Abstract
Human genetic studies support an inverse causal relationship between leukocyte telomere length (LTL) and coronary artery disease (CAD), but directionally mixed effects for LTL and diverse malignancies. Clonal hematopoiesis of indeterminate potential (CHIP), characterized by expansion of hematopoietic cells bearing leukemogenic mutations, predisposes both hematologic malignancy and CAD. TERT (which encodes telomerase reverse transcriptase) is the most significantly associated germline locus for CHIP in genome-wide association studies. Here, we investigated the relationship between CHIP, LTL, and CAD in the Trans-Omics for Precision Medicine (TOPMed) program (n = 63,302) and UK Biobank (n = 47,080). Bidirectional Mendelian randomization studies were consistent with longer genetically imputed LTL increasing propensity to develop CHIP, but CHIP then, in turn, hastens to shorten measured LTL (mLTL). We also demonstrated evidence of modest mediation between CHIP and CAD by mLTL. Our data promote an understanding of potential causal relationships across CHIP and LTL toward prevention of CAD.
Genetic analyses support opposing bidirectional causality for CHIP and LTL toward possible therapeutic intervention for CAD.
INTRODUCTION
Telomeres consist of repetitive DNA sequences with associated protective proteins (1), which stabilize chromosomes by several mechanisms (2). Shortening of telomeres during successive mitoses aims to protect the remaining chromosomal DNA. Reverse transcription by the telomerase complex mitigates telomere attrition in cells requiring frequent division such as hematopoietic stem cells (3–5). However, with aging, telomeres continue to shorten, and protective mechanisms are less efficient, leading to accumulation of senescent cells with shortened telomeres providing a fertile substrate for genomic instability (6, 7). Senescent cells also acquire a proinflammatory senescence-associated secretory phenotype (SASP), which promotes aging-related cardiovascular disease (8, 9).
While Mendelian randomization (MR) studies consistently support a causal relationship between shorter leukocyte telomere length (LTL) and coronary artery disease (CAD) (10, 11), the relationship between LTL and cancer is more complex (10–12). In vitro studies indicate that short telomeres promote genomic instability, thereby leading to malignancies (2, 13), and most tumor cells have shortened telomeres (14, 15). Mendelian disorders characterized by severe telomere shortenings, such as pediatric bone marrow failure and dyskeratosis congenita, manifest with premature aging, organ damage, and high rates of malignant blood disorders (16, 17). However, MR studies indicate that longer genetically imputed LTL (gLTL) may cause an increased incidence of various malignancies, including lung adenocarcinoma, glioma, melanoma, or leukemia (10–12, 18, 19).
Clonal hematopoiesis of indeterminate potential (CHIP), characterized by clonally expanded hematopoietic cells bearing leukemogenic mutations [most commonly in DNA methyltransferase 3 alpha (DNMT3A), Tet methylcytosine dioxygenase 2 (TET2), ASXL transcriptional regulator 1 (ASXL1), and Janus kinase 2 (JAK2)] without clinical hematologic disorders, represents a premalignant condition (20). CHIP strongly predicts future risk for myeloid malignancy and human and murine data indicate that CHIP is a causal risk factor for CAD as well (21–26). In cross-sectional analyses, CHIP correlates with shorter measured LTL (mLTL), after adjustment for age (27). However, similar to aforementioned cancer studies, in genome-wide association analyses of CHIP, the most significant risk allele resides in the TERT locus (28) (encoding telomerase reverse transcriptase) and is associated with longer mLTL (19). Whether and how CHIP and LTL have causal association are unknown, and whether this relationship influences CHIP-associated risk for CAD is unknown.
Here, we investigated the relationships between LTL, CHIP, and CAD to address these questions using mLTL and gLTL in the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) program (n = 63,302) and the UK Biobank (n = 47,080) (19, 29). After estimating the associations across mLTL, CHIP, and CAD, we assessed these associations for evidence supporting bidirectional causality using MR framework with gLTL. Last, we estimated the mediation effect of mLTL for the CHIP-associated CAD risk.
RESULTS
Baseline characteristics
We detected CHIP and estimated LTL with blood DNA-derived whole-genome sequencing (WGS) from the U.S. NHLBI TOPMed program (29) and whole-exome sequencing (WES) from the UK Biobank (30). In TOPMed, CHIP calls and LTL estimates (mLTL) were obtained from WGS analyses previously published (28, 31). We also obtained telomere measurements by classical methods (19) and compared them with WGS/WES-based estimation.
In TOPMed, after excluding kinship through second-degree relatives and those who have discordant information, 63,302 individuals had both indices measured, of whom 36,507 (57.7%) were female. The mean age was 54.3 years old (SD 18.0) at the time of blood draw, and 31,294 (48.6%) were of European ancestry (Fig. 1, fig. S1, and table S1). CHIP calls from the first WES samples released from UK Biobank were obtained (26, 30) with some modifications (see Materials and Methods), yielding 47,080 individuals after excluding related individuals (second degree) and discordance between genetic sex and self-reported sex with overlapped quantitative polymerase chain reaction (qPCR)–based telomere measurement (19). Among the UK Biobank participants included, 25,648 (54.4%) were female. The mean age was 56.5 years old (SD 8.0), and 43,907 (93.3%) were of European ancestry. In total, 3284 (TOPMed: 5.1%) and 2206 (UK Biobank: 4.7%) individuals had evidence of at least one CHIP-related mutation. Of these, 2862 (TOPMed: 4.5%) and 1008 (UK Biobank: 2.1%) individuals had variant allele frequency (VAF) greater than 0.10, a threshold previously associated with increased CAD incidence (table S1) (23, 24, 26). Here, we define VAF as the largest size of the clones in the same individual with multiple CHIP clones. Considering the improved detection of CHIP using WES, we included CHIP with VAF > 5% in subsequent meta analyses. All the CHIP detected in TOPMed had VAF > 5%, while 382 individuals detected as having CHIP in UK Biobank had VAF ≦ 5%.
