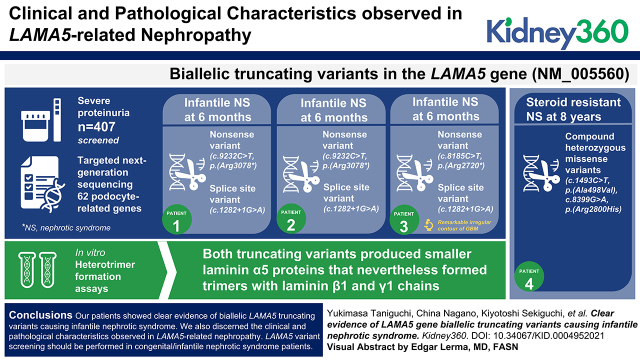
Key Points
LAMA5 gene biallelic variants have been identified in only seven patients so far, and no functional analysis had been conducted for all but one.
We report three patients with LAMA5 biallelic truncating variants manifesting infantile nephrotic syndrome and in vitro heterotrimer assays.
We report one patient with SRNS with biallelic LAMA5 missense variants.
Keywords: clinical nephrology, LAMA5, nephrotic syndrome, pathology, SRNS
Visual Abstract
Abstract
Background
Pathogenic variants in single genes encoding podocyte-associated proteins have been implicated in about 30% of steroid-resistant nephrotic syndrome (SRNS) patients in children. However, LAMA5 gene biallelic variants have been identified in only seven patients so far, and most are missense variants of unknown significance. Furthermore, no functional analysis had been conducted for all but one of these variants. Here, we report three patients with LAMA5 gene biallelic truncating variants manifesting infantile nephrotic syndrome, and one patient with SRNS with biallelic LAMA5 missense variants.
Methods
We conducted comprehensive gene screening of Japanese patients with severe proteinuria. With the use of targeted next-generation sequencing, 62 podocyte-related genes were screened in 407 unrelated patients with proteinuria. For the newly discovered LAMA5 variants, we conducted in vitro heterotrimer formation assays.
Results
Biallelic truncating variants in the LAMA5 gene (NM_005560) were detected in three patients from two families. All patients presented with proteinuria within 6 months of age. Patients 1 and 2 were siblings possessing a nonsense variant (c.9232C>T, p.[Arg3078*]) and a splice site variant (c.1282 + 1G>A) that led to exon 9 skipping and a frameshift. Patient 3 had a remarkable irregular contour of the glomerular basement membrane. She was subsequently found to have a nonsense variant (c.8185C>T, p.[Arg2720*]) and the same splice site variant in patients 1 and 2. By in vitro heterotrimer formation assays, both truncating variants produced smaller laminin α5 proteins that nevertheless formed trimers with laminin β1 and γ1 chains. Patient 4 showed SRNS at the age of 8 years, and carried compound heterozygous missense variants (c.1493C>T, p.[Ala498Val] and c.8399G>A, p.[Arg2800His]).
Conclusions
Our patients showed clear evidence of biallelic LAMA5 truncating variants causing infantile nephrotic syndrome. We also discerned the clinical and pathologic characteristics observed in LAMA5-related nephropathy. LAMA5 variant screening should be performed in patients with congenital/infantile nephrotic syndrome.
Introduction
Nephrotic syndrome is characterized by heavy proteinuria, hypoalbuminemia, edema, and hyperlipidemia. Most children with nephrotic syndrome have steroid-sensitive nephrotic syndrome. However, 10%–20% of patients with nephrotic syndrome have steroid-resistant nephrotic syndrome (SRNS) and are at risk to progress to end stage kidney disease (ESKD) (1).
FSGS is the most frequently observed pathologic finding in SRNS and often develops into ESKD in children. Hereditary FSGS is a genetically heterogeneous condition that is associated with >60 podocyte-related genes. Identifying genetic bases for SRNS/FSGS in patients could allow discontinuation of immunosuppressive therapy and provide further information about prognosis.
Whole-exome sequencing has revealed new genes associated with SRNS, thus expanding the genetic heterogeneity of the disease. Chatterjee et al. reported a patient with biallelic LAMA5 missense variants associated with FSGS for the first time (2). However, those are variants of unknown significance, and there are no reports of any in vitro functional or in vivo pathologic analyses. Likewise, Braun et al. identified homozygous variants of unknown significance in LAMA5 in five pediatric patients with nephrotic syndrome from three families using whole-exome sequencing (3). They also have not reported further in vitro functional or in vivo pathologic analyses. Therefore, the role of LAMA5 pathogenic variants in causing SRNS/FSGS is still unclear. The LAMA5 variant p.(Arg286Leu) is clearly pathogenic, and causes complex syndromic developmental defects along with FSGS and ESKD (4).
The glomerular basement membrane (GBM) is an important component of the kidney’s glomerular filtration barrier. The GBM contains members of the four major classes of basement membrane proteins: type IV collagen, laminin, heparan sulfate proteoglycan, and nidogen (5). The GBM's major laminin isoform is laminin-521 (LM-521), a cross-shaped heterotrimeric glycoprotein composed of the laminin α5, β2, and γ1 chains (6). The LAMA5 gene encodes laminin α5, which is a widely expressed chain (7) found in many embryonic and adult basement membranes (8).
We have characterized in detail three infantile patients with nephrotic syndrome and biallelic LAMA5 truncating variants and one patient with SRNS and a LAMA5 missense variant. These patients provided clear evidence for the first time that biallelic truncating variants in LAMA5 can cause early-onset nephrotic syndrome.
Materials and Methods
Study Participants
After receiving informed consent, we obtained clinical data and blood samples from individuals with severe proteinuria in Japan. Study approval was obtained from the Institutional Review Board of Kobe University Graduate School of Medicine (approval number 301). Patients were enrolled between January 2016 and January 2021. We performed targeted next-generation sequencing (NGS) in 407 families.
Genetic Analysis
Genomic DNA was extracted from peripheral blood using the Quick Gene Mini 80 system (Wako Pure Chemical Industries, Ltd., Tokyo, Japan) and used for targeted sequencing and Sanger sequencing. Targeted sequencing using NGS was conducted for genes responsible for inherited kidney disease (listed in Supplemental Table 1). The sample library for NGS analysis was prepared using SureSelect XT2 custom capture library 0.5–2.9 Mb (Agilent Technologies, Santa Clara, CA, USA), in accordance with the manufacturer’s workflow. Briefly, 150 ng of genomic DNA was used for a restricted reaction and hybridized using SureSelect XT Low Input reagents. All indexed DNA samples were amplified by PCR and sequenced using the Miseq platform (Illumina, San Diego, CA, USA). The results were analyzed using SureCall 3.0 (Agilent Technologies).
