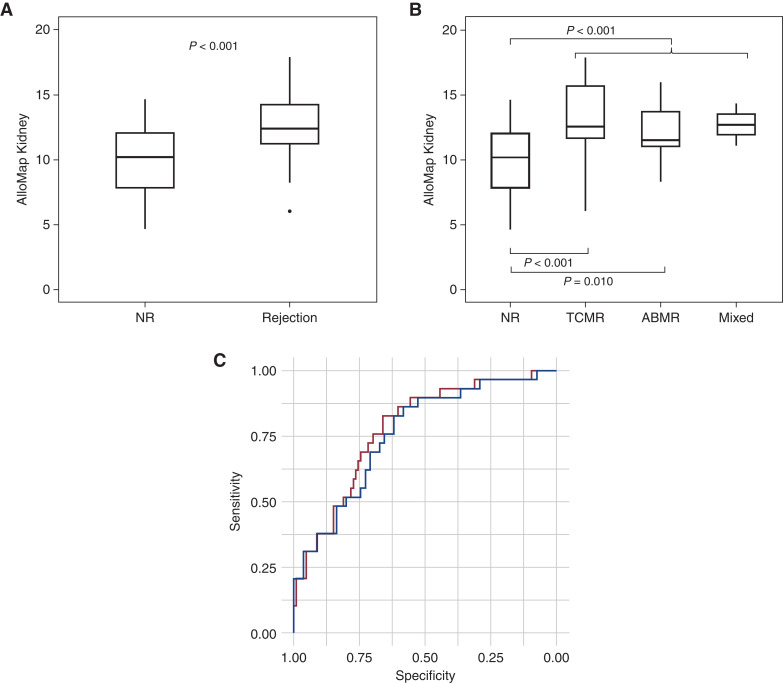
To further validate the performance of AlloMap Kidney, a set of samples from a center not included in the DART study was also evaluated. The Quiescence cohort in this set included eight NR samples, and the Rejection cohort contained 11 rejection samples (two ABMR, seven TCMR, two mixed rejection). In this set, AlloMap Kidney scores were significantly different between Quiescence (NR) and Rejection groups (P=0.03; Figure 2A). Both the TCMR and the ABMR samples had elevated scores relative to the NR group (Figure 2B). The TCMR group consisted of five IIA rejections and two borderline rejections, with a median (IQR) AlloMap Kidney score of 11.85 (11.26–12.67). The AUC for discriminating samples with rejection in this sample set was 0.796 (95% CI, 0.57 to 1; Figure 2C).
Figure 2.
AlloMap Kidney differentiates quiescence from rejection in a single-center validation set. (A) Biopsy specimen–defined rejection (n=11) is significantly different from biopsy specimen–defined no rejection (n=8). (B) All three rejection groups are elevated relative to NR (NR, n=8; TCMR, n=7; ABMR, n=2; mixed rejection, n=2). (C) Receiver operating characteristic plot for the second validation set NR versus rejection (TCMR, ABMR, and mixed rejection). Unpaired t test.
We also assessed the performance of AlloMap Kidney in the combined validation sets to provide an analysis with a larger number of rejection samples. In the combined analysis, the scores for the NR group (median, 10.19; IQR, 7.64–12.09) were significantly lower than the scores for the Rejection cohort (median, 12.43; IQR, 11.12–14.29); P<0.001. All three Rejection groups showed elevated scores: for TCMR (n=14), median (IQR) of 12.55 (11.52–16.25); for ABMR (n=12), median (IQR) of 11.48 (10.95–14.06); and for mixed rejection (n=3), median (IQR) of 12.72 (11.12–14.33) (Figure 3B). Analysis of the combined independent validation sets resulted in an AUC of 0.779 (95% CI, 0.69 to 0.87) for quiescence versus rejection cohorts (Figure 3C).
Figure 3.
The combined validation sets discriminate quiescence from rejection. (A) Quiescence (n=55) versus rejection (n=29) for the full combined datasets. (B) NR versus each type of rejection (NR, n=55; TCMR, n=14; ABMR, n=12, mixed rejection, n=3). (C) Receiver operating characteristic plot of the combined data. Red, quiescence (n=106) versus rejection, AUC=0.779; blue, NR versus rejection, AUC=0.776. Unpaired t test.