Fig. 1. Analytical procedure in this study.
TOPMed (N = 63,302) and UK Biobank (N = 47,054) are the study cohorts. Mutect2 detected CHIP-associated mutations. Telomere length was estimated by TelSeq in TOPMed and qPCR (T/S ratio) in UK Biobank. We performed observational study and causal inference by bidirectional MR between LTL and CHIP. CHIP was associated with shorter LTL in the observational study. Germline genetic factors that increase CHIP development were associated with shorter LTL, whereas germline genetic factors that increase LTL were associated with developing CHIP. Mediation analysis adjusted for the measurable confounders detected the mediation effect of LTL on CHIP. CHIP, clonal hematopoiesis of indeterminate potential; LTL, leukocyte telomere length; TOPMed, Trans-Omics for Precision Medicine; UKBB, UK Biobank.
Comparison of telomere length estimation across methods
We compared available measurements of LTL (mLTL) across methods. The LTL estimation by TelSeq (32) using WGS was available from the previous work (31). We obtained LTL measurement by Southern blot in a subset of Women’s Health Initiative (WHI) (n = 686), which is also a part of TOPMed. We estimated LTL by TelSeq using 49,738 WES CRAM files in UK Biobank as previously done using WGS data in TOPMed. qPCR-based measurement in UK Biobank was available from the previous work. Consistent with restricted sequencing inherent to WES, the estimated absolute LTL (mean ± SD: 0.83 ± 0.13 kb) from WES in UK Biobank was much shorter than those estimated from WGS in TOPMed (mean ± SD: 3.27 ± 1.01 kb) or from conventional LTL measured by Southern blot (mean ± SD: 6.87 ± 0.62 kb) measured in WHI (fig. S2, A and B). Thus, TelSeq estimations were batch-corrected with the first nine principal components (PCs) generated by NGS-PCA (https://github.com/PankratzLab/NGS-PCA), which uses read coverage information. All LTL estimations were standardized to assess the relative value. We assessed 20 genetic variants previously reported as associated with mLTL (11) whether they were similarly associated with LTL estimation used in this study. All tested measurement correlated with those variants similarly with the previous report (TOPMed TelSeq: R2 = 0.83, P = 1.35 × 10−6; UK Biobank TelSeq: R2 = 0.71, P = 1.57 × 10−6; UK Biobank telomeric DNA/single copy gene (T/S) ratio: R2 = 0.83, P = 1.39 × 10−8) with effect deflation only in TelSeq-based measurement using WES in UK Biobank (fig. S3). Thus, we focus on TOPMed TelSeq and UK Biobank T/S ratio hereafter. Age similarly correlated with mLTL in TOPMed WGS (β = −0.026, P < 2 × 10−16) and UK Biobank T/S ratio (β = −0.025, P < 2 × 10−16) after adjustment with sex, TOPMed study (if applicable), sequencing center or batch, and the first 11 genetic PCs.
Shorter mLTL is associated with increased CHIP prevalence and increased CAD incidence
We performed association studies between mLTL, CHIP, and CAD separately in TOPMed and UK Biobank, followed by meta-analyses. Consistent with prior reports (27, 33), CHIP was associated with shorter mLTL in meta-analysis results after adjustment for age, sex, ever smoking, body mass index (BMI), study, sequencing center, and the first 11 genetic PCs [β = −0.10; 95% confidence interval (CI), −20.13 to −0.07; P (heterogeneity) = 0.0053] (fig. S4A). CHIP with VAF > 0.10 was associated with shorter mLTL with less heterogeneity across studies [β = −0.15; 95% CI, −0.18 to −0.11; P (heterogeneity) = 0.35] (Fig. 2A and fig. S4B), likely because of reduced sensitivity of smaller CHIP clones in WGS and robust inverse correlation between VAF and mLTL as shown in subsequent analysis. Prior cell-based studies have shown that Dnmt3a loss of function increases telomere length (34), Tet2 loss of function decreases telomere length (35), and p53 Tumor protein p53 (TP53) protects telomeres from DNA damage (36). Thus, we estimated the effect size of each mutated gene on mLTL (Fig. 2A, fig. S5, and table S2). We focused on CHIP with VAF > 10% for this meta-analysis because of its less heterogeneity across cohorts. DNMT3A did not show a significant association with mLTL, whereas TET2, ASXL1, PPM1D, JAK2, and TP53 were significantly associated with shorter mLTL (Fig. 2A). Multiple CHIP mutations in the same individuals had an additive effect on shorter mLTL (Fig. 2B and fig. S6A). Each additional CHIP-related mutation yielded an effect size of −0.13 when meta-analyzed across both cohorts [95% CI, −0.16 to −0.09; P (heterogeneity) = 0.35] (fig. S6B). Among those with CHIP, increasing VAF strongly correlated with shorter mLTL after adjustment in both cohorts [β = −1.16/1% of VAF; 95% CI, −1.45 to −0.87; P (heterogeneity) = 0.35] (Fig. 2C and fig. S7).
