Key Points
A computable phenotype combines routinely collected data elements from the EHR with logic elements to identify a condition of interest.
This validated computable phenotype has strong classification characteristics to identify individuals with primary nephrotic syndrome.
This computable phenotype for primary nephrotic syndrome can facilitate future research of these rare diseases.
Keywords: glomerular and tubulointerstitial diseases, computable phenotype, nephrotic syndrome, validation
Visual Abstract
Abstract
Background
Primary nephrotic syndromes are rare diseases which can impede adequate sample size for observational patient-oriented research and clinical trial enrollment. A computable phenotype may be powerful in identifying patients with these diseases for research across multiple institutions.
Methods
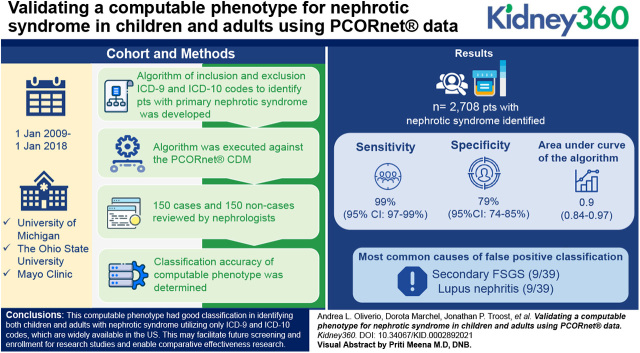
A comprehensive algorithm of inclusion and exclusion ICD-9 and ICD-10 codes to identify patients with primary nephrotic syndrome was developed. The algorithm was executed against the PCORnet CDM at three institutions from January 1, 2009 to January 1, 2018, where a random selection of 50 cases and 50 noncases (individuals not meeting case criteria seen within the same calendar year and within 5 years of age of a case) were reviewed by a nephrologist, for a total of 150 cases and 150 noncases reviewed. The classification accuracy (sensitivity, specificity, positive and negative predictive value, F1 score) of the computable phenotype was determined.
Results
The algorithm identified a total of 2708 patients with nephrotic syndrome from 4,305,092 distinct patients in the CDM at all sites from 2009 to 2018. For all sites, the sensitivity, specificity, and area under the curve of the algorithm were 99% (95% CI, 97% to 99%), 79% (95% CI, 74% to 85%), and 0.9 (0.84 to 0.97), respectively. The most common causes of false positive classification were secondary FSGS (nine out of 39) and lupus nephritis (nine out of 39).
Conclusion
This computable phenotype had good classification in identifying both children and adults with primary nephrotic syndrome utilizing only ICD-9 and ICD-10 codes, which are available across institutions in the United States. This may facilitate future screening and enrollment for research studies and enable comparative effectiveness research. Further refinements to the algorithm including use of laboratory data or addition of natural language processing may help better distinguish primary and secondary causes of nephrotic syndrome.
Introduction
Nephrotic syndrome is defined as the confluence of heavy proteinuria (>3.5 g per day), hypoalbuminemia, and edema (1). This syndrome is further classified by kidney histopathologic findings, including minimal change disease (MCD), FSGS, and membranous nephropathy (MN). It is well recognized that this syndrome and these histologic findings can occur secondarily to systemic disorders, viruses, malignancies, or medications, or may occur in isolation, termed primary nephrotic syndrome. In addition, even when excluding presumed secondary causes of nephrotic syndrome, those remaining have heterogeneous disease features spanning their molecular mechanisms (2,3), histopathology (4), responsiveness to treatment, and outcomes (5,6).
Primary nephrotic syndromes are rare diseases: MCD is estimated to affect 0.23–15.6 per 100,000 children and 0.06 per 100,000 adults per year, FSGS 0.2–1.1 per 100,000 per year, and MN 1.2 per 100,000 per year (7). Their rarity and heterogeneity contribute to sample size limitations, thus performing rigorous comparative effectiveness and outcomes research is challenging. However, this work is important and necessary for patients and their caregivers, who prioritize outcomes of kidney health and survival, while articulating that their lived experiences, symptoms, and functionality remain important in the assessment of successful disease management (8).
A computable phenotype is a clinical condition, characteristic, or set of clinical features that can be determined solely from the data in electronic health records (EHRs) using an algorithm and ancillary data sources without requiring chart review or interpretation by a clinician (9). Routinely collected data elements from the EHR combined with logic elements, such as AND, OR, and IF, are used to identify a clinical condition or characteristic of interest from a given data source. Applications of machine learning models with natural language processing can also develop probabilistic predictions for phenotyping, which can identify patients with disease by supervised learning from example patients (10). Valid computable phenotype algorithms for rare conditions such as nephrotic syndrome could identify large, robust, and representative retrospective cohorts that otherwise would be cumbersome to recruit prospectively. Additionally, this could improve feasibility assessment and site selection for clinical trials. Phenotypes can be replicated across data sources and health care organizations to generate consistent cohort identification, which can streamline enrollment in registries and facilitate comparative effectiveness research.