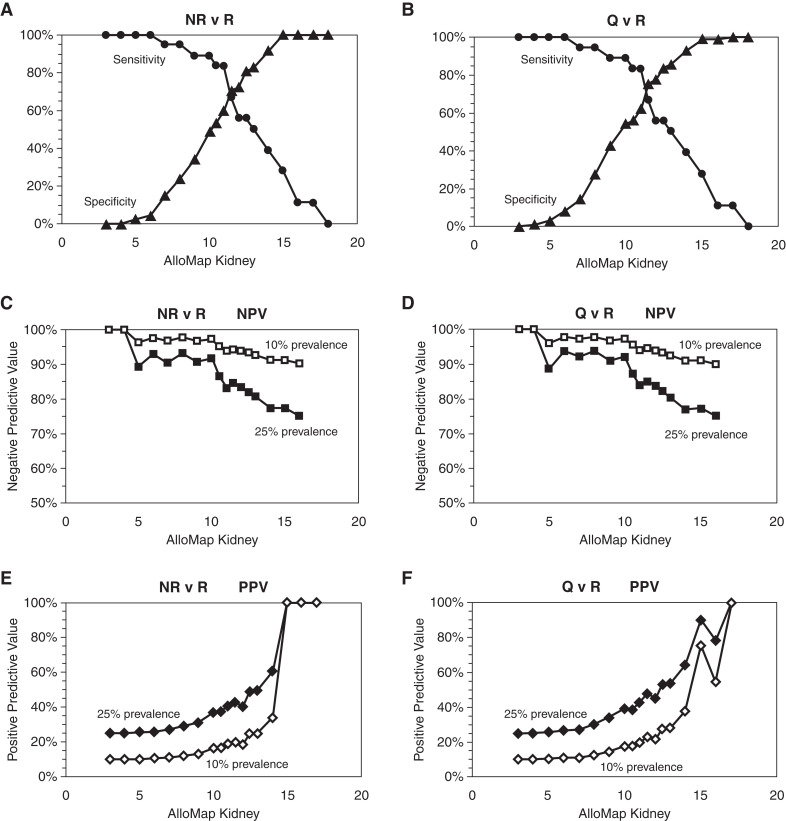
NPV and positive predictive value (PPV) were determined at prevalence levels of 10% and 25%, representing the estimated prevalence of rejection on first-year surveillance and clinically indicated biopsies, respectively (4,9). The single-center sample set contained only NR in the Quiescent cohort; therefore, the performance of the classifier to differentiate the full quiescent group from biopsy sample–defined rejection was assessed on the DART validation set. Figure 4 shows the plots of the sensitivity, specificity, PPV, and NPV of the classifier. For all performance metrics, the data are shown with either the full Quiescence cohort (HS, NR, and pNR) or with only the NR group. Sensitivity did not change because the Rejection cohort remained the same, but specificity, NPV, and PPV are dependent on the choice of quiescence cohort samples. The threshold used for this binary performance characterization was 11.5, at which the AlloMap Kidney score achieved the maximum accuracy for sensitivity and specificity (Figure 4). At the 11.5 score, AlloMap Kidney had a PPV of 23% and an NPV of 95% at 10% prevalence, and a PPV of 48% and an NPV of 87% at 25% prevalence to discriminate rejection from quiescence.
Figure 4.
AlloMap Kidney performance can be assessed across the range of scores. (A) Sensitivity (circles) and specificity (triangles) for NR versus rejection. (B) Sensitivity (circles) and specificity (triangles) for quiescence (Q) versus rejection (R). (C) Negative predictive value (NPV) for NR versus rejection. (D) NPV for quiescence versus rejection. (E) Positive predictive value (PPV) for NR versus rejection. (F) PPV for quiescence versus rejection. For NPV and PPV, 25% prevalence is shown in filled symbols, 10% prevalence in open symbols.