Fig. 2. CHIP prevalence and VAF are associated with shorter LTL.
The associations of CHIP with LTL were assessed by linear regression model in both TOPMed and UK Biobank and then meta-analyzed by fixed-effect model. Both models were adjusted with age, sex, ever smoking, BMI, the first 11 genetic PCs, study within TOPMed, and sequencing center or batch (study was only applicable to TOPMed). The prevalence of CHIP with greater than 10% VAF associations was evaluated for overall and each mutated gene (A) and for each number of mutated genes in the same individuals (B). (C) The correlation between LTL and VAF among the population with CHIP from both TOPMed and UK Biobank pooled analysis is displayed. A subset in TOPMed with age 40 to 70 was included in the analysis to align with the age distribution in UK Biobank. As we could not include the population without CHIP in the analysis of (C), we added a red dashed line representing the average LTL in the population without CHIP. ***P < 0.001, after Bonferroni’s correction if applicable. DDR, DNA damage repair; VAF, variant allele frequency.
We next assessed the association of mLTL with CAD using subsets of the cohorts with information on incident CAD in TOPMed (n = 38,645) and in UK Biobank (n = 47,080). Individuals who experienced CAD before the blood draw used to determine CHIP status were excluded from TOPMed (n = 417) and UK Biobank (n = 1753) analyses. Follow-up duration was calculated starting the time at the blood draw. Incident CAD was defined in both TOPMed and UK Biobank by ischemic heart disease events, including myocardial infarction and coronary revascularization (table S3).
We used Cox proportional hazard models to evaluate the association between mLTL and CAD including multivariable adjustment with covariates age, sex, ever smoking, BMI, hypercholesterolemia, the first 11 genetic PCs, study within TOPMed, and sequencing batch or center. Missing covariates excluded 8345 (including 5362 COPDGene participants without blood lipids and 124 with other missing covariates) from 27,521 TOPMed participants with available incident CAD follow-up and 1919 UK Biobank individuals from the analysis (fig. S1). Of the remaining 19,176 TOPMed and 43,408 UK Biobank individuals, 3283 TOPMed (17.1%) and 2396 UK Biobank (5.5%) participants developed CAD during the follow-up duration [mean (SD) duration, 12.0 (5.8) years in TOPMed and 10.0 (1.5) years in UK Biobank]. (The number of individuals included in the final analysis in each TOPMed cohort is shown in table S4.) Shorter mLTL was associated with increased CAD risk [hazard ratio (HR) = 1.08; 95% CI, 1.05 to 1.11; P (heterogeneity) = 0.94] (fig. S8A) as previously reported (37, 38).
We replicated previous findings that CHIP is associated with increased CAD incidence across the UK Biobank and TOPMed cohorts [HR = 1.12; 95% CI, 1.04 to 1.20; P (heterogeneity) = 0.074] (fig. S9 and table S5). Given the heterogeneous association of CHIP with LTL by mutated gene, we also assessed whether each CHIP gene is differently associated with the incidence of CAD. We observed consistent directionality between the CAD-CHIP relationship and the LTL-CHIP relationship in some CHIP-related genes including DNMT3A, TET2, ASXL1, and JAK2, while PPM1D, SF3B1, and TP53 were inconsistent.
As we observed a significant association between CHIP, CAD, and LTL, we conducted sensitivity analyses for the observations above conditioning by each other (tables S5 and S6). We observed independent association of CHIP and previous CAD on LTL when both CHIP and previous CAD are evaluated in linear regression model with or without interaction term. Similarly, independent association was observed for LTL and CHIP on incidence of CAD when both CHIP and LTL are included in Cox proportional hazard model with or without interaction term. The significant association between CAD and LTL was robust among those without CHIP (fig. S8B). We observed consistent association between LTL and CHIP by each mutated CHIP-related gene after adjustment with previous CAD (table S7).
MR studies indicate that longer LTL causes CHIP acquisition
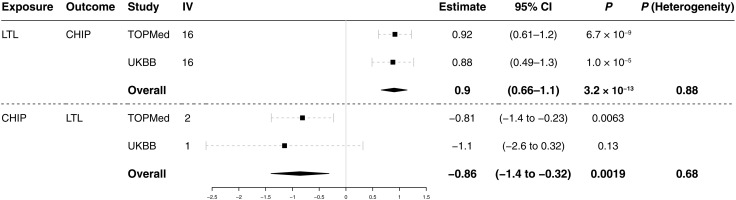
We performed one-sample MR study to infer whether LTL has a causal effect on CHIP acquisition. Using TOPMed individual-level data, the association between gLTL and CHIP was tested to infer causality in MR framework. We found instrumental variables (IVs) from an independent GWAS of mLTL (11). Twenty independent single-nucleotide polymorphisms (SNPs) with P < 10−8 were pruned as 10 Mb apart and in linkage disequilibrium resulting in 16 IVs (table S8). Two-stage least-square regression suggested a causal association of LTL with CHIP (Estimate = 0.92; 95% CI, 0.61 to 1.2; P = 6.7 × 10−9) (Fig. 3). The estimate here is the logarithm of odds ratio normalized per 1 SD of LTL. As a sensitivity analysis, we performed one-sample MR using UK Biobank and meta-analyzed with TOPMed. The putative causal association of LTL with CHIP was consistent across cohorts [Estimate = 0.90; 95% CI, 0.66 to 1.1; P (heterogeneity) = 0.88].