PCORnet, the National Patient-Centered Clinical Research Network, incorporates data from millions of patients across nine clinical research networks and two health plan research networks, including more than 300 hospitals. Data are maintained locally in participating health systems and have a shared structure (common data model [CDM]) that facilitates application of a computable phenotype (11). Computable phenotypes developed and utilized for research within PCORnet include those for type II diabetes mellitus (12,13) and heart failure (14). Other examples of computable phenotypes in action include the Chronic Conditions Data Warehouse (15), which utilizes administrative claims from Centers for Medicare and Medicaid Services, and the Clinical Classifications Software developed as part of the Healthcare Cost and Utilization Project (16). Prior computable phenotypes developed for glomerular diseases have been shown to be reliable and valid, but focused on the pediatric population and utilized data elements that are not universally available, including provider type and Systematized Nomenclature of Medicine Clinical Terms codes (17).
We sought to develop and validate a computable phenotype to identify both adult and pediatric patients with primary nephrotic syndrome, utilizing universally available data elements to reliably capture prevalent patients in the PCORnet CDM.
Materials and Methods
Patient Definition and Computable Phenotype Development
Electronic medical record data was originally extracted for 12,233 patients seen for glomerular disease and proteinuria at the University of Michigan from 2009 to 2014, with a broad range of diagnoses. A comprehensive list of intelligent medical object (IMO) codes from patient problem lists and International Classification of Diseases, 9th Revision, Clinical Modification (ICD-9-CM) codes was generated from this population, and the list of codes was reviewed by three investigators. The codes were cross-referenced with biopsy-proven kidney diseases attributed to primary nephrotic syndrome, or for patients without biopsy the clinician’s diagnosis in the chart was reviewed. Preliminary inclusion codes were established for nephrotic syndrome utilizing those that are largely defined by the pathologic lesion determined by kidney biopsy. However, these codes do not specify if the nephrotic syndrome is idiopathic or secondary to another underlying condition, thus exclusion codes were selected for conditions known to cause secondary nephrotic syndrome (e.g., diabetes, lupus, viral illnesses). Inclusion and exclusion criteria were refined on the basis of these codes, existing literature, and clinical rationale, then the codes and an initial algorithm were iteratively revised to optimize sensitivity and specificity in two health systems. Subsequently, as part of the Renal Collaborative Research Group within PCORnet, these codes were reviewed by a team of clinician scientists in 2018. Tools for mapping ICD-9-CM to ICD-10-CM codes were utilized so that ICD-10-CM codes could be added to facilitate searching beyond 2015 (18). IMO codes were removed to improve the generalizability of the algorithm, due to lack of universal availability of IMO codes in EHR warehouses.
The final list of codes and subsequent computable phenotype, which defined inclusion to the primary nephrotic syndrome patients, and those that defined exclusion, are listed in Supplemental Table 1 and the algorithm scripts using SQL code logic can be accessed at https://github.com/AoliverioUM/nephroticsyndrome-computablephenotype. Patient cases with primary nephrotic syndrome were defined across all ages as subjects that were seen for at least one encounter of any kind, had ≥2 nephrotic syndrome codes, and did not have exclusion codes for diabetes mellitus, systemic lupus erythematosus, amyloidosis, or other less common causes of secondary nephrotic syndrome, such as HIV, viral hepatitis, GN, obstructive uropathy, or mitochondrial metabolism disorders at any time. Two or more nephrotic syndrome codes were required to minimize false positives. Additionally, for individuals aged <20 years, codes of nephrotic syndrome not otherwise specified were also allowable for inclusion, given the majority of children with nephrotic syndrome are steroid sensitive, and consequently a kidney biopsy is pursued less frequently in this demographic. Patient noncases were randomly selected individuals not meeting case criteria who were seen within the same calendar year and were within 5 years of age of the cases.
Data Extraction
PCORnet data are maintained locally with a shared structure in the CDM (19,20). The developed computable phenotype was executed in SQL against the PCORnet CDM at three academic institutions (University of Michigan, Ann Arbor, MI; The Ohio State University, Columbus, OH; Mayo Clinic, Rochester, MN, Jacksonville, FL, and Scottsdale/Phoenix, AZ) from January 1, 2009 to January 1, 2018. From the data extraction, a random selection of 50 cases and 50 noncases was generated at each institution for chart validation.
Data coordination was conducted at Arbor Research for Health and the University of Michigan. An independent review board deemed the study exempt from institutional review board approval for data coordination. Institutional review board approval for this study with a waiver of individual patient consent was obtained at each individual study site where charts were abstracted.
Validation
As the gold-standard comparator, a nephrologist at each institution performed a comprehensive chart review of each generated case and noncase to determine concordance with the algorithm. When available, kidney biopsy data and nephrologist-written documentation were carefully reviewed for all cases detected by the computable phenotype. Covariates collected included date of case capture by the computable phenotype, ESKD, and transplant status at time of capture, details of kidney biopsy, and, in patients with discordance between algorithm-defined and nephrologist-defined primary nephrotic syndrome, the potential causes of discordance. After individual chart review, all three sites participated in consensus conference to facilitate any necessary additional adjudication to reach a consensus nephrologist definition.