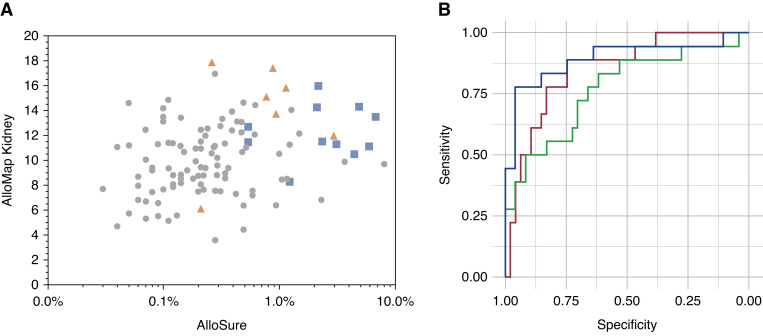
For samples in the DART study, plasma was also collected to measure dd-cfDNA using AlloSure. AlloSure is highly associated with graft injury (9); the signal is hypothesized to be different from that of the AlloMap Kidney signature, although both are correlated with rejection. Figure 5A shows the data for all quiescence and rejection cohorts (mixed rejection was included in the ABMR group). There was a weak correlation between AlloSure and AlloMap Kidney (R=0.15, P=0.23). However, samples with AlloSure results of ≥1% had higher AlloMap Kidney scores. Several TCMR samples with AlloSure between 0.5% and 1% had very high AlloMap Kidney scores, suggesting a role for AlloMap Kidney to inform on which of these intermediate scores likely correlate with rejection (11). To examine the potential of coupled testing in post-transplant care, a combined score was derived with equal weighting of AlloSure and AlloMap Kidney. The range of this combined score can be envisioned along the diagonal from lower left in the plot in Figure 5A to the upper right. These data were used to generate an ROC plot for the combined score, which was compared to the ROC plots for AlloSure or AlloMap Kidney alone in the same sample set (Figure 5B). These data showed a superior performance for the combined use of AlloMap Kidney and AlloSure versus AlloMap Kidney alone (P=0.005).
Figure 5.
The combination of AlloMap Kidney with AlloSure has greater discriminating ability than either alone. (A) Plot of all quiescence and rejection points from the validation set for both AlloMap Kidney (y axis) and AlloSure (x axis). Circles are all types of quiescence (NR, HS, pNR). Orange triangles, TCMR; blue squares, ABMR. (B) Receiver operating characteristic plots of a linear combination of AlloMap Kidney and AlloSure scores as shown in (A). Blue is the combined score (AUC=0.894), AlloMap Kidney alone is shown in green (AUC=0.768), and AlloSure alone is shown in red (AUC=0.85).
Discussion
Factors predisposing the development of active rejection have been extensively studied, with the recipient immune system proving to be a key intermediary in many relevant processes, including ischemia-reperfusion injury, infection, and response to immunosuppression. Uncontrolled inflammation in kidney allografts leads to the chronic damage and progressive fibrosis that accounts for the majority of long-term allograft loss (28,29). Genetic predictors of active rejection have also been described in recent years, some of which implicate immune activity. Taken in concert, these data suggest that monitoring gene expression in peripheral blood immune cells may lead to earlier or more sensitive detection of active rejection (19,20,30).
The objective of this study was to validate a classifier that discriminates immune quiescence from kidney allograft rejection. Rather than novel discovery from the whole transcriptome, this classifier was developed using a candidate gene approach using the AlloMap Heart genes, which have been implicated in immune responses or regulation and, therefore, presented a rational starting point for developing a quiescence signature for kidney transplantation. Furthermore, we elected to measure whole blood gene transcripts rather than looking specifically at the PBMC subset, the method used by the AlloMap Heart assay. Collection of whole blood is significantly less complicated than PBMC collection, enabling a more streamlined workflow at the collection site. After accurately measuring the expression of these candidate genes, we identified five that could discriminate kidney rejection using a training sample set from the DART study that was collected in PAXgene tubes. This classifier was tested using two validation sets: the primary validation set comprised independent patients from DART, and the second validation set was from a single center. Both sample sets demonstrate the validity of the classifier to discriminate biopsy sample–defined rejection from quiescence. Classifier scores were statistically significantly different between the rejection cohort and the quiescence cohort, and between the quiescence cohort and either TCMR or ABMR. Results in the three subgroups of quiescence (HS, pNR, NR) were each statistically different from those seen in rejection. In both validation sets, the AUC demonstrated excellent diagnostic performance. The classifier had an NPV of >95% in a surveillance population (10% prevalence) on the basis of a score of 11.5, chosen for maximal sensitivity and specificity.