Fig. 3. Bidirectional one-sample MR studies indicated the positive causal effect of LTL on CHIP and the inverse causal effect of CHIP on LTL.
Bidirectional one-sample MR was performed to assess the causal effect of both LTL on CHIP and CHIP on LTL. The summary statistics for LTL GWAS in Li et al. (11) was used for IV discovery for LTL on CHIP and TOPMed for CHIP on LTL. IVs were clumped if <10 Mb apart and in linkage disequilibrium (R2 > 0.001 calculated in European ancestry from the 1000 Genome Project). IVs were further assessed by Steiger test to mitigate the effect of reverse causation resulting in 16 and 2 IVs, respectively. TOPMed and UK Biobank were used as the test cohort for both CHIP on LTL and LTL on CHIP and meta-analyzed. Used IVs and cohorts for each analysis are summarized in tables S4, S8, and S9. CI, confidence interval; IV, instrumental variable; MR: Mendelian randomization.
We performed two-sample MR as a replication analysis. We used previous European LTL summary statistics (11) as the exposure (LTL) cohort (n = 78,592) and the European subset of the UK Biobank as the outcome (CHIP) cohort (n = 43,906). The same 16 IVs were used as one-sample MR. The positive potential causal effect of LTL on CHIP was shown in the conventional two-sample MR approach [inverse variance weighted (IVW) method; Estimate = 1.06; 95% CI, 0.39 to 1.74; P = 1.89 × 10−3] (fig. S10). The global test by MR-PRESSO (Mendelian Randomization Pleiotropy Residual Sum and Outlier) (39) suggested significant horizontal pleiotropy before outlier exclusion (P < 1.0 × 10−4) and detected the TERT and ATM loci as outliers (table S8). While the leave-one-out analysis showed that the TERT locus variant had the most significant effect among IVs, the analysis remained robust (fig. S11). Outlier exclusion supported a significant causal association of LTL on CHIP by MR-PRESSO in two-sample MR (Estimate= 0.79; 95% CI, 0.24 to 1.34; P = 0.015) (fig. S10) without significant statistical evidence of horizontal pleiotropy (Global test, P = 0.15). Reevaluation of all the models after outlier exclusion showed stable estimates across methods indicating robust causal inference (fig. S12).
Although the Sargan test indicated endogeneity of used IVs (P < 2.2 × 10−16), we do not have an appropriate strategy to find pleiotropy in one-sample MR. Hence, the one-sample MR study in TOPMed was reexamined after exclusion of outliers detected by MR-PRESSO in the two-sample MR study. Outlier exclusion still demonstrated significant causal association (Estimate = 0.48; 95% CI, 0.13 to 0.83; P = 0.0062) without statistically significant evidence of pleiotropy (Sargan test, P = 0.83).
We next evaluated the relationship of gLTL with the occurrence of acquired genome-wide singleton single-nucleotide substitution. Using WGS from a subset of the TOPMed study population (n = 56,266), we tabulated per-individual genome-wide somatic mutations. Outlier-excluded 14 IVs discovered in the previous section were used for MR study. MR analyses supported a causal relationship between longer LTL and increased somatic mutations in one-sample MR study (Fig. 4). Next, we assessed the effect of gLTL for COSMIC (catalogue of somatic mutations in cancer) signature version 2 (https://cancer.sanger.ac.uk/cosmic/signatures_v2). Failure of DNA double-strand break repair by homologous recombination (signature 3) and other signatures with unknown etiologies (signatures 8, 17, 19, and 30) associated with longer gLTL in MR analyses (fig. S13). These observations suggested that longer LTL promotes CHIP acquisition by accelerating mutagenesis. The TERT locus variant is associated prominently with mutational occurrence in line with the pleiotropic effect detected in MR studies (fig. S14).
Fig. 4. Effect of mLTL and gLTL for mutation occurrence.
Effect estimates of (A) measured LTL (mLTL) and (B) gLTL (one-sample MR using 14 IVs) on singleton mutation occurrence. The vcf files were generated by Mutect2 from 56,266 CRAM files in TOPMed with appropriate filters and single-base substitutions were extracted, stratified by trinucleotide context. IVs were selected as two-sample MR for LTL (Fig. 3) with outlier exclusion. Effect estimates with P < 0.05 are colored. * denotes false discovery rate < 0.05.
MR studies support the idea that CHIP causes shortened telomeres
Given the inverse correlation between CHIP and mLTL and previous reports that some CHIP-related genes affect telomere length, we performed one-sample MR for CHIP on LTL with TOPMed. IVs were used from a previously reported GWAS of CHIP in TOPMed (28). There are three loci reported to have genome-wide significance (P < 1 × 10−8). To avoid the possible bias from reverse causality, we filtered discovered IVs using the Steiger test that identified the TERT locus as having a significantly higher correlation with mLTL than CHIP (P = 0.0105) concordantly with the well-known causal effect of TERT on telomere length; hence, SNPs at KPNA4/TRIM59 and TET2 loci were valid IVs. The significance of each variant supported the robust association with exposure, the first assumption of MR (table S9). No statistical evidence of endogeneity for IVs used was shown by the Sargan test (P = 0.306). The MR analysis with two IVs was consistent with an inverse causal effect of CHIP on LTL (Estimate = −0.81; 95% CI, −1.40 to −0.23; P = 0.0063) (Fig. 3). Single-IV analysis demonstrated consistent effect sizes across two IVs (fig. S15). As a sensitivity analysis, we performed one-sample MR using UK Biobank and meta-analyzed with TOPMed. Meta-analyzed one-sample MR in UK Biobank and TOPMed was consistent with an inverse causal effect of CHIP on LTL [Estimate = −0.86; 95% CI, −1.40 to −0.32; P (heterogeneity) = 0.68] (Fig. 3).