Transcript Analysis
Total RNA was isolated from urine-derived cells using an RNeasy Mini Kit (QIAGEN Hilden, Germany). Urine-derived cells were cultured in accordance with a previously reported protocol (9). Total RNA was reverse-transcribed into cDNA using Ecodry Premix (Double Primed) (Takara Bio Inc., Kusatsu, Japan), and PCR was performed. The primers used are shown in Supplemental Table 2.
Immunofluorescence Analysis of Laminin α5
Double immunostaining for laminin α5 and laminin β2 or type IV collagen α5 chain was performed using frozen kidney tissues. Each sample was fixed with acetone for 10 minutes, followed by heat-induced epitope retrieval. After blocking with 10% goat serum (Vector Laboratories, Burlingame, CA, USA), samples were incubated with primary mAbs against the laminin α5 chain (AMAb91124) (Atlas Antibodies AB, Stockholm, Sweden), laminin β2 chain (sc-20777, H-300) (Santa Cruz Biotechnology, Dallas, CA, USA), or type IV collagen α5 chain (H53) (Shigei Medical Research Institute, Okayama, Japan) overnight. Bound mAbs were detected using specific secondary antibodies, Alexa Fluor 546 goat anti-rabbit IgG and Alexa Fluor 488 goat anti-rat IgG, for 90 minutes. Finally, samples were incubated with DAPI for 5 minutes to stain nuclei.
Construction of Expression Vectors
Expression vectors for human laminin α5 (LMα5-pcDNA3.1), β1 (LMβ1-pCEP4), and FLAG-tagged γ1 (LMγ1FLAG-pcDNA3.1) were prepared as described previously (10–12). An expression vector for a human laminin α5 chain mutant with mouse Ig κ-chain (Ig-κ) leader sequence and 10×His-tag was prepared as follows. A cDNA encoding the Ig-κ leader sequence and 10×His-tag sequence (LMα5His-pcDNA3.1) was prepared by extension PCR using LMα5-pcDNA3.1 as a template. The primers used are shown in Supplemental Table 3.
The resultant cDNA fragment was inserted into the HindIII and SacII sites of LMα5-pcDNA3.1 using GeneArt Seamless Cloning and Assembly Kit (Thermo Fisher Scientific). Expression vectors for two human LAMA5 truncation mutants, Arg2720* and Arg3078*, were prepared by extension PCR using a LMα5His-pcDNA3.1 as a template. Resultant cDNA fragments were inserted into the BbrCI and AscI sites of LMα5His-pcDNA3.1 using GeneArt Seamless Cloning and Assembly Kit. The DNA sequence of the resulting vectors was confirmed using an ABI PRISM 3130 Genetic Analyzer.
In Vitro Secretion of Wild-Type LM511 and Its Chain-Termination Mutants
Wild-type LM511 and its mutants were transiently expressed in the FreeStyle 293 Expression System (Thermo Fisher Scientific), according to the manufacturer's instructions. Briefly, 293-F cells were simultaneously transfected with expression vectors for laminin α5His, β1, and γ1FLAG and grown for 3 days. To inhibit the proteolytic cleavage between laminin globular (LG) domain 3 and 4, 200 µg/ml heparin was added to the culture medium at 4 hours after transfection. At 72–75 hours after transfection, the cells were collected and washed with D-PBS without divalent cations. Cell pellets were lysed with lysis buffer (50 mM HEPES-NaOH [pH 7.4], 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.1% [w/v] sodium dodecyl sulfate, 1% [w/v] sodium deoxycholate, 1% [v/v] Triton X-100, and protease inhibitor cocktail, [FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan]). The conditioned media and cell lysates were clarified by centrifugation and analyzed by SDS-PAGE under reducing conditions, followed by immunoblotting with mAbs against horseradish peroxidase (HRP)–conjugated PentaHis (for laminin α5 chain, QIAGEN). Bound antibodies were visualized using ECL Western Blotting Detection Reagents (Cytiva). Band densities were quantified using ImageJ packaged in Fiji (13).
To confirm the heterotrimerization of LMα5 chain-termination mutants in LM511, the conditioned media were incubated with anti–human laminin α5 mAb 5D6 (14) and Protein G Sepharose 4 Fast Flow (Cytiva, Tokyo, Japan) or anti–FLAG M2 agarose (Sigma) overnight at 4°C. After centrifugation, resultant immunoprecipitates were separated by SDS-PAGE under reducing conditions, followed by immunoblotting with mAbs HRP-conjugated PentaHis (for laminin α5), DG10 (for laminin β1), C12SW, and anti–FLAG mAb M2 (for laminin γ1). After reaction with HRP-conjugated donkey anti–mouse IgG pAb (Jackson ImmunoResearch, PA, USA), the antigen-antibody complexes were visualized using ECL Western Blotting Detection Reagents (Cytiva).
Results
Patients
A total of 407 unrelated patients (216 males and 191 females) were included for genetic analysis by targeted sequencing using NGS. LAMA5 (NM_005560.4) gene variants were detected in four patients in three families. The genetic and concise clinical information for patients 1 and 2 was already published in our previous report (15), but no precise clinical and pathologic findings were described. Table 1 summarizes the characteristics of our four patients and the seven patients from previous reports with LAMA5 variants.
Table 1.