The limitations of this study include the number of rejection samples and limited scope of the patient population. To generate training and validation sets from completely independent patients, we were limited to only 18 rejection samples in each set, randomly selected to generate the two independent sets. Despite these small numbers, independent validation sets of TCMR and ABMR, and of mixed rejection, demonstrated excellent diagnostic performance in all comparisons. Further, a second independent validation set recapitulated the performance of the classifier in the primary validation set. Although three different types of quiescence samples (NR, pNR, and HS) were included in this study, there may be other subsets of the target patient population that should be independently characterized on this classifier in future studies, such as infection, interstitial fibrosis and tubular atrophy, drug toxicity, BKV nephropathy, and recurrent or de novo glomerular disease. Each of these could have been undiagnosed in the HS set, which serves as an indicator of a surveillance population at large. However, clinicians may still wish to biopsy to identify some of these pathologies in patients with high suspicion. The results presented here represent assessment of the status of the allograft at the time of the AlloMap Kidney testing, rather than long-term graft survival. A correlation with long-term outcomes necessarily requires large, prospective studies and does not detract from the demonstrated capability of AlloMap Kidney to provide noninvasive assessment of current rejection status. Lastly, our approach necessarily limited the scope of possible genes and, therefore, may not have included the best genes for discrimination of rejection from immune quiescence. With further sample availability from ongoing studies, we anticipate a robust sample set to potentially expand the clinical utility of the assay with expanded gene sets.
One strength of this study is that we started with a well-validated gene set and test condition. The gene set from the Food and Drug Administration–cleared AlloMap Heart test has proven robust and clinically relevant for >15 years in heart transplantation (17). In contrast, within kidney transplantation, previously developed gene expression profiling assays have limited overall use in more narrow clinical indications, such as replacing surveillance biopsies or assessment of fibrosis (19,21).
In contrast to the AlloMap Kidney measure of immune activity, the use of AlloSure to quantify dd-cfDNA provides insight into molecular injury in the allograft. Prior experience with combined testing in heart transplantation suggested that gene expression measurement in immune cells provides a complementary signal that can provide added insight into allograft rejection. Gene expression profiling may discriminate between types of allograft injury, including drug toxicity that can lead to injury in the absence of rejection. Therefore, there is potential added value when the two tests are combined. Indeed, when the DART validation set was analyzed using both AlloMap Kidney and AlloSure, a higher diagnostic performance was observed. Only a single rejection sample was below the nominal threshold for both tests. The details of the pathology report for this sample were independently reviewed and, despite being called TCMRIA by the pathologist at the treating center, the biopsy specimen only showed focal mild tubulitis. On the basis of Banff 2019, this biopsy specimen would not even meet the criteria for borderline TCMR; it would be called acute tubular injury on the basis of the pathology report. A post hoc reanalysis of the AlloMap Kidney AUC with this sample in the NR set produced a value of 0.83. Future studies will better define how these complementary signals can enhance diagnostic assessment and management of recipients of kidney transplants. In the primary validation cohort, AlloMap Kidney scores appear higher in TCMR than in ABMR, whereas AlloSure scores have been reported higher with ABMR (9). The coupled use of these assays may allow noninvasive discrimination of the type of rejection. The high NPV of AlloMap Kidney makes it an ideal assay for integration into routine post-transplant care, allowing minimization of biopsies in the same manner as AlloMap Heart has in heart transplantation for >15 years. This may be complemented by the AlloSure signal indicating injury, thus improving the PPV. Future analysis of larger datasets of paired data will provide detailed performance of the use of both tests together.