As another sensitivity analysis, we performed two-sample MR study for CHIP on LTL. As SNP at TET2 locus was African ancestry specific, SNP at KPNA4/TRIM59 locus was the only valid IV among European ancestry. We used the European subset of TOPMed for exposure (CHIP; n = 27,402) and UK Biobank for outcome (LTL, n = 412,308). CHIP consistently has inverse causation on LTL (Estimate = −0.13; 95% CI, −0.17 to −0.10; P = 8.6 × 10−15; fig. S16). Furthermore, we flipped the cohorts (UK Biobank CHIP, n = 43,906; TOPMed LTL, n = 27,402), and meta-analyzed with the previous analysis, which suggested that CHIP consistently has inverse causation on LTL [Estimate = −0.13; 95% CI, −0.17 to −0.091; P (heterogeneity) = 0.89; fig. S16].
Mediation analysis of LTL for CHIP-associated CAD risks
We assessed the mediation effect of mLTL on CHIP-associated CAD risk in UK Biobank. The proportion of mediation effect of mLTL in the total effect of CHIP to CAD was estimated as 3.4% (95% CI, 1.3 to 8.3%; P ≤ 2 × 10−16) using the “mediation” package (40) in R (R Foundation for Statistical Computing, Vienna, Austria) (Table 1). Both mediator and outcome models were adjusted for age, sex, BMI, ever smoking, previous type 2 diabetes mellitus, previous hypercholesterolemia, previous hypertension, sequencing batch, and the first 11 genetic PCs. We performed a replication analysis using the WHI cohort subset of the TOPMed cohort (n = 3734), because sufficient covariate information was available to adjust models precisely. Both mediator and outcome models were adjusted for WHI inverse probability weight (to account for the nonrandom selection of women for WGS in WHI), history of hormone therapy, and history of hysterectomy in addition to the covariates in UK Biobank. CAD was defined as the composite of myocardial infarction and coronary revascularization. The proportion of mediation effect of mLTL in the total effect of CHIP to CAD was estimated as 6.4% (95% CI, 0.88 to 19%; P = 0.02) in WHI.
Table 1. Mediation analysis showed mediation effect of LTL for CHIP-associated CAD risk.
The mediation effect of LTL for CHIP-associated CAD risk was estimated by mediation package in R. A mediation effect of 0 indicates that LTL does not mediate the CHIP-associated CAD risks, and a mediation effect of 1 indicates that LTL mediates all of the CHIP-related CAD risks. The P value reflects whether the proportion of the mediation effect on the CHIP-related CAD risks is 0% versus not 0%. Both mediator and outcome models are adjusted by age, sex, ever smoking, BMI, prevalent type 2 diabetes, prevalent hypercholesterolemia, prevalent hypertension, sequencing batch, and the first 11 genetic PCs in UK Biobank, and age at blood draw, ever smoking, race, dyslipidemia, hypertension, BMI, WHI inverse probability weight (to account for the nonrandom selection of women for WGS in WHI), history of hormone therapy, history of hysterectomy, and the first 11 genetic PCs in WHI. CAD, coronary artery disease; CHIP, clonal hematopoiesis of indeterminate potential; LTL, leukocyte telomere length; WHI, Women’s Health Initiative.
|
Proportion of
mediation effect of LTL for CHIP- associated CAD risk (95% CI) |
P | |
| UK Biobank (n = 43,408) |
0.034 (0.013–0.083) | <2 × 10−16 |
| WHI (n = 3734) | 0.064 (0.0088–0.19) | 0.02 |
DISCUSSION
We used observational and MR studies to examine how processes regulating LTL and CHIP acquisition interrelate and how they influence CAD risk. Consistent with prior observational epidemiologic analyses, CHIP and mLTL were inversely correlated. Bidirectional MR supported the hypotheses that longer LTL promotes CHIP acquisition, while CHIP in turn shortens LTL potentially among affected cells. Although this inverse bidirectional causal relationship is complicated and partially counterintuitive, the first part (from LTL to CHIP) is consistent with recent reports that described similar relationships in related phenotypes such as myeloproliferative neoplasm (MPN) and clonal somatic copy number alterations (41), and the later part (from CHIP to LTL) is consistent with observational studies. While both CHIP and shorter mLTL have been independently associated with CAD, mediation analysis indicated that a modest fraction of CHIP-associated CAD risk may be mediated by resultant LTL shortening.
Our findings have several implications for the understanding of CHIP, LTL, and CAD. First, we replicated modest overall observational inverse association between CHIP and mLTL, which was previously shown in independent cohorts (27, 33). The association varies by mutated genes, indicating gene-specific mechanisms promoting LTL shortening. DNMT3A mutations did not show significant association with mLTL, while JAK2 mutations, which have been previously reported as strongly correlated with shorter mLTL (42) in patients with MPN, had the most robust association with shorter mLTL in the context of CHIP.