Characteristics of 11 patients
| ID | Age of Follow-up, yr | Sex | Age at Onset | Age at ESKD | Biopsy Findings | Ethnic Origin | Diagnosis | Gene Variants | dbSNP | ACMG | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | Neph236 | 13 | M | 3 mo | 1 yr | ND | Japanese | CNS | c.1282+1G>A c.9232C>T, p.(Arg3078*) |
rs1168208691 rs369268267 |
Pathogenic |
| Patient 2 | Neph236 sister | 5 | F | 4 mo | 3.5 yr | ND | Japanese | INF | c.1282+1G>A c.9232C>T, p.(Arg3078*) |
Pathogenic | |
| Patient 3 | Neph341 | 6 | F | 6 mo | 3.3 yr | DMS | Japanese | INF | c.1282+1G>A c.8158C>T, p.(Arg2720*) |
Pathogenic | |
| Patient 4 | Neph428 | 9 | F | 8 yr | ND | FSGS | Japanese | SRNS | c.1493C>T, p.(Ala498Val) c.8399G>A, p.(Arg2800His) |
rs139957521 rs142801594 |
Likely pathogenic |
| (2) | 07-0430-00016 | 38 | F | 27 yr | ND | FSGS | Black | Proteinuria | Ser1469Ala Val2440Ile |
VUS | |
| (3) | A4389-21 | M | 3 yr | No | ND | Turkey | SSNS | c.2239C>T, p.(Arg747Trp) c.2239C>T, p.(Arg747Trp) |
rs370940497 | VUS | |
| (3) | A4389-22 | M | 22 mo | No | ND | Turkey | SSNS | c.2239C>T, p.(Arg747Trp) c.2239C>T, p.(Arg747Trp) |
rs370940497 | VUS | |
| (3) | B150-21 | M | 4 yr | No | FSGS | Egypt | SRNS | c.3002A>G, p.(Glu1001Gly) c.3002A>G, p.(Glu1001Gly) |
VUS | ||
| (3) | B150-22 | F | 1.5 yr | 6 yr | FSGS | Egypt | SRNS | c.3002A>G, p.(Glu1001Gly) c.3002A>G, p.(Glu1001Gly) |
VUS | ||
| (3) | B1284-21 | M | 4 yr | No | ND | Arabic | SSNS/SDNS | c.8842G>A, p.(Gly2948Ser) c.8842G>A, p.(Gly2948Ser) |
rs529211517 | VUS | |
| (4) | M | 2 yr | 2 yr | FSGS | Italian | SRNS | c.857G>T, p.(Arg286Leu) c.857G>T, p.(Arg286Leu) |
Pathogenic |
M, male; F, female; dbSNP, the Single Nucleotide Polymorphism Database; ACMG, the American College of Medical Genetics and Genomics; ND, no data; INF, infantile nephrotic syndrome; DMS, diffuse mesangial sclerosis; SSNS, steroid-sensitive nephrotic syndrome; SDNS, steroid-dependent nephrotic syndrome; VUS, variants of unknown significance.
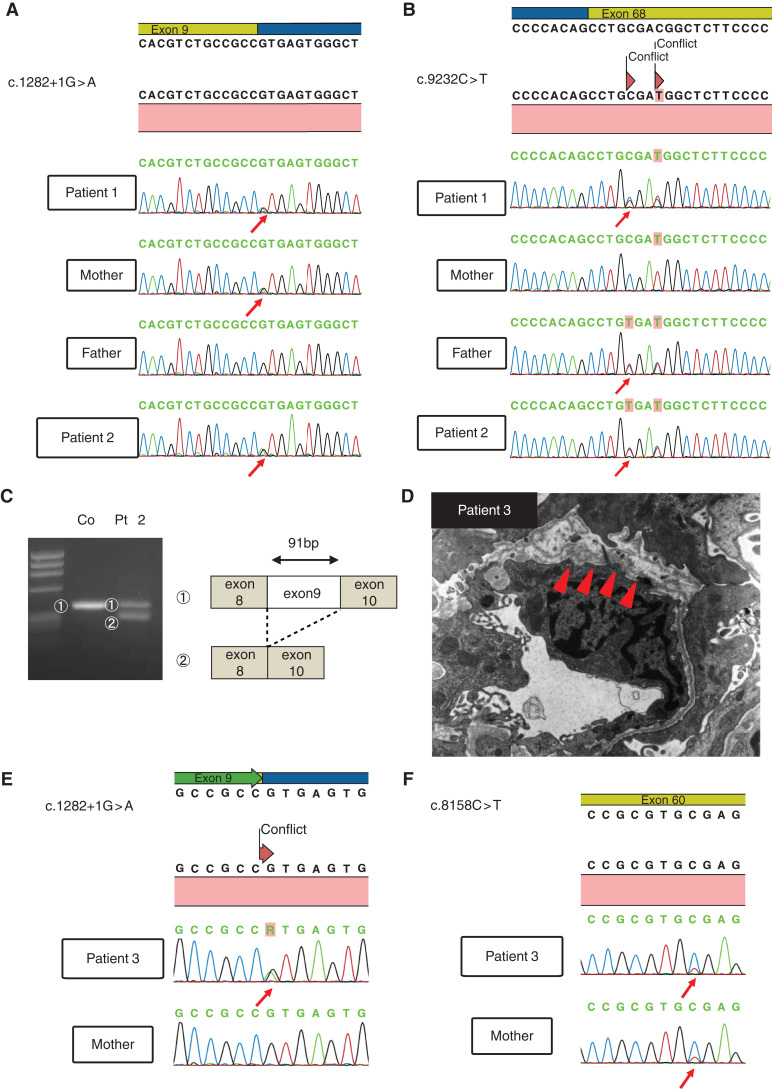
Patient 1 was a 13-year-old boy. At 3 months of age, his edema was detected by chance when he had a fever. Laboratory findings were as follows: serum total protein 3.9 g/dl; serum albumin 1.8 g/dl; serum creatinine, 0.55 mg/dl; and cholesterol 940 mg/dl. Urinalysis showed heavy proteinuria. He was diagnosed with nephrotic syndrome. Because his renal function was gradually deteriorating, peritoneal dialysis was started at the age of 1 year. We conducted genetic testing using targeted sequencing of the genes known to be responsible for inherited kidney disease and detected compound heterozygous variants in LAMA5. The variants include a heterozygous G to A substitution at base position 1282+1 in intron 9 (IVS9+1G>A) and a heterozygous C to T substitution at base position 9232 in exon 68, which replaces the amino acid arginine with a stop codon at codon 3078 (p.Arg3078*) (Figure 1, A and B). IVS9+1G>A led to the entire exon 9 (91 bp) being skipped at the transcript level (Figure 1C), meaning this is also a truncating variant. The patient’s parents each carried a different variant in heterozygous form.
Figure 1.
Genetic and pathologic analyses of patients 1, 2, and 3 reveal variants in LAMA5 and glomerular basement membrane (GBM) defects in patient 3. (A), (B) LAMA5 gene sequencing analysis of patients 1 and 2 and their parents. A splice site variant (c.1282+1G>A) and a nonsense variant (c.9232C>T) were detected in patients 1 and 2 (the sister). The nonsense variant is linked to a nearby variant (c.9235C>T). Each parent carried a different one of these variants in heterozygous form. Red arrows indicate the heterozygous variants in the chromatograms. (C) RT-PCR analysis of LAMA5 RNA from patient 2’s urinary cells using primers in exons flanking the c.1282+1G>A variant reveals a full length and a shorter amplicon. Sanger sequencing (Supplemental Figure 1) indicates the shorter product lacks exon 9, indicating exon skipping resulting in a frameshift. (D) Electron microscopic analysis of patient 3′s kidney biopsy reveals irregular appearance of the GBM (arrowheads) and podocyte foot process effacement. (E), (F) Genetic analysis of patient 3 and her mother. The same splice site variant found in patients 1 and 2 (c.1282+1G>A) and a novel nonsense variant (c.8185C>T) were detected in patient 3. The mother carried the nonsense variant.