Immune activity biomarkers can strengthen the high NPV of existing markers, allowing the confidence to rule out pathology by identifying those who are immunoquiescent. These types of markers also open the prospect of managing immunomodulation. Reducing medication dose for patients who are adequately immunosuppressed, and increasing the dose in those patients who are not, may lead to improved outcomes for both patient populations. The combination of allograft injury (AlloSure dd-cfDNA) and immune activation markers (AlloMap Kidney GEP) also take us in the direction of noninvasive characterization of underlying pathology and one step closer to offering a true liquid biopsy for monitoring allograft health. With uncontrolled inflammation playing such a fundamental role across the spectrum of allograft loss, noninvasive characterization of both injury and gene expression testing presents an attractive paradigm shift toward transplant precision medicine.
Disclosures
E. Akalin reports receiving research funding from Angion, Astellas, CareDx, and National Institutes of Health (NIH); and having consultancy agreements with, receiving honoraria from, and serving as a scientific advisor for or member of CareDx and Immucor. D.C. Brennan reports having consultancy agreements with CareDx, Medeor, Sanofi, and Veloxis; receiving research funding from CareDx and Natera; receiving honoraria from CareDx, Sanofi, and Veloxis; and serving as a scientific advisor for or member of the editorial boards of Transplantation and UpToDate. D.C. Brennan, J.S. Bromberg, R. Weir, S. Bunnapradist, R. Delos Santos, and A. Langone report being DART investigators. J.S. Bromberg reports receiving research funding from Angion, Astellas, CareDx, Natera, Novartis, and Quark; having consultancy agreements with Eurofins; and serving as a scientific advisor for, or member of, NIH and Transplantation. S. Bunnapradist reports receiving research funding from Alexion, Angion, Astellas, CareDx, and Merck; receiving honoraria from Bristol Myers Squibb (BMS), CareDx, Sanofi, and Veloxis; serving on speakers bureau for BMS, CareDx, Natera, Veloxis, and Vitaeris; and having consultancy agreements with CareDx. R. Delos Santos reports having other interests/relationships with the American Society of Transplantation (AST) Conflict of Interest Committee and AST Transplant Nephrology Fellowship Training Accreditation Program Review Committee; receiving research funding from CareDx, Merck, and Veloxis; having ownership interest in Pfizer; and receiving honoraria from UpToDate. S. Dholakia reports having ownership interest in CareDx. S. Dholakia, X. Jin, H. Xu, and R.N. Woodward report being employees of CareDx. A. Djamali reports having consultancy agreements with, receiving honoraria from, and serving as a scientific advisor for or member of CareDx and CSL; and receiving research funding from CareDx and Takeda. X. Jin reports having ownership interest in CareDx. M.R. Weir reports having consultancy agreements (all are modest, <$10,000) with, receiving honoraria from, and serving as a scientific advisor for or member of AstraZeneca, Boehringer-Ingelheim, Bayer, CareDx, Janssen, Merck, Novo Nordisk, and Vifor Pharma. R.N. Woodward reports having ownership interest in CareDx. H. Xu reports having ownership interest in CareDx and Proxim Diagnostics, and having consultancy agreements with Proxim Diagnostics.
Funding
The DART study was funded by CareDx. Collection of the second validation set samples and data was funded by Einstein-Montefiore Abdominal Transplant Center.
Acknowledgments
We thank David Lew for laboratory work on the initial training dataset. Dr. David Hiller, Dr. Divya Kilam, Dr. Kunbin Qu, and Dr. Ping Shi contributed to the statistical analysis; Dr. Lihong Bu provided pathology review; and Dr. Grigoriy Shekhtman and Kristina Jensen provided critical editing of the manuscript.
Author Contributions
E. Akalin, D.C. Brennan, J.S. Bromberg, S. Dholakia, R.N. Woodward, and H. Xu conceptualized the study; E. Akalin, R.N. Woodward, and H. Xu provided supervision; S. Dholakia, X. Jin, and H. Xu were responsible for validation; S. Dholakia and R.N. Woodward wrote the original draft; X. Jin, R.N. Woodward, and H. Xu were responsible for data curation and methodology; X. Jin and H. Xu were responsible for investigation; R.N. Woodward was responsible for resources; R.N. Woodward and H. Xu were responsible for formal analysis and project administration; and all authors reviewed and edited the manuscript.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0005062021/-/DCSupplemental.