Second, we observe a bidirectional causal relationship between LTL and CHIP, advancing our understanding of the malignancy–telomere length association. As described earlier, prior studies have shown complex relationships between LTL and cancer risk (18). Several models were proposed, including a heterogeneous multihit theory (43) and the biphasic effect of TERT promoter mutation throughout tumor development (14). CHIP provides an opportunity to focus on an incipient step of malignant cell development. Our results suggest that longer LTL may promote CHIP acquisition through increasing mutagenesis. One potential model could be that longer telomeres support the longevity of the cells, thus augmenting opportunities to acquire somatic mutations over time, while telomeres begin accelerated shortening once the cell cycle accelerates owing to driver mutation acquisition (Fig. 5). Consistent with this model, we observed that increased clone size, a readout of increased cellular cycles, is correlated with shorter mLTL (44). In the setting of shortened telomere Mendelian syndromes, shortened telomeres promote genomic instability and subsequent acquisition and retention of neoplastic driver mutations (45). This may be consistent with the hypothesis that CHIP-associated LTL shortening may hasten subsequent malignancy (Fig. 6). Further assessment of longitudinal mLTL follow-up among the CHIP-positive population would be desired.
Fig. 5. Proposed model to explain “telomere paradox” in CHIP.
People with longer gLTL have a higher incidence of mutagenesis and, thus, have a higher chance to acquire CHIP-associated mutations (middle). The cells that acquired CHIP have a shorter telomere such that mean mLTL decreases as the clone expands (bottom). This model can explain the “paradox” that genetically longer LTL is associated with higher incidence of CHIP, which has measured shorter LTL on average. HSPC, hematopoietic stem cell.
Fig. 6. Estimated change of mean LTL in each scenario.
Schematic representation of estimated change of mLTL in each scenario speculated from our study. The slope after CHIP acquisition may differ by CHIP-related gene and mutation.
Third, CHIP-associated CAD risk may be partly attributed to subsequent LTL shortening. Prior cell-based, murine, and human genetic analyses have prioritized the NLR family pyrin domain containing 3 (NLRP3) inflammasome pathway in CHIP-associated CAD risk (23–26, 28). In the present work, we orthogonally implicate LTL in both the genesis of CHIP, a new CAD risk factor, and its clinical consequences. Our study is underpowered for gene-specific analyses but notably did not observe an association between DNMT3A CHIP and mLTL alteration. Consistent with this observation, prior work suggests that hematopoietic stem cell loss of Tet2 leads to shortened telomeres, whereas loss of Dnmt3a leads to telomere preservation (35, 46). Such differences may also partly explain gene-specific differences in CAD risk (24–26). While interrupting CHIP-mediated LTL shortening may be a viable strategy to mitigate CHIP-associated CAD risk, this general strategy may be limited to the overall modest estimated mediating effect. However, given the heterogeneity observed, this strategy may be more efficiently applied to non-DNMT3A CHIP.
Key limitations must be considered in the interpretation of our study findings. First, limited CHIP GWAS availability prevented sensitivity analyses for conventional two-sample MR approaches in the causal inference of CHIP on LTL. The ongoing effort of accumulating CHIP cases would address this issue. Second, the cross-sectional observational nature of our analyses limits inference regarding causal temporal relationships. Longitudinal analyses of LTL, CHIP, and incident diseases as well as experimental models are needed to confirm our hypotheses. Third, the mediation effect estimate of mLTL for CHIP-associated CAD may be limited by conflicting bidirectional causal effect. We found that MR study for CAD and CHIP is more challenging because the following known observations indicate potential violation for MR assumptions: (i) well-established MR-imputed causal association from LTL to CAD, (ii) the known biological causality of TERT for telomere length, and (iii) the TERT locus is the top hit in CHIP GWAS. Increased sample size for CHIP GWAS with careful selection of the IVs will be necessary to conduct an appropriate MR study for CAD and CHIP.
In conclusion, we showed a bidirectional relationship between LTL and CHIP, shedding light on the mechanisms by which telomere length contributes to age-related disorders. The mediation effect of LTL on CHIP-related CAD incidence suggests the plausibility of developing harmonized therapies for both blood cancer and cardiovascular diseases.
MATERIALS AND METHODS
Experimental design
Given the known association between CHIP, LTL, and CAD, we intended to understand the causality and mediation effect to seek the possibility to develop preventative therapies for CAD through the intervention for CHIP and/or LTL. We studied observational and causal relationships between CHIP and LTL and then assessed the mediation effect on CAD. For this purpose, we leveraged two large-scale cohorts UK Biobank and TOPMed for observational studies, MR studies for causal inference, and mediation analyses.
Study population
From NHLBI’s TOPMed program, WGS of blood DNA performed in participating studies were used for CHIP detection and LTL estimation in recent studies (28, 29). The included studies are largely observational cohorts and have previously been described in detail. Cohorts included in this study are reported in table S4. We excluded samples that had conflicting information for sex and excess kinship within second degree determined by kinship-based inference for GWAS (KING) coefficient > 0.0884 (fig. S1).
The UK Biobank is a population-based cohort of >500,000 UK adult residents recruited between 2006 and 2010 and followed prospectively via linkage to national health records (30). At the baseline study visit, participants underwent phlebotomy and provided detailed information about medical history and medication use. In the present study, the UK Biobank cohort included adults aged 40 to 70 years at blood draw with available WES. Follow-up in the UK Biobank occurred through March 2020 for inpatient diagnosis. We excluded samples with consent retraction, excess heterogeneity or missingness in genotyping array, discordance between reported and genetically imputed sex, and excess kinship within second degree determined by KING coefficient > 0.0884 (fig. S1). We used the samples from which all information used was available.