Statistical Analyses
The classification accuracy including sensitivity, specificity, positive and negative predictive value, and F1 score of the computable phenotype was determined. Meta-analytic summary statistics for sensitivity, specificity, and area under the curve were calculated using a hierarchical summary receiver operating characteristic model (21). A sensitivity analysis was performed to examine the classification accuracy of the algorithm in adult patients only (≥20 years of age). Descriptive statistics were used to describe each of the classification groups (true positives, false positives, false negatives, true negatives).
Results
At the time of execution of the computable phenotype against the PCORnet CDM, there were 1,365,050 individual patients in the University of Michigan CDM, 1,072,626 in the Ohio State University CDM, and 1,867,416 in the Mayo Clinic CDM. The algorithm generated 666 patients with primary nephrotic syndrome at University of Michigan, 321 at Ohio State University, and 1721 at Mayo Clinic. Classification statistics by study center are shown in Table 1. Of 150 patients identified as having primary nephrotic syndrome by the computable phenotype, 111 were determined to be true positives by nephrologist review. Using meta-analytic summary statistics, the overall sensitivity of the computable phenotype was 99% (95% confidence interval [95% CI], 97% to 99%), specificity of 79% (95% CI, 74% to 85%), and area under the curve of 0.9 (95% CI, 0.84 to 0.97). Given a minority of patients captured by our computable phenotype were <20 years old (n=37 across all three centers) and a computable phenotype for glomerular diseases including nephrotic syndrome in pediatric patients utilizing PEDSnet is available (17), a sensitivity analysis of the computable phenotype classification statistics solely in adults aged ≥20 years was performed. The performance of the computable phenotype in adults was similar and is shown in Supplemental Table 2.
Table 1.
Performance characteristics of the computable phenotype at three academic PCORnet centers
| Center | True Positive | False Positive | False Negative | True Negative | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value | Accuracy | F1 Score | Area Under Curve (95% Confidence Interval) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| University of Michigan | 42 | 8 | 1 | 49 | 0.98 | 0.86 | 0.84 | 0.98 | 0.91 | 0.90 | 0.92 (0.87 to 0.97) |
| The Ohio State University | 35 | 15 | 0 | 50 | 1.00 | 0.77 | 0.70 | 1.00 | 0.85 | 0.82 | 0.88 (0.83 to 0.94) |
| Mayo Clinic | 34 | 16 | 0 | 50 | 1.00 | 0.76 | 0.68 | 1.00 | 0.84 | 0.81 | 0.88 (0.83 to 0.93) |
Clinical characteristics and diagnoses for each classification category (true positive, false positive, true negative, false negative are described in more detail in Tables 2 and 3). The most common misclassifications were due to secondary FSGS and membranous lupus nephritis (World Health Organization Class V). Compared with patients with true positive nephrotic syndrome, false positive individuals were not more likely to be on dialysis (10% versus 10%, respectively) or have a kidney transplant (21% versus 23%, respectively) at the time of capture by the computable phenotype than those that were correctly classified as having primary nephrotic syndrome. The vast majority of both true positives and false positives were seen by nephrologists within the health care system queried, 99% and 97%, respectively. Among the 150 noncases identified by the computable phenotype, only 8% had seen a nephrologist within the health care system. The sole false negative generated by the algorithm was a patient with FSGS and a kidney transplant. Of the 149 true negatives, two patients were on dialysis at the time of capture and one had received a kidney transplant.
Table 2.
Diagnosis and demographic characteristics of true and false positives identified by the computable phenotype
| Characteristics | True Positives n=111 |
False Positives n=39 |
|---|---|---|
| Nephrologist-validated diagnosis, n (%) | ||
| MCD | 18 (16) | 0 |
| FSGS | 38 (34) | 0 |
| MN | 45 (41) | 0 |
| SSNS/SRNS | 4 (4) | 0 |
| Othera | 6 (5) | 0 |
| Secondary FSGS | 0 | 9 (23) |
| Lupus nephritis | 0 | 9 (23) |
| Diabetic kidney disease | 0 | 4 (10) |
| Focal global glomerulosclerosis | 0 | 2 (5) |
| Vasculitis | 0 | 2 (5) |
| Hypertensive nephrosclerosis | 0 | 1(3) |
| Other secondary causeb | 0 | 9 (23) |
| Unknown | 0 | 3 (8) |
| ESKD status, n (%) | ||
| Dialysis | 11 (10) | 4 (10) |
| Kidney transplant | 25 (23) | 8 (21) |
| Age, yr, mean (SD) | 43.3 (20.9) | 44.9 (19.2) |
| Nephrology encounter at site, n (%) | 110 (99) | 38 (97) |
| Time from nephrology encounter to computable phenotype capture, days, median (IQR) | 396 (0–1792) | 543 (16–2279) |
| Biopsy status, n (%) | N/A | |
| Documented and reviewed | 83 (75) | |
| Not documented or not performed | 28 (25) |
MCD, minimal change disease; MN, membranous nephropathy; SSNS/SRNS, steroid sensitive nephrotic syndrome, steroid resistant nephrotic syndrome; IQR, interquartile range.