The patient’s mother was phenotypically healthy and did not have proteinuria. The patient’s father had hypoplastic kidney, but he did not have proteinuria and did not reach ESKD.
Patient 2 was a 5-year-old girl. She is patient 1’s younger sister. At 4 months of age, edema and hypoalbuminemia were detected. Urinalysis showed heavy proteinuria, and she was diagnosed with nephrotic syndrome. She did not have any extrarenal abnormalities. Peritoneal dialysis was started at the age of 3.5 months. She carried the same variants as patient 1.
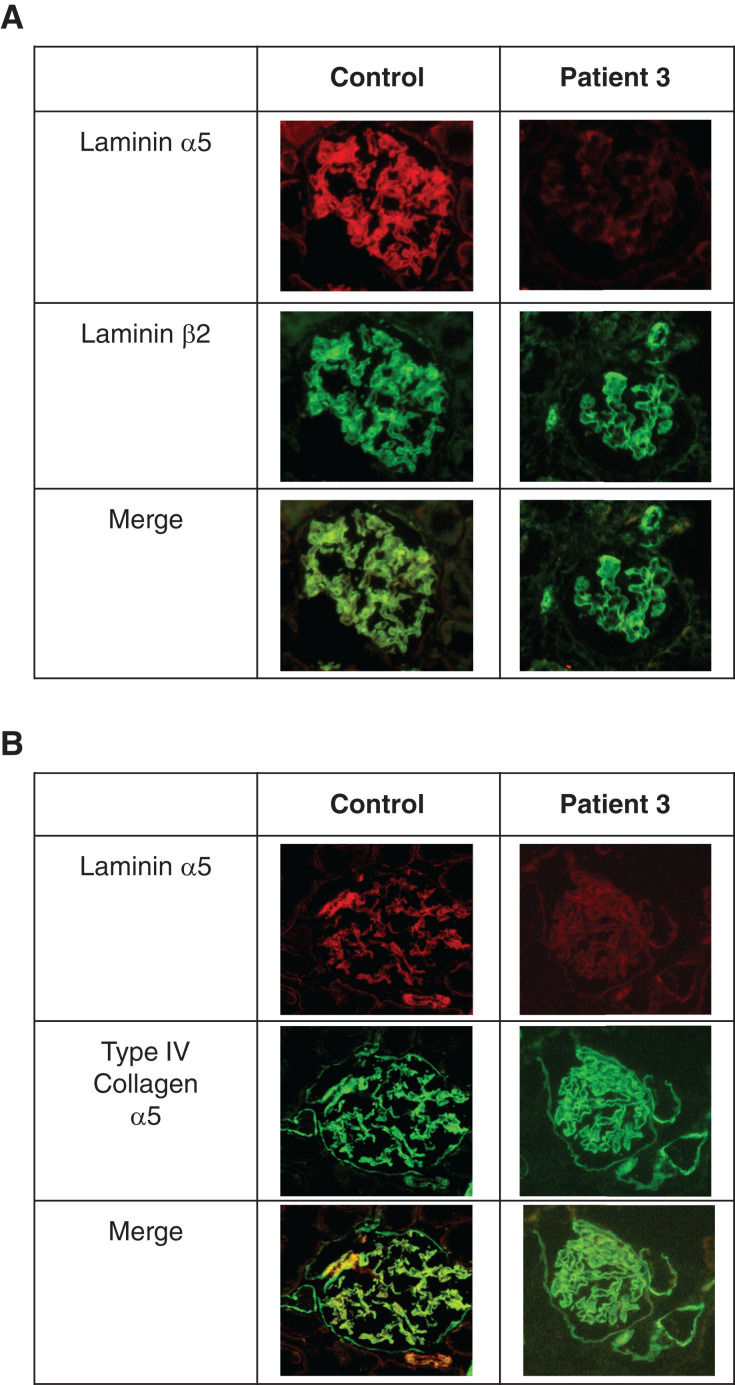
Patient 3 was a 6-year-old girl in whom hypoalbuminemia was detected by chance when she had been infected with the virus at 6 months of age. She had congenital cataract and hypoplastic kidney. There was no family history of kidney diseases. She was diagnosed with infantile nephrotic syndrome and started steroid therapy. However, she did not respond to immunosuppressive therapy. At 11 months of age, she underwent a renal biopsy. Pathologically, light microscopy demonstrated diffuse mesangial sclerosis, and electron microscopy demonstrated thinning and irregular structure of the GBM (Figure 1D). She reached ESKD at 3 years and 3 months of age. We conducted targeted sequencing analysis using NGS and found compound heterozygous variants in LAMA5. The variants include a heterozygous G to A substitution at base position 1282+1 in intron 9 (the same as patients 1 and 2), and a heterozygous C to T substitution at base position 8158 in exon 60, which replaces the amino acid arginine with a stop codon at codon 2720 (p.Arg2720*) (Figure 1, E and F). We conducted immunostaining analysis of laminin α5. In the control, laminin α5 showed the characteristic linear GBM pattern; however, patient 3 showed weakly positive staining for laminin α5, although laminin β2 and collagen α5 (IV) showed normal patterns (Figure 2, A and B). This suggests that mutant laminin α5 could form laminin-521 heterotrimers, although levels in the GBM appear reduced.
Figure 2.
Immunofluorescence analysis reveals reduced levels of laminin α5 in the GBM of patient 3. (A) Immunostaining for laminin α5 and laminin β2 in control and patient 3 shows the characteristic linear GBM patterns in the control, but patient 3 showed a very weak linear pattern for laminin α5. (B) Immunostaining for laminin α5 and collagen IV α5 in control and patient 3 shows the characteristic linear GBM patterns in the control, but patient 3 showed a very weak linear pattern for laminin α5.