Kinship inference and genetic PCs were centrally calculated in both cohorts. For the two-sample MR studies, the European ancestry (n = 27,402 in TOPMed and n = 43,906 in UK Biobank) subset was used to avoid bias from population structures in both cohorts.
Because the WHI represents one of the largest TOPMed cohorts and had exposure, detailed covariates, and outcome data available, we used WHI for the replication of mediation analysis for mLTL on CHIP-associated CAD risks. Briefly, the WHI is a prospective study of women recruited at 40 centers throughout the United States between 1993 and 1998 (47). Participants were enrolled in the clinical trial(s) (of hormone therapy, calcium/vitamin D supplementation, and/or dietary modification) or the observational study. Unrelated women in WHI who underwent >30× WGS using blood drawn at age 50 to 70 years as part of the TOPMed program were included in the present analysis. To avoid the effects of the study intervention on outcomes, we excluded women in the WHI hormone therapy trial with blood draw ≥2 years after the screening visit (n = 483) as previously described (48).
Sequencing and CHIP calling
UK Biobank WES of whole blood–derived DNA was performed using Illumina NovaSeq 6000 platform at the Regeneron Sequencing Center (Tarrytown, NY) as described previously (30). TOPMed WGS to an average depth of 38× was performed using whole blood–derived DNA, PCR-free library construction, and Illumina HiSeq X technology as described elsewhere (29).
CHIP mutations were called previously in TOPMed (28) and UK Biobank (26). CHIP mutations were reevaluated after the error-corrected release of WES in UK Biobank in June 2020. Briefly, CHIP mutations were detected with GATK MuTect2 software (49) with parameters as previously described (28). Samples were annotated as having CHIP if Mutect2 identifies one or more of a prespecified list of pathogenic somatic variants (23, 24). Common germline variants and sequencing artifacts were excluded as before. Each study includes both the presence of (i) any CHIP and (ii) CHIP with VAF > 0.1, as larger CHIP clones above this threshold have previously been more strongly associated with adverse clinical outcomes (24, 26).
Estimating LTL from NGS of blood-derived DNA
The estimation of mean LTL was performed using TelSeq (32) previously in TOPMed with WGS for 109,648 participants by the TOPMed Hematology and Hemostasis working group (31). Given imperfect capture, TelSeq is expected to be able to estimate LTL from WES (32). We applied an analogous method to WES in UK Biobank with k = 7, while k = 12 was used for TOPMed WGS. Read coverage was calculated by Mosdepth (50), and PCA was conducted for read coverage. Estimated LTL was log-transformed, and linear regression was performed using the first nine PCs. Standardized residuals were used as the relative value of estimated LTL within each study for mean = 0 and SD = 1 for each cohort.
Telomere measurement by Southern blotting and qPCR
We measured LTL in a subset of WHI by Southern blotting for direct comparison with TelSeq using WGS. DNA was extracted from baseline (or year 1) blood samples by the 5-prime method (5 PRIME Inc., Gaithersburg, MD) and sent in batches over a 1-year period to the Center of Human Development and Aging laboratory at Rutgers for LTL measurement. The laboratory conducting the LTL measurements was blinded to all characteristics of participants. DNA integrity was assessed visually after ethidium bromide–stained 1% agarose gel electrophoresis (200 V for 2 hours). We required DNA to appear as a single compact crown-shaped band that migrated in parallel with the other samples on the gel. Telomere length in kilobases was measured by the mean length terminal restriction fragments using the Southern blot method as previously described (51). Each sample was run in duplicate on different gels and mean LTL was used for statistical analyses. The average interassay coefficient of variation for blinded pair sets was 2.0%. Individuals with LTL values exceeding 3 SDs from the sample mean were excluded from the analyses, n = 3. LTL in UK Biobank participants was measured by qPCR-based T/S ratio as described previously (19).
Statistical analysis
Observational epidemiology
Baseline continuous variables were compared between populations with large clone size CHIP (VAF ≧ 0.1) and small clone size CHIP (VAF < 0.1), and without CHIP using analysis of variance (ANOVA), and categorical variable associations were estimated using the chi-square test. For the association analyses, linear regression models were used for continuous outcomes, and logistic regression models were used for binary outcomes.
Cox proportional hazard models were used in survival analyses with CAD as the outcome. Cox proportionality assumption was assessed by Schoenfeld. Models were adjusted for age, sex, ever smoking, BMI, hypercholesterolemia, the first 11 genetic PCs, study within TOPMed, and sequencing center (study and sequencing center are only applicable to TOPMed). Quadratic age was used as a covariate in all the models throughout this study where applicable. Age, sex, and sequencing center were stratified, and frailty model was introduced for TOPMed study to comply with the Cox proportionality assumption in TOPMed.
Meta-analyses were performed by the fixed-effect model using the “meta” package (52) (version 4.18-0) in R. Two-sided P < 0.05 was considered statistically significant. Analyses were conducted using R 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria).
Mendelian randomization
We performed bidirectional MR studies between CHIP and LTL. All procedures were consistent with the current recommendations for MR studies (53). IVs detected in previously reported CHIP (28) and LTL (11) GWAS studies were used for the causal inference by two-stage least-squares regression in one-sample MR and its analogous IVW method in two-sample MR. Genome-wide significance (P < 5 × 10−8) was considered as the criteria for the IV assumption of robust relevance (the first assumption of MR). Given the limited availability of GWAS for CHIP, we used SNPs detected in multiethnic analysis in the MR for CHIP on LTL. The associations of the SNPs with CHIP were recalculated in the European ancestry subsets in each cohort. IVs were pruned into independent loci <10 Mb apart and in the linkage equilibrium (R2 > 0.001 calculated in European ancestry from the 1000 Genome Project) using the “TwoSampleMR” package (54) (version 0.5.4) in R. Used IVs are reported in tables S8 and S9 for CHIP on LTL and LTL on CHIP, respectively. Estimates were normalized per 1 SD LTL in both directions.