Supplemental methods. Download Supplemental Appendix 1, PDF file, 237 KB (236.1KB, pdf)
Conversion of AlloMap Kidney classifier trained on qPCR data to produce the same results from RNA-seq data. Download Supplemental Figure 1, PDF file, 237 KB (236.1KB, pdf)
Comparison of AlloMap Kidney data from manual and automated extraction methods. Download Supplemental Figure 2, PDF file, 237 KB (236.1KB, pdf)
Overview of the AlloMap genes. Download Supplemental Table 1, PDF file, 237 KB (236.1KB, pdf)
List of the AlloMap reference genes. Download Supplemental Table 2, PDF file, 237 KB (236.1KB, pdf)
References
- 1.Mas VR, Mueller TF, Archer KJ, Maluf DG: Identifying biomarkers as diagnostic tools in kidney transplantation. Expert Rev Mol Diagn 11: 183–196, 2011. 10.1586/erm.10.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo DJ, Kaplan B, Kirk AD: Biomarkers for kidney transplant rejection. Nat Rev Nephrol 10: 215–225, 2014. 10.1038/nrneph.2013.281 [DOI] [PubMed] [Google Scholar]
- 3.Bontha SV, Maluf DG, Mueller TF, Mas VR: Systems biology in kidney transplantation: The application of multi-omics to a complex model. Am J Transplant 17: 11–21, 2017. 10.1111/ajt.13881 [DOI] [PubMed] [Google Scholar]
- 4.Hart A, Smith JM, Skeans MA, Gustafson SK, Wilk AR, Robinson A, Wainright JL, Haynes CR, Snyder JJ, Kasiske BL, Israni AK: OPTN/SRTR 2016 Annual Data Report: Kidney. Am J Transplant 18[Suppl 1]: 18–113, 2018. 10.1111/ajt.14557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan TA, Chandran S, Burger IM, Zhang CA, Goldstein RB: Complications of ultrasound-guided renal transplant biopsies. Am J Transplant 16: 1298–1305, 2016. 10.1111/ajt.13622 [DOI] [PubMed] [Google Scholar]
- 6.Reschen ME, Mazzella A, Sharples E: A retrospective analysis of the utility and safety of kidney transplant biopsies by nephrology trainees and consultants. Ann Med Surg (Lond) 28: 6–10, 2018. 10.1016/j.amsu.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plattner BW, Chen P, Cross R, Leavitt MA, Killen PD, Heung M: Complications and adequacy of transplant kidney biopsies: A comparison of techniques. J Vasc Access 19: 291–296, 2018. 10.1177/1129729817747543 [DOI] [PubMed] [Google Scholar]
- 8.Cravedi P, Mannon RB: Noninvasive methods to assess the risk of kidney transplant rejection. Expert Rev Clin Immunol 5: 535–546, 2009. 10.1586/eci.09.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, Matas AJ, Mehta S, Mannon RB, Sharfuddin A, Fischbach B, Narayanan M, Jordan SC, Cohen D, Weir MR, Hiller D, Prasad P, Woodward RN, Grskovic M, Sninsky JJ, Yee JP, Brennan DC; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators : Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol 28: 2221–2232, 2017. 10.1681/ASN.2016091034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan SC, Bunnapradist S, Bromberg JS, Langone AJ, Hiller D, Yee JP, Sninsky JJ, Woodward RN, Matas AJ: Donor-derived cell-free DNA identifies antibody-mediated rejection in donor specific antibody positive kidney transplant recipients. Transplant Direct 4: e379, 2018. 10.1097/TXD.0000000000000821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stites E, Kumar D, Olaitan O, John Swanson S, Leca N, Weir M, Bromberg J, Melancon J, Agha I, Fattah H, Alhamad T, Qazi Y, Wiseman A, Gupta G: High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant 20: 2491–2498, 2020. 