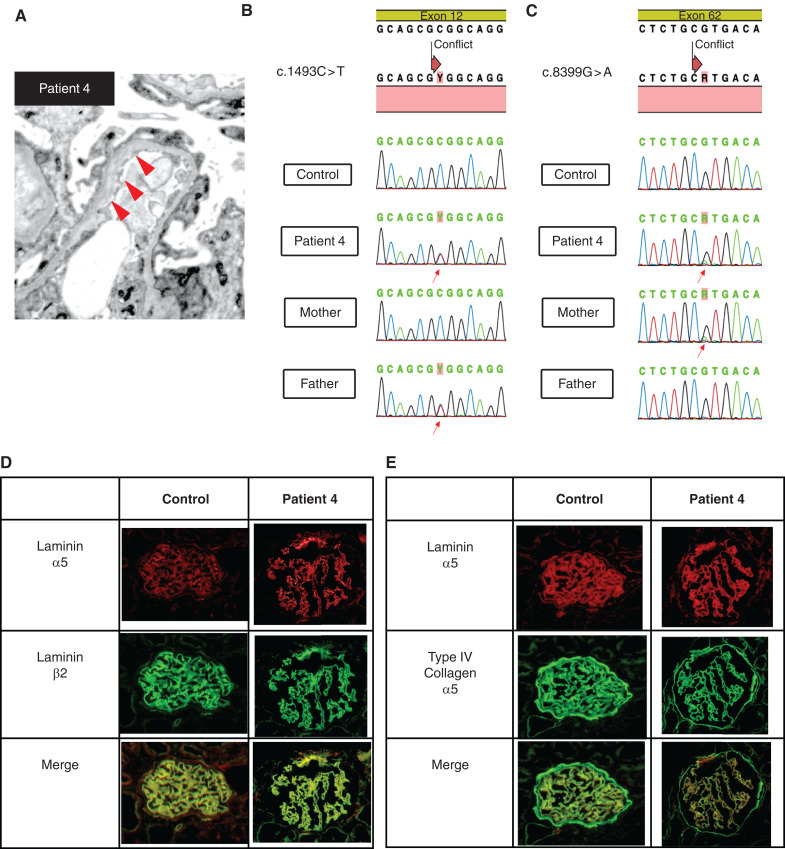
Patient 4 was a 9-year-old girl. At 8 years old, she was referred to the hospital because of proteinuria, which had been identified in a school-based urine screening program. Urinalysis showed heavy proteinuria. She did not have any extrarenal abnormalities. She was diagnosed with nephrotic syndrome and started steroid therapy. She underwent a renal biopsy because of SRNS. Pathologically, light microscopy demonstrated FSGS cellular variant, and electron microscopy demonstrated thinning and irregular structure of the GBM (Figure 3A). We conducted targeted sequencing analysis using NGS. She had compound heterozygous missense variants in LAMA5. The variants include a heterozygous C to T substitution at base position 1493 in exon 12, which replaces the amino acid alanine with valine at codon 498 (p.Ala498Val) and a heterozygous G to A substitution at base position 8399 in exon 62, which replaces the amino acid arginine with histidine at codon 2800 (p.Arg2800His) (Figure 3, B and C). We conducted immunostaining analysis of laminin α5. In the control, laminin α5 shows a characteristic linear GBM pattern; patient 4 showed the same degree of expression of laminin α5 as control (Figure 3, D and E). The patient’s parents each carried a different variant in heterozygous form. They were phenotypically healthy and did not have proteinuria.
Figure 3.
Genetic and pathologic analyses of patient 4 and her parents reveals LAMA5 missense variants and a GBM defect. (A) Electron microscopy shows an irregular appearance of the GBM. (B), (C) Genetic analysis of LAMA5 reveals compound heterozygous missense variants c.1493C>T (p.Ala498Val) and c.8399G>A (p.Arg2800His). Each parent carried a different one of these variants in heterozygous form. Red arrows indicate the heterozygous variants in the chromatograms. (D) Immunofluorescence analysis reveals normal levels of laminin α5 in the GBM of patient 4. Immunostaining for laminin α5 and laminin β2 in control and patient 4 shows the characteristic strong, linear GBM patterns. (E) Immunostaining for laminin α5 and collagen IV α5 in control and patient 4 shows the characteristic strong, linear GBM patterns.
In Vitro Analysis
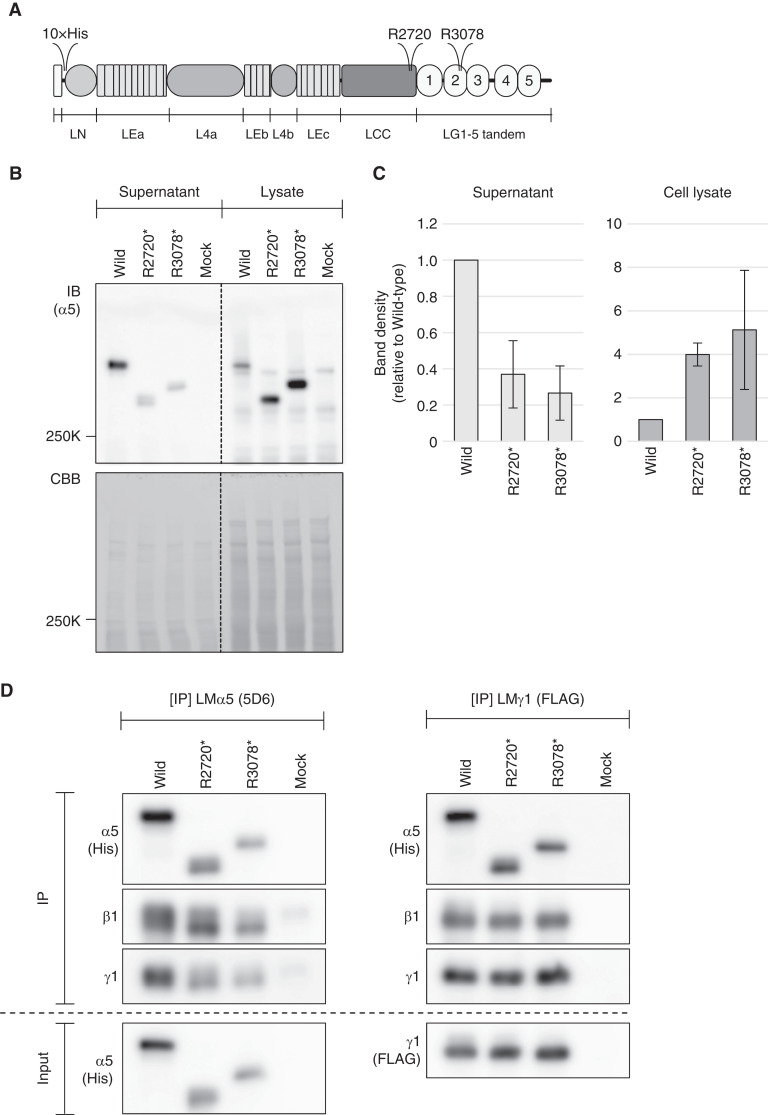
To investigate the potential pathogenicity of the two nonsense variants, LMα5-Arg2720* and -Arg3078* (Figure 4A), we assayed their synthesis and secretion in the context of the LM-511 trimer. We expressed either wild-type or mutant tagged human LMα5 along with tagged LMβ1 and γ1 to produce either wild-type or mutant LM-511 trimers by cotransfecting 293-F cells with three expression vectors: LMα5His-pcDNA3.1, LMβ1-pCEP4, and LMγ1FLAG-pcDNA3.1. Conditioned media were separated by SDS-PAGE, followed by immunoblotting using the mAb against the 5×His tag at the amino-terminus of LMα5. This analysis showed that both truncating mutants, LMα5-Arg2720* and -Arg3078*, exhibited a decreased trimer secretion level by about 40% and 30%, respectively, compared with wild-type (Figure 4, B and C). Consistent with these results, the signal intensities in cell lysates were increased by 4–5 fold for the mutants compared with wild-type (Figure 4, B and C).