Other primary etiologies included four patients with congenital nephrotic syndrome, one membranoproliferative glomerulonephritis, and one lipoprotein glomerulonephropathy.
Other secondary etiologies included two patients with interstitial nephritis, and one each of secondary nephrotic syndrome due to lymphoma, Alport syndrome, obstructive nephropathy, acute mediated rejection in transplant, immune complex glomerulonephritis, hemolytic uremic syndrome, and Kawasaki’s disease.
Table 3.
Diagnosis and demographic characteristics of true and false negatives identified by the computable phenotype
| Characteristics | True Negatives n=149 |
False Negatives n=1 |
|---|---|---|
| Nephrologist-validated diagnosis, n (%) | ||
| MCD | 0 | 0 |
| FSGS | 0 | 1 |
| MN | 0 | 0 |
| SSNS/SRNS | 0 | 0 |
| ESKD status | ||
| Dialysis | 2 | 0 |
| Kidney transplant | 1 | 1 |
| Age, yr, mean (SD) | 44.3 (20.3) | 29 |
| Nephrology encounter at site, n (%) | 11 (7) | 1 (100) |
MCD, minimal change disease; MN, membranous nephropathy; SSNS/SRNS: steroid sensitive nephrotic syndrome, steroid resistant nephrotic syndrome.
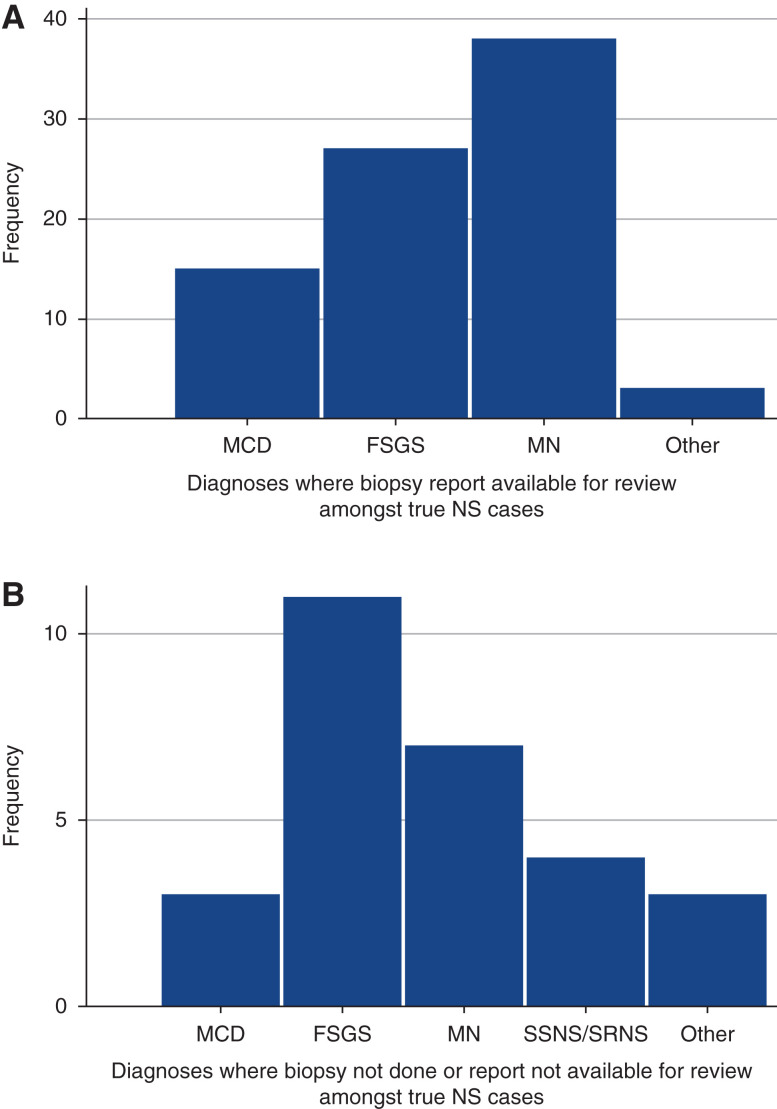
Of the true positives, 75% (83 out of 111) of patients had biopsy reports available for review in the medical record, whereas 25% (28 out of 111) did not. The frequency of each unique nephrotic syndrome diagnosis, by biopsy status, is shown in Figure 1. MN was the most commonly identified diagnosis with a biopsy report available, followed by FSGS. FSGS was the most common diagnosis when a true positive primary nephrotic syndrome patient was confirmed by chart validation, but a biopsy report was not available for review.
Figure 1.