10.1111/ajt.15822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grskovic M, Hiller DJ, Eubank LA, Sninsky JJ, Christopherson C, Collins JP, Thompson K, Song M, Wang YS, Ross D, Nelles MJ, Yee JP, Wilber JC, Crespo-Leiro MG, Scott SL, Woodward RN: Validation of a clinical-grade assay to measure donor-derived cell-free DNA in solid organ transplant recipients. J Mol Diagn 18: 890–902, 2016. 10.1016/j.jmoldx.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 13.Xie WY, Kim K, Goussous N, Drachenberg CB, Scalea JR, Weir MR, Bromberg JS: Causes of renal allograft injury in recipients with normal donor-derived cell-free DNA. Transplant Direct 7: e679, 2021. 10.1097/TXD.0000000000001135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf-Doty TK, Mannon RB, Poggio ED, Hinojosa RJ, Hiller D, Bromberg JS, Brennan DC: Dynamic response of donor-derived cell-free DNA following treatment of acute rejection in kidney allografts. Kidney360 2: 729–736, 2021. 10.34067/KID.0000042021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mamlouk O, Nair R, Iyer SP, Edwards A, Neelapu SS, Steiner RE, Adkins SA, Hawkins M, Saini N, Devashish K, Strati P, Mandayam S, Ahmed S: Safety of CAR T-cell therapy in kidney transplant recipients. Blood 137: 2558–2562, 2021. 10.1182/blood.2020008759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng MC, Eisen HJ, Mehra MR, Billingham M, Marboe CC, Berry G, Kobashigawa J, Johnson FL, Starling RC, Murali S, Pauly DF, Baron H, Wohlgemuth JG, Woodward RN, Klingler TM, Walther D, Lal PG, Rosenberg S, Hunt S; CARGO Investigators : Noninvasive discrimination of rejection in cardiac allograft recipients using gene expression profiling. Am J Transplant 6: 150–160, 2006. 10.1111/j.1600-6143.2005.01175.x [DOI] [PubMed] [Google Scholar]
- 17.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, Cappola TP, Kao A, Anderson AS, Cotts WG, Ewald GA, Baran DA, Bogaev RC, Elashoff B, Baron H, Yee J, Valantine HA; IMAGE Study Group : Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med 362: 1890–1900, 2010. 10.1056/NEJMoa0912965 [DOI] [PubMed] [Google Scholar]
- 18.Deng MC: The AlloMap™ genomic biomarker story: 10 years after. Clin Transplant 31: e12900, 2017. 10.1111/ctr.12900 [DOI] [PubMed] [Google Scholar]
- 19.Kurian SM, Williams AN, Gelbart T, Campbell D, Mondala TS, Head SR, Horvath S, Gaber L, Thompson R, Whisenant T, Lin W, Langfelder P, Robison EH, Schaffer RL, Fisher JS, Friedewald J, Flechner SM, Chan LK, Wiseman AC, Shidban H, Mendez R, Heilman R, Abecassis MM, Marsh CL, Salomon DR: Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant 14: 1164–1172, 2014. 10.1111/ajt.12671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Khatri P, Sigdel TK, Tran T, Ying L, Vitalone MJ, Chen A, Hsieh S, Dai H, Zhang M, Naesens M, Zarkhin V, Sansanwal P, Chen R, Mindrinos M, Xiao W, Benfield M, Ettenger RB, Dharnidharka V, Mathias R, Portale A, McDonald R, Harmon W, Kershaw D, Vehaskari VM, Kamil E, Baluarte HJ, Warady B, Davis R, Butte AJ, Salvatierra O, Sarwal MM: A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant 12: 2710–2718, 2012. 10.1111/j.1600-6143.2012.04253.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Yi Z, Keung KL, Shang H, Wei C, Cravedi P, Sun Z, Xi C, Woytovich C, Farouk S, Huang W, Banu K, Gallon L, Magee CN, Najafian N, Samaniego M, Djamali A, Alexander SI, Rosales IA, Smith RN, Xiang J, Lerut E, Kuypers D, Naesens M, O’Connell PJ, Colvin R, Menon MC, Murphy B: A peripheral blood gene expression signature to diagnose subclinical acute rejection. J Am Soc Nephrol 30: 1481–1494, 2019. 