Figure 4.
Secretion efficiencies of wild-type LM-511 and its laminin α5 chain-termination mutants. (A) Domain structure of the laminin α5 chain. The sites of truncation in patients 1 and 2 (R3078) and in patient 3 (R2720) are positioned in laminin globular (LG) domain 2 and in the carboxyl-terminal region of the laminin coiled-coil (LCC) domain, respectively. (B) The amount of wild-type and mutant LM-511 expressed in and secreted from cells. Equal amounts of conditioned media and cell lysates were applied to SDS-PAGE under reducing conditions. Laminin α5 chains are detected with HRP-conjugated anti-5×His tag mAb (upper panel). Proteins blotted on polyvinylidene difluoride membranes were visualized with Coomassie brilliant blue (CBB) and used as loading controls (lower panel). (C) Band densities in (B) were quantified using ImageJ software. Data were expressed as average±SD. (D) α5-R2720* and -R3078* mutants heterotrimerize with laminin β1 and γ1. 6×His tagged laminin α5 and the two chain-termination mutants were coexpressed with laminin β1 and FLAG tagged γ1 chains. Conditioned media were immunoprecipitated with anti-laminin α5 mAb (left panel) or anti-FLAG M2 mAb agarose (right panel). Immunoprecipitates were analyzed by SDS-PAGE under reducing conditions, followed by immunoblotting with mAbs against 5×His tag, β1, and γ1. Conditioned media were used as input samples.
To further explore whether secreted LMα5-Arg2720* and -Arg3078* mutants can trimerize with laminin β1 and γ1, we performed immunoprecipitation using mAbs against the human laminin α5 chain (5D6) and the FLAG tag at the amino-terminus of the laminin γ1 chain. Immunoblotting analyses showed that laminin β1 and γ1 were detected in mAb 5D6 immunoprecipitates (Figure 4D, left panel). When the γ1 chain was immunoprecipitated with anti-FLAG mAb, laminin α5 and β1 chains were detected in the precipitates (Figure 4D, right panel). These results indicate that both α5-Arg2720* and -Arg3078* mutants are able to assemble with laminin β1 and γ1 to yield a trimer, but compared with wild-type, secretion efficiencies of the mutant LM-511s were decreased by an unidentified intracellular quality control mechanism. This is in good agreement with our in vivo findings that the level of laminin α5 was decreased in glomeruli of patient 3.
Discussion
This paper presents three infantile patients with nephrotic syndrome with LAMA5 gene biallelic truncating variants and one patient with SRNS with LAMA5 biallelic missense variants. For our patients, we performed in vitro heterotrimer formation assays and demonstrated defects in the function of truncating variants. This is the first report to show clear evidence that LAMA5 pathogenic variants can lead to nephrotic syndrome in the absence of major developmental anomalies. Our patients suggest the need for awareness of LAMA5 gene screening in patients with early-onset proteinuria.
Three of our patients (patients 1, 2, and 3) possess biallelic truncating variants. One was a nonsense variant, and the other was a consensus donor splice-site variant that led to skipping of exon 9 (91 bp) and a frameshift. Therefore, both should be considered disease-causing pathogenic variants. Although one of our patients (patient 4) had only missense variants, pathologically and clinically these variants should also be considered disease causing. Until now, several pathogenic and likely pathogenic variants, including splice sites and frameshifts, have been reported in Clinvar database. Limited to variants for which phenotypic information was available, only seven patients who are proteinuric with biallelic LAMA5 variants were reported, and all of them were missense variants. The first patient was an adult female with proteinuria beginning at age 27, as reported by Chatterjee et al. (2). Recently, using whole-exome sequencing, homozygous missense variants in LAMA5 were identified in five pediatric patients with nephrotic syndrome in three families (3). All five patients had nephrotic syndrome with onset before 4 years of age. One of them did not respond to immunosuppressive treatment and reached ESKD at age 6, but in contrast, other patients responded to immunosuppressive treatment. This indicates significant phenotypic heterogeneity in this disease, which resembles somewhat the situation for LAMB2 gene variants (16).
Our three patients with truncations (patients 1, 2, and 3) showed more severe phenotypes than the patients with missense variants, but not lethality. Lama5 null mice die at late fetal stages with multiple development defects (17), and variant of Lama5 specifically in podocytes causes nephrotic syndrome (8). One reported homozygous variant in human LAMA5 (Arg286Leu) that impairs laminin polymer formation causes a complex syndromic disorder in addition to FSGS and ESKD (4), and the overall features (including syndactyly) are consistent with the phenotypes of the Lama5 null mouse. That patients 1, 2, and 3 with biallelic truncating variants survived the fetal period and manifested either no or mild extrarenal symptoms suggests the presence of truncated LMα5 proteins maintains significant LM-521 functions. Our in vitro biochemical analysis using recombinant truncated proteins explained this phenotype.
Laminins are a group of cross-shaped heterotrimeric proteins, each consisting of α, β, and γ subunits joined together through a coiled coil domain (16). Three mutants we characterized were able to form trimers because the coiled-coil domain was preserved in each truncating variant. On the basis of the results of our in vitro trimer formation assay, mutant LM-521 trimers should form, although the laminin α5 proteins would be truncated due to the nonsense variants. Although secretion of the mutant trimers into patient 3’s GBM was observed, the level was low. This weak accumulation of incomplete laminin α5 likely saved the lives of our patients.