Frequency of true positive primary nephrotic syndrome diagnoses, by biopsy review status. (A) n=83 patients with true positive primary nephrotic syndrome with biopsies reviewed by chart validation. Other diagnoses consisted of one of each: congenital nephrotic syndrome, lipoprotein glomerulopathy, and membranoproliferative glomerulonephritis. (B) n=28 patients with true positive primary nephrotic syndrome, where biopsies were not performed or unable to be reviewed by chart validation. Other diagnoses consisted of three patients with congenital nephrotic syndrome. MCD, minimal change disease; MN, membranous nephropathy; SSNS, steroid sensitive nephrotic syndrome; SRNS, steroid resistant nephrotic syndrome; NS, nephrotic syndrome.
Discussion
A good computable phenotype is explicit, reproducible, reliable, and valid (22). This study demonstrates that a reliable and valid computable phenotype for primary nephrotic syndrome with an overall sensitivity of 99%, specificity of 79%, and area under the curve of 0.9 can be created using data elements from ICD-9-CM and ICD-10-CM codes alone, which are readily available across health systems in the United States. This improves the generalizability of this computable phenotype when compared with others, which have relied on IMO or Systematized Nomenclature of Medicine Clinical Terms codes as well. Using three academic institutions in the PCORnet CDM, this computable phenotype captured a total of 2708 patients with primary nephrotic syndrome; on the basis of classification statistics determined by this study, this is estimated to represent 559 prevalent patients with true primary nephrotic syndrome at the University of Michigan, 225 at The Ohio State University, and 1170 patients at Mayo Clinic over 8 years—a significant achievement, given the rarity of primary nephrotic syndrome and the fraction of the CDM that was utilized for validation.
Further refinements to the computable phenotype developed in our study may also be feasible to improve specificity. Distinguishing primary and secondary FSGS can be difficult for clinicians and similarly proved a challenge to the computable phenotype. Secondary FSGS and focal global glomerulosclerosis collectively were a significant source of misclassification of primary nephrotic syndrome by the computable phenotype. In addition, 11 patients who were true positives were validated as primary FSGS, despite the lack of a biopsy report available for review. Although degree of foot process effacement may vary between primary and secondary maladaptive types of FSGS (e.g., obesity-related FSGS), the clinical history of overt sudden onset nephrotic syndrome, response to immunosuppression, and absence of causes of secondary FSGS allowed reviewers to adequately support a diagnosis of primary FSGS (23). Similarly, refinement of the computable phenotype for improved specificity may be possible through incorporating medication data elements, laboratory values, or utilizing natural language processing to distinguish between primary and secondary FSGS. Natural language processing is a field of computer science and artificial intelligence that performs computational analysis of human language and is often used together with machine learning. It has emerged as a technique that can be used to identify patients with specific diagnoses by analysis of electronic medical record documentation and has been used with success in liver (24) and pulmonary disease (25). This approach may also be useful in distinguishing MN secondary to lupus nephritis. Strengthening our exclusion codes to include those for diabetes irrespective of the presence of kidney disease may also help minimize false positives. However, this may also increase false negatives in patients with true concurrent disease that could be relevant for future study (for example, when immunosuppression induces diabetes mellitus).
One limitation of our computable phenotype validation study was the inability to selectively draw noncases from nephrology clinics at the study sites; the CDM at the time of our analysis did not contain data on specific providers or clinics within a given institution. Given that primary nephrotic syndromes are rare diseases, drawing noncases from a general health care population likely inflated our sensitivity and negative predictive value statistics. For this reason, we also assessed the F1 score of the computable phenotype. An F1 score is a useful measure in classification when seeking to balance precision (the share of the predicted patients who are positive that are correct) and recall (the share of the actual positive patients that are predicted correctly) when there is a large number of true negatives, as expected in this population. The F1 score of the computable phenotype ranged from 0.81 to 0.9 at the individual study sites; an F1 score of 1 is considered perfect and our results indicate the computable phenotype has an acceptable balance of precision and recall. Another limitation is that our algorithm was also developed initially within two health systems and then revised and tested specifically for PCORnet; however, there was some variation in classification statistics between study sites. This likely represents differences in coding practices across institutions. There may also be geographic differences; although all three primary research sites are located in the Midwest, all Mayo Clinic clinical sites (Minnesota, Florida, and Arizona) contributed data to the validation study, which could have lent greater variation in provider coding practices. Our study did use ICD-9-CM and ICD-10-CM codes exclusively to help with generalizability and the PCORnet CDM to allow implementation across hundreds of hospitals, but we cannot be certain our computable phenotype will perform as well in other large datasets or EHRs. Finally, 50 patients from each site were selected at random from all ages. Given the low numbers of pediatric patients in our validation set, we are unable to differentially examine how the computable phenotype compares in adults versus children. However, a computable phenotype for pediatric glomerular disease has been rigorously evaluated in PEDSnet previously and serves as another excellent resource for researchers (17).
Despite these limitations, the computable phenotype will improve practitioners’ abilities to identify patients with primary nephrotic syndrome for clinical trials. This will facilitate rapid and realistic assessments of site feasibility and improve enrollment for much-needed research to improve health outcomes in these rare diseases. Use of our computable phenotype and EHR data more broadly throughout PCORnet also has the potential to strengthen pragmatic comparative effectiveness research and observational research by using “real-world” data.