10.1681/ASN.2018111098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Loon E, Gazut S, Yazdani S, Lerut E, de Loor H, Coemans M, Noël LH, Thorrez L, Van Lommel L, Schuit F, Sprangers B, Kuypers D, Essig M, Gwinner W, Anglicheau D, Marquet P, Naesens M: Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: A multicentre, prospective study. EBioMedicine 46: 463–472, 2019. 10.1016/j.ebiom.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Loon E, Giral M, Anglicheau D, Lerut E, Dubois V, Rabeyrin M, Brouard S, Roedder S, Spigarelli MG, Rabant M, Bogaerts K, Naesens M, Thaunat O: Diagnostic performance of kSORT, a blood-based mRNA assay for noninvasive detection of rejection after kidney transplantation: A retrospective multicenter cohort study. Am J Transplant 21: 740–750, 2021. 10.1111/ajt.16179 [DOI] [PubMed] [Google Scholar]
- 24.Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, Nankivell BJ, Halloran PF, Colvin RB, Akalin E, Alachkar N, Bagnasco S, Bouatou Y, Becker JU, Cornell LD, Duong van Huyen JP, Gibson IW, Kraus ES, Mannon RB, Naesens M, Nickeleit V, Nickerson P, Segev DL, Singh HK, Stegall M, Randhawa P, Racusen L, Solez K, Mengel M: The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant 18: 293–307, 2018. 10.1111/ajt.14625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Q, Vijaya Satya R, Lewis M, Randad P, Wang Y: Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics 16: 589, 2015. 10.1186/s12864-015-1806-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong L, Scott S, Grskovic M, Dholakia S, Woodward R: The evolution and innovation of donor-derived cell-free DNA testing in transplantation. J Med Diagn Meth 9: 302, 2020. 10.35248/2168-9784.2020.9.302 [DOI] [Google Scholar]
- 27.Dedrick RL: Understanding gene expression patterns in immune-mediated disorders. J Immunotoxicol 4: 201–207, 2007. 10.1080/15476910701385562 [DOI] [PubMed] [Google Scholar]
- 28.Naesens M, Khatri P, Li L, Sigdel TK, Vitalone MJ, Chen R, Butte AJ, Salvatierra O, Sarwal MM: Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int 80: 1364–1376, 2011. 10.1038/ki.2011.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moreso F, Ibernon M, Gomà M, Carrera M, Fulladosa X, Hueso M, Gil-Vernet S, Cruzado JM, Torras J, Grinyó JM, Serón D: Subclinical rejection associated with chronic allograft nephropathy in protocol biopsies as a risk factor for late graft loss. Am J Transplant 6: 747–752, 2006. 10.1111/j.1600-6143.2005.01230.x [DOI] [PubMed] [Google Scholar]
- 30.Dorr C, Wu B, Guan W, Muthusamy A, Sanghavi K, Schladt DP, Maltzman JS, Scherer SE, Brott MJ, Matas AJ, Jacobson PA, Oetting WS, Israni AK: Differentially expressed gene transcripts using RNA sequencing from the blood of immunosuppressed kidney allograft recipients. PLoS One 10: e0125045, 2015. 10.1371/journal.pone.0125045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download Supplemental Appendix 1, PDF file, 237 KB (236.1KB, pdf)
Conversion of AlloMap Kidney classifier trained on qPCR data to produce the same results from RNA-seq data. Download Supplemental Figure 1, PDF file, 237 KB (236.1KB, pdf)
Comparison of AlloMap Kidney data from manual and automated extraction methods. Download Supplemental Figure 2, PDF file, 237 KB (236.1KB, pdf)
Overview of the AlloMap genes. Download Supplemental Table 1, PDF file, 237 KB (236.1KB, pdf)
List of the AlloMap reference genes. Download Supplemental Table 2, PDF file, 237 KB (236.1KB, pdf)