Renal pathologic findings showed irregular architecture of the GBM with similarity to Alport or Pierson syndrome in patients 3 and 4. These were the novel findings of LAMA5-related nephrosis. Variant of the mouse Lama5 gene results in varying degrees of proteinuria and rates of progression to nephrotic syndrome (8). The GBM of proteinuric mice appeared thickened, with a moth-eaten appearance (8). These mouse pathologic findings were in concordance with our patients. We also performed immunofluorescence analysis that showed weak deposition of laminin α5, but it was not negative in patient 3. This result was somewhat surprising to us, because we did not expect biallelic truncating variants to produce laminin α5 able to form laminin-521 trimers. However, our in vitro trimer formation assay clearly proved that the two truncated laminin α5 variants can trimerize. Patient 4 showed a laminin α5 expression level similar to the control. This result is in concordance with the clinical course of preserved renal function in patient 4. In limitation, we did not quantify the fluorescent intensity.
The age of onset in patients 1–3 was under 1 year. In congenital and infantile nephrotic syndrome patients, monogenetic disease is typically suspected. In the PodoNet study, genetic disease was identified in 24% of patients with SRNS; the most commonly mutated genes were NPHS2, WT1, and NPHS1 (18). In Japan, genetic components were identified in 30% of patients who were proteinuric; the most commonly mutated genes were WT1, NPHS1, and INF2. However, comprehensive genetic testing was performed before the publication of a report of pathogenic homozygous LAMA5 variants in pediatric nephrotic syndrome in 2019 (3). Among the patients diagnosed with congenital nephrotic syndrome or infantile nephrotic syndrome for which the causative gene has not been identified, LAMA5 biallelic truncated variants, such as in three of our four patients, may be the hidden cause. It is therefore necessary to consider LAMA5 as one of the causative genes of congenital/infantile nephrotic syndrome.
We have reported three patients with infantile nephrotic syndrome with LAMA5 biallelic truncating variants and one patient with SRNS with LAMA5 missense variants. Our patients showed clear evidence that LAMA5 biallelic variants can cause SRNS. We think it is likely that patients with nephrotic syndrome with these types of LAMA5 variants are so rare because only specific types of LAMA5 variants are compatible with life.
Disclosures
J. Miner reports having consultancy agreements with Alpha Insights, AstraZeneca, Bridge Bio, Deerfield Management, Janssen Biotech Inc., GLG Council, Kurma, Mantra Bio, National Institutes of Health, Retrophin, and The Planning Shop; reports receiving research funding from Chinook Therapeutics and Reneo Pharmaceuticals; reports receiving honoraria from Japanese Society of Pharmacology, NephCure Kidney International, Western Michigan University Medical School, and University of Kansas Medical Center; reports patents and inventions with Angion, Eli Lilly, Genentech, Kerafast, and Maze Therapeutics; reports being a scientific advisor or member of Journal of Clinical Investigation Consulting Editor, Kidney International Editorial Board, Matrix Biology Editorial Board, and Matrix Biology Plus Editorial Board; and reports other interests/relationships with the Alport Syndrome Foundation (Scientific Advisory Research Network) and the American Society for Matrix Biology (President-Elect). K. Iijima reports having consultancy agreements with JCR Pharmaceuticals Co., Kyowa Hakko Kirin Co., Ono Pharmaceutical Co., Sanofi K.K., Takeda Pharmaceutical Co., and Zenyaku Kogyo Co.; reports receiving research funding from Air Water Medical Inc., Astellas Pharma Inc., Eisai Co., JCR Pharmaceutical Co., Mochida Pharmaceutical Co., Nihon Pharmaceutical Co. Otsuka Pharmaceutical Co., Shionogi & Co., Zenyaku Kogyo Co.; reports receiving honoraria from Astellas Pharma Inc., Chugai Pharmaceutical Co., Integrated Development Associates Co., Kyowa Hakko Kirin Co., Shionogi & Co., and Zenyaku Kogyo Co.; reports being a scientific advisor or member of Clinical Journal of the American Society of Nephrology and the Pediatric Nephrology Editorial Board. K. Nozu reports receiving research funding from Chugai Pharmaceutical Company, Dainippon Sumitomo Pharmaceutical Company, Kyowa Kirin Pharmaceutical Company, and Shionogi Inc.; reports patents and inventions with Daiichi Sankyo Pharmaceutical Company; reports being a scientific advisor or member of Toa Eiyo Ltd.; and reports receiving lecture fees from Chugai Pharmaceutical Co., Daiichi Sankyo Company, Kyowa Kirin Pharmaceutical Co., Novartis Pharma Co., and Sumitomo Dainippon Pharma Co. K. Sekiguchi reports having an ownership interest in and receiving research funding from Matrixome, Inc.; and reports patents and inventions with Osaka University. T. Horinouchi reports receiving research funding from Otsuka Pharmaceutical Co. Y. Taniguchi reports being a Project Leader at Matrixome Inc. All remaining authors have nothing to disclose.
Funding
This study was supported by Grants-in-Aid for Scientific Research (KAKENHI) from the Ministry of Education, Culture, Sports, Science and Technology of Japan grant 18K15713 (to C.N.), the Collaborative Research Program of Institute for Protein Research, Osaka University grant CR-21-06 (to Y.T. and K.S.), and the National Institutes of Health grants R01DK078314 and R01DK058366 (to J.H.M.).
Footnotes
See editorial, “Pathogenic LAMA5 Variants and Kidney Disease,” on pages 1876–1879.