In summary, we developed and validated a reliable and accurate computable phenotype for identification of primary nephrotic syndromes in children and adults using ICD-9-CM and ICD-10-CM codes. To better improve our understanding of these rare diseases, we must utilize all available tools, from genome to phenome (26), and EHR data can generate large-scale observational clinical data, a better understanding of how patients are treated and respond to treatments, and the complications they face, augmenting our base of knowledge.
Disclosures
C. Tran reports being a scientific advisor or member of Frontiers in Pediatrics Review Editor on the Editorial Board of Pediatric Nephrology. D. Gipson reports having consultancy agreements, through the University of Michigan, with AstraZeneca, Boehringer Ingelheim, Roche/Genentech, and Vertex Pharmaceuticals; reports receiving research funding, through the University of Michigan, from Atrium Health Medical Foundation, Boehringer Ingelheim, Centers for Disease Control, Food and Drug Administration, Goldfinch Bio, National Institutes of Health, Novartis, Reata, and Travere; and reports being a scientific advisor or member of the American Society of Pediatric Nephrology, American Society of Nephrology, and International Society of Pediatric Nephrology. H. Desmond reports receiving research funding through a percentage of salary from the University of Michigan, funded by Boehringer Ingelheim. I. Ayoub reports receiving honoraria from the American College of Rheumatology; reports being a scientific advisor or member of the Journal of Clinical Nephrology (Editorial Board) and the Lupus Foundation of America (advisory board). L. Mariani reports having consultancy agreements with, and receiving honoraria from, Calliditas Therapeutics Advisory Board, CKD Advisory Committee, Reata Pharmaceuticals, and Travere Therapeutics Advisory Board; reports receiving research funding from Boehringer Ingelheim; reports receiving honoraria from American Society of Nephrology Board Review Course and Update; and reports being a scientific advisor or member of Calliditas Therapeutics, Reata Pharmaceuticals, and Travere Therapeutics. M. Denburg reports having consultancy agreements with Trisalus Life Sciences (spouse); reports having an ownership interest in In-Bore LLC (spouse) and Precision Guided Interventions LLC (spouse); reports receiving research funding from Mallinckrodt; reports being a scientific advisor or member of NKF Delaware Valley Medical Advisory Board and Trisalus Life Sciences Scientific Advisory Board (spouse); and reports other interests/relationships with the American Society of Pediatric Nephrology Research and Program Committees and the National Kidney Foundation Pediatric Education Planning Committee. M. Matheny reports consultancy agreements with National Institutes of Health- Veterans Affairs- Department of Defense Pain Management Grant Consortium (PMC3); reports being a scientific advisor or member of Scientific Merit Review Board Study Section, VA Health Services Research and Development (HSR&D), Informatics and Methods Section, the Steering Committee of Indianapolis VA HSR&D Center of Innovation; and the Steering Committee of VA HSR&D VA Information Resource Center. S. Almaani reports having consultancy agreements with Aurinia Pharmaceuticals and Kezar Life Sciences; reports receiving research funding from Gilead Sciences; reports being a scientific advisor or member Clinical Nephrology Editorial Board; and reports speakers bureau from Aurinia Pharmaceuticals. S. Massengill reports having consultancy agreements with Guidepoint Group; reports being a scientific advisor or member of the Editorial Board of online Glomerular Disease Journal (Karger Publishers). All remaining authors have nothing to disclose.
Funding
This work was supported by NephCure Kidney Network for Patients with Nephrotic Syndrome, Denburg (PI) 07/01/18-03/30/19 grant PPRN-1306-04903, PPRN Phase II; National Institute of Diabetes and Digestive and Kidney Diseases grants 5T32DK007378-39 (June 2018–2019), 5KL2TR002241-04 (March 2020–September 2020), K23 DK123413 (September 2020–) (to A.L. Oliverio), and K08 DK115891 (to L.H. Mariani); and the National Center for Advancing Translational Sciences for the Michigan Institute for Clinical and Health Research (UL1TR002240) (to J.P. Troost).
Footnotes
See editorial, “Optimizing the Electronic Health Record for Clinical Research: Has the Time Come?” on pages 1880–1881.
Author Contributions
M. Denburg, D. Gipson, L. Mariani, and M. Matheny conceptualized the study; S. Almaani, I. Ayoub, H. Desmond, D. Gipson, J. Greco, D. Marchel, L. Mariani, S. Massengill, A. Oliverio, and C. Tran were responsible for data curation; D. Gipson, L. Mariani, A. Oliverio, and J. Troost were responsible for formal analysis; M. Denburg and L. Mariani were responsible for funding acquisition; M. Denburg, C. Dorn, D. Gipson, M. Matheny, and J. Troost were responsible for the methodology; H. Desmond and A. Oliverio were responsible for the project administration; C. Dorn and M. Matheny were responsible for software; D. Gipson and L. Mariani provided supervision; L. Mariani was responsible for the resources and visualization; S. Almaani, I. Ayoub, H. Desmond, J. Greco, D. Marchel, L. Mariani, A. Oliverio, and C. Tran were responsible for validation; A. Oliverio was responsible for the project administration, and wrote the original draft; S. Almaani, I. Ayoub, M. Denburg, H. Desmond, C. Dorn, D. Gipson, J. Greco, D. Marchel, L. Mariani, M. Matheny, S. Massengill, A. Oliverio, C. Tran, and J. Troost reviewed and edited the manuscript.
Supplemental Material
This article contains the following supplemental material online at http://kidney360.asnjournals.org/lookup/suppl/doi:10.34067/KID.0002892021/-/DCSupplemental.
Nephrotic Syndrome Inclusion Codes (all individuals). Download Supplemental Table 1, PDF file, 206 KB (205.7KB, pdf)
Performance characteristics of the computable phenotype at 3 academic PCORnet centers, excluding individuals <20 years old. Download Supplemental Table 2, PDF file, 206 KB (205.7KB, pdf)
References
- 1.Group KDIGO : KDIGO clinical practice guideline for glomerulonephritis. Available at https://kdigo.org/wp-content/uploads/2017/02/KDIGO-Glomerular-Diseases-Guideline-2021-English.pdf. Accessed November 22, 2021 [Google Scholar]
- 2.Friedman DJ, Pollak MR: APOL1 nephropathy: From genetics to clinical applications. Clin J Am Soc Nephrol 16: 294–303, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merchant ML, Barati MT, Caster DJ, Hata JL, Hobeika L, Coventry S, Brier ME, Wilkey DW, Li M, Rood IM, Deegens JK, Wetzels JF, Larsen CP, Troost JP, Hodgin JB, Mariani LH, Kretzler M, Klein JB, McLeish KR: Proteomic analysis identifies distinct glomerular extracellular matrix in collapsing focal segmental glomerulosclerosis. J Am Soc Nephrol 31: 1883–1904, 2020. 10.1681/ASN.2019070696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariani LH, Martini S, Barisoni L, Canetta PA, Troost JP, Hodgin JB, Palmer M, Rosenberg AZ, Lemley KV, Chien HP, Zee J, Smith A, Appel GB, Trachtman H, Hewitt SM, Kretzler M, Bagnasco SM: Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant 33: 310–318, 2018. 10.1093/ndt/gfw443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troost JP, Trachtman H, Spino C, Kaskel FJ, Friedman A, Moxey-Mims MM, Fine RN, Gassman JJ, Kopp JB, Walsh L, Wang R, Gipson DS: Proteinuria reduction and kidney survival in focal segmental glomerulosclerosis. Am J Kidney Dis 77: 216–225, 2021. 10.1053/j.ajkd.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laurin LP, Gasim AM, Derebail VK, McGregor JG, Kidd JM, Hogan SL, Poulton CJ, Detwiler RK, Jennette JC, Falk RJ, Nachman PH: Renal survival in patients with collapsing compared with not otherwise specified FSGS. Clin J Am Soc Nephrol 11: 1752–1759, 2016. 10.2215/CJN.13091215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGrogan A, Franssen CF, de Vries CS: The incidence of primary glomerulonephritis worldwide: A systematic review of the literature. Nephrol Dial Transplant 26: 414–430, 2011. 10.1093/ndt/gfq665 [DOI] [PubMed] [Google Scholar]
- 8.Carter SA, Gutman T, Logeman C, Cattran D, Lightstone L, Bagga A, Barbour SJ, Barratt J, Boletis J, Caster D, Coppo R, Fervenza FC, Floege J, Hladunewich M, Hogan JJ, Kitching AR, Lafayette RA, Malvar A, Radhakrishnan J, Rovin BH, Scholes-Robertson N, Trimarchi H, Zhang H, Azukaitis K, Cho Y, Viecelli AK, Dunn L, Harris D, Johnson DW, Kerr PG, Laboi P, Ryan J, Shen JI, Ruiz L, Wang AY, Lee AHK, Fung S, Tong MK, Teixeira-Pinto A, Wilkie M, Alexander SI, Craig JC, Tong A; SONG-GD Investigators : Identifying outcomes important to patients with glomerular disease and their caregivers. Clin J Am Soc Nephrol 15: 673–684, 2020. 10.2215/CJN.13101019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collaboratory NHCSR : Electronic health records-based phenotyping, 2020. Available at: https://sites.duke.edu/rethinkingclinicaltrials/ehr-phenotyping/. Accessed November 11, 2020
- 10.Koola JD, Davis SE, Al-Nimri O, Parr SK, Fabbri D, Malin BA, Ho SB, Matheny ME: Development of an automated phenotyping algorithm for hepatorenal syndrome. J Biomed Inform 80: 87–95, 2018. 10.1016/j.jbi.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrest CB, McTigue KM, Hernandez AF, Cohen LW, Cruz H, Haynes K, Kaushal R, Kho AN, Marsolo KA, Nair VP, Platt R, Puro JE, Rothman RL, Shenkman EA, Waitman LR, Williams NA, Carton TW: PCORnet® 2020: Current state, accomplishments, and future directions. J Clin Epidemiol 129: 60–67, 2021. 10.1016/j.jclinepi.2020.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiese AD, Roumie CL, Buse JB, Guzman H, Bradford R, Zalimeni E, Knoepp P, Morris HL, Donahoo WT, Fanous N, Epstein BF, Katalenich BL, Ayala SG, Cook MM, Worley KJ, Bachmann KN, Grijalva CG, Rothman RL, Chakkalakal RJ: Performance of a computable phenotype for identification of patients with diabetes within PCORnet: The patient-centered clinical research network. Pharmacoepidemiol Drug Saf 28: 632–639, 2019. 