Author Contributions
K. Iijima and H. Nagase were responsible for the data curation; Y. Aoto, T. Horinouchi, S. Ishik, A. Kondo, S. Nagai, R. Rossanti, N. Sakakibara, K. Sekiguchi, and T. Yamamura were responsible for the investigation; H. Sakaguchi, N. Sugawara, A. Tashiro, and C. Umeda were responsible for the resources; K. Nozu provided supervision; C. Nagano and Y. Taniguchi wrote the original draft; and J. Miner reviewed and edited the manuscript.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0004952021/-/DCSupplemental
Genes targeted by next-generation sequencing analysis. Download Supplemental Table 1, PDF file, 685 KB (684.1KB, pdf)
The primers for PCR. Download Supplemental Table 2, PDF file, 685 KB (684.1KB, pdf)
The primer sets for extension PCR. Download Supplemental Table 3, PDF file, 685 KB (684.1KB, pdf)
Direct sequencing for transcript analysis. Download Supplemental Figure 1, PDF file, 685 KB (684.1KB, pdf)
References
- 1.Preston R, Stuart HM, Lennon R: Genetic testing in steroid-resistant nephrotic syndrome: Why, who, when and how? Pediatr Nephrol 34: 195–210, 2017. 10.1007/s00467-017-3838-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatterjee R, Hoffman M, Cliften P, Seshan S, Liapis H, Jain S: Targeted exome sequencing integrated with clinicopathological information reveals novel and rare mutations in atypical, suspected and unknown cases of Alport syndrome or proteinuria. PLoS One 8: e76360, 2013. 10.1371/journal.pone.0076360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun DA, Warejko JK, Ashraf S, Tan W, Daga A, Schneider R, Hermle T, Jobst-Schwan T, Widmeier E, Majmundar AJ, Nakayama M, Schapiro D, Rao J, Schmidt JM, Hoogstraten CA, Hugo H, Bakkaloglu SA, Kari JA, El Desoky S, Daouk G, Mane S, Lifton RP, Shril S, Hildebrandt F: Genetic variants in the LAMA5 gene in pediatric nephrotic syndrome. Nephrol Dial Transplant 34: 485–493, 2019. 10.1093/ndt/gfy028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones LK, Lam R, McKee KK, Aleksandrova M, Dowling J, Alexander SI, Mallawaarachchi A, Cottle DL, Short KM, Pais L, Miner JH, Mallett AJ, Simons C, McCarthy H, Yurchenco PD, Smyth IM: A mutation affecting laminin alpha 5 polymerisation gives rise to a syndromic developmental disorder. Development 147: dev189183, 2020. 10.1242/dev.189183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funk SD, Lin MH, Miner JH: Alport syndrome and Pierson syndrome: Diseases of the glomerular basement membrane. J Int Soc Matrix Biol 71–72: 250–261, 2018. 10.1016/j.matbio.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miner JH, Patton BL, Lentz SI, Gilbert DJ, Snider WD, Jenkins NA, Copeland NG, Sanes JR: The laminin alpha chains: Expression, developmental transitions, and chromosomal locations of α1-5, identification of heterotrimeric laminins 8-11, and cloning of a novel α3 isoform. J Cell Biol 137: 685–701, 1997. 10.1083/jcb.137.3.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miner JH, Lewis RM, Sanes JR: Molecular cloning of a novel laminin chain, α5, and widespread expression in adult mouse tissues. J Biol Chem 270: 28523–28526, 1995. 10.1074/jbc.270.48.28523 [DOI] [PubMed] [Google Scholar]
- 8.Goldberg S, Adair-Kirk TL, Senior RM, Miner JH: Maintenance of glomerular filtration barrier integrity requires laminin α5. J Am Soc Nephrol 21: 579–586, 2010. 10.1681/ASN.2009091004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazzeri E, Ronconi E, Angelotti ML, Peired A, Mazzinghi B, Becherucci F, Conti S, Sansavini G, Sisti A, Ravaglia F, Lombardi D, Provenzano A, Manonelles A, Cruzado JM, Giglio S, Roperto RM, Materassi M, Lasagni L, Romagnani P: Human urine-derived renal progenitors for personalized modeling of genetic kidney disorders. J Am Soc Nephrol 26: 1961–1974, 2015. 10.1681/ASN.2014010057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ido H, Harada K, Futaki S, Hayashi Y, Nishiuchi R, Natsuka Y, Li S, Wada Y, Combs AC, Ervasti JM, Sekiguchi K: Molecular dissection of the α-dystroglycan- and integrin-binding sites within the globular domain of human laminin-10. J Biol Chem 279: 10946–10954, 2004. 10.1074/jbc.M313626200 [DOI] [PubMed] [Google Scholar]
- 11.Hayashi Y, Kim KH, Fujiwara H, Shimono C, Yamashita M, Sanzen N, Futaki S, Sekiguchi K: Identification and recombinant production of human laminin α4 subunit splice variants. Biochem Biophys Res Commun 299: 498–504, 2002. 10.1016/S0006-291X(02)02642-6 [DOI] [PubMed] [Google Scholar]
- 12.0.012w?>Ido H, Ito S, Taniguchi Y, Hayashi M, Sato-Nishiuchi R, Sanzen N, Hayashi Y, Futaki S, Sekiguchi K: Laminin isoforms containing the γ3 chain are unable to bind to integrins due to the absence of the glutamic acid residue conserved in the C-terminal regions of the γ1 and γ2 chains. J Biol Chem 283: 28149–28157, 2008. 10.1074/jbc.M803553200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A: Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682, 2012. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiwara H, Kikkawa Y, Sanzen N, Sekiguchi K: Purification and characterization of human laminin-8. Laminin-8 stimulates cell adhesion and migration through α3β1 and α6β1 integrins. J Biol Chem 276: 17550–17558, 2001. 10.1074/jbc.M010155200 [DOI] [PubMed] [Google Scholar]
- 15.Nagano C, Yamamura T, Horinouchi T, Aoto Y, Ishiko S, Sakakibara N, Shima Y, Nakanishi K, Nagase H, Iijima K, Nozu K: Comprehensive genetic diagnosis of Japanese patients with severe proteinuria. Sci Rep 10: 270, 2020. 10.1038/s41598-019-57149-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matejas V, Hinkes B, Alkandari F, Al-Gazali L, Annexstad E, Aytac MB, Barrow M, Bláhová K, Bockenhauer D, Cheong HI, Maruniak-Chudek I, Cochat P, Dötsch J, Gajjar P, Hennekam RC, Janssen F, Kagan M, Kariminejad A, Kemper MJ, Koenig J, Kogan J, Kroes HY, Kuwertz-Bröking E, Lewanda AF, Medeira A, Muscheites J, Niaudet P, Pierson M, Saggar A, Seaver L, Suri M, Tsygin A, Wühl E, Zurowska A, Uebe S, Hildebrandt F, Antignac C, Zenker M: Mutations in the human laminin β2 (LAMB2) gene and the associated phenotypic spectrum. Hum Mutat 31: 992–1002, 2010. 10.1002/humu.21304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miner JH, Cunningham J, Sanes JR: Roles for laminin in embryogenesis: Exencephaly, syndactyly, and placentopathy in mice lacking the laminin α5 chain. J Cell Biol 143: 1713–1723, 1998. 10.1083/jcb.143.6.1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trautmann A, Lipska-Ziętkiewicz BS, Schaefer F: Exploring the clinical and genetic spectrum of steroid resistant nephrotic syndrome: The PodoNet Registry. Front Pediatr 6: 200, 2018. 10.3389/fped.2018.00200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes targeted by next-generation sequencing analysis. Download Supplemental Table 1, PDF file, 685 KB (684.1KB, pdf)
The primers for PCR. Download Supplemental Table 2, PDF file, 685 KB (684.1KB, pdf)
The primer sets for extension PCR. Download Supplemental Table 3, PDF file, 685 KB (684.1KB, pdf)
Direct sequencing for transcript analysis. Download Supplemental Figure 1, PDF file, 685 KB (684.1KB, pdf)