10.1002/pds.4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachmann KN, Roumie CL, Wiese AD, Grijalva CG, Buse JB, Bradford R, Zalimeni EO, Knoepp P, Dard S, Morris HL, Donahoo WT, Fanous N, Fonseca V, Katalenich B, Choi S, Louzao D, O’Brien E, Cook MM, Rothman RL, Chakkalakal RJ: Diabetes medication regimens and patient clinical characteristics in the national patient-centered clinical research network, PCORnet. Pharmacol Res Perspect 8: e00637, 2020. 10.1002/prp2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tison GH, Chamberlain AM, Pletcher MJ, Dunlay SM, Weston SA, Killian JM, Olgin JE, Roger VL: Identifying heart failure using EMR-based algorithms. Int J Med Inform 120: 1–7, 2018. 10.1016/j.ijmedinf.2018.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Center for Medicare & Medicaid Services : Chronic conditions data warehouse, 2021. Available at: https://www2.ccwdata.org/web/guest/condition-categories. Accessed February 18, 2021
- 16.Agency for Healthcare Research and Quality : HCUP Clinical Classifications Software (CCS) for ICD-9-CM, 2017. Available at: https://www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp. Accessed November 12, 2020
- 17.Denburg MR, Razzaghi H, Bailey LC, Soranno DE, Pollack AH, Dharnidharka VR, Mitsnefes MM, Smoyer WE, Somers MJG, Zaritsky JJ, Flynn JT, Claes DJ, Dixon BP, Benton M, Mariani LH, Forrest CB, Furth SL: Using electronic health record data to rapidly identify children with glomerular disease for clinical research. J Am Soc Nephrol 30: 2427–2435, 2019. 10.1681/ASN.2019040365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agency for Healthcare Research and Quality : Impact of ICD-10-CM/PCS on research using administrative databases, 2016. Available at: https://www.hcup-us.ahrq.gov/reports/methods/methods.jsp. Accessed April 2, 2021
- 19.Collins FS, Hudson KL, Briggs JP, Lauer MS: PCORnet: Turning a dream into reality. J Am Med Inform Assoc 21: 576–577, 2014. 10.1136/amiajnl-2014-002864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS: Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 21: 578–582, 2014. 10.1136/amiajnl-2014-002747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macaskill P: Empirical Bayes estimates generated in a hierarchical summary ROC analysis agreed closely with those of a full Bayesian analysis. J Clin Epidemiol 57: 925–932, 2004. 10.1016/j.jclinepi.2003.12.019 [DOI] [PubMed] [Google Scholar]
- 22.Richesson RSM: Electronic health records-based phenotyping, 2020. Available at: https://sites.duke.edu/rethinkingclinicaltrials/ehr-phenotyping/#intro-definitions. Accessed November 12, 2020
- 23.De Vriese AS, Sethi S, Nath KA, Glassock RJ, Fervenza FC: Differentiating primary, genetic, and secondary FSGS in adults: A clinicopathologic approach. J Am Soc Nephrol 29: 759–774, 2018. 10.1681/ASN.2017090958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang EK, Yu CY, Clarke R, Hackbarth A, Sanders T, Esrailian E, Hommes DW, Runyon BA: Defining a patient population with cirrhosis: An automated algorithm with natural language processing. J Clin Gastroenterol 50: 889–894, 2016. 10.1097/MCG.0000000000000583 [DOI] [PubMed] [Google Scholar]
- 25.Weiner M, Dexter PR, Heithoff K, Roberts AR, Liu Z, Griffith A, Hui S, Schelfhout J, Dicpinigaitis P, Doshi I, Weaver JP: Identifying and characterizing a chronic cough cohort through electronic health records. Chest 159: 2346–2355, 2020 [DOI] [PubMed] [Google Scholar]
- 26.Mariani LH, Kretzler M: Pro: ‘The usefulness of biomarkers in glomerular diseases’. The problem: Moving from syndrome to mechanism: Individual patient variability in disease presentation, course and response to therapy. Nephrol Dial Transplant 30: 892–898, 2015. 10.1093/ndt/gfv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nephrotic Syndrome Inclusion Codes (all individuals). Download Supplemental Table 1, PDF file, 206 KB (205.7KB, pdf)
Performance characteristics of the computable phenotype at 3 academic PCORnet centers, excluding individuals <20 years old. Download Supplemental Table 2, PDF file, 206 KB (205.7KB, pdf)