To mitigate the influence of reverse causality, we performed Steiger filtering to remove variants that have significantly greater association with the outcome than exposure. Some variants may be associated with exposure via first outcome and secondary exposure. These variants should not be included as instruments for causal inference because of the violation of the exclusion-restriction assumption (the third assumption of MR). Previous reports showed that TERT locus is the leading predisposition for both CHIP and LTL; thus, this locus may have significant effects on both directions. Steiger filtering calculates the variant-exposure and variant-outcome correlations and removes variants where the variant-outcome correlation is significantly greater than the variant-exposure correlation. To perform Steiger filtering, the correlations of variants with exposure and outcome were calculated. For the continuous trait (LTL), the squared correlation of each variant with LTL was calculated as the R2 from the association of the trait with the variant. Cox and Snell pseudo R was calculated and squared for the binary measurement (CHIP). Steiger test was performed using the r.test function in R package “psych” (version 2.0.12) (55). To apply Steiger filtering, variants with R2exposure < R2outcome and P < 0.05 were removed.
Sensitivity analyses
Weighted median, weighted mode, and MR-Egger were calculated using the “MendelianRandomization” package (56) (version 0.5.0) in R if multiple IVs were available. Because these methods use different assumptions to estimate the causal effect and are influenced differently by biases, the stable estimates across methods indicate robust causal inference (53). Variant effect heterogeneity assessment and outlier detection were performed via MR-PRESSO residual sum of squares using the “MRPRESSO” package (version 1.0) in R, which shows improved false-positive rates (39). Further assessment of the reliance on a particular variant was performed by IVW-based leave-one-out analysis to detect potential outliers. All the models were reevaluated after removing those outliers and further assessed by heterogeneity test using MR-PRESSO. A scatterplot of each variant’s effect on exposure versus outcome and a funnel plot to visualize the balance of horizontal pleiotropy by weak instruments were generated by “TwoSampleMR” package (version 0.5.4) in R (54).
Given the large scale of sample size and abundance of CHIP cases with available individual-level data, we bidirectionally performed one-sample MR in TOPMed and UK Biobank and then meta-analyzed. F statistics were used to support robust associations between IVs and exposure, which is the first assumption of MR (relevance). The second and third assumptions of MR require exogeneity of the IVs, which means that IVs are not associated with outcome via horizontal pleiotropy (exchangeability and exclusion restriction). Sargan test statistically tests the association of outcome with the residuals from the association of the exposure with the IVs. When the exogeneity is violated, the null hypothesis will be declined. We performed the Sargan test to check the exchangeability and exclusion restriction assumptions in our model. When the Sargan test was violated, we tried to exclude endogenous (not exogenous) IVs by outlier exclusion. Because we do not have an appropriate method to detect outliers in one-sample MR, we excluded outliers detected in two-sample MR when using the same IVs. The model was reevaluated after outlier exclusion. Age at blood draw, sex, TOPMed study, sequencing center or batch, and the first 11 genetic PCs to control for population structure were included as covariates. Effect estimates for continuous exposure and outcome (LTL) were normalized to 1 SD.
Mediation analysis of mLTL on CHIP-associated CAD
We performed mediation analysis to evaluate the proportional contribution of mLTL to the association between CHIP and CAD using the R mediation package (40) (version 4.5.0) in UK Biobank (n = 43,408) and WHI (n = 3734). For UK Biobank, the covariates used in both mediation and outcome models included age, sex, ever smoking, hypercholesterolemia, hypertension, BMI, type 2 diabetes, and the first 11 genetic PCs. For WHI, covariates in both the mediation and outcome model were age at blood draw, ever smoking, race, dyslipidemia, hypertension, BMI, WHI inverse probability weight (to account for the nonrandom selection of women for WGS in WHI), history of hormone therapy, history of hysterectomy, and the first 11 genetic PCs. Each mediation analysis model was run using 100 simulations with a quasi-Bayesian approach to estimate confidence intervals (57).
Acknowledgments
We acknowledge the studies and participants who provided biological samples and data for TOPMed and UK Biobank. The acknowledgments for each study in TOPMed are listed in table S4. Although WGS for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the NHLBI, the views expressed in this manuscript are those of the authors and do not necessarily represent the views of the NHLBI, the NIH, or the U.S. Department of Health and Human Services. Secondary use of the TOPMed data was approved by the Massachusetts General Hospital institutional review board (protocol 2016P001308). All participants in the UK Biobank study provided written informed consent, and the ethics approval for the UK Biobank study was obtained as described before (58). UK Biobank Resource was used under application number 7089. Secondary use of the UK Biobank data was approved by the Massachusetts General Hospital institutional review board (protocol 2013P001840).
Funding: This work was supported by the following: NHLBI 3R01HL-117626-02S1, contract no. HHSN268201800002I (core support including centralized genomic read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center); NHLBI R01HL-120393, U01HL-120393, contract HHSN268201800001I (core support including phenotype harmonization, data management, sample-identity QC, and general program coordination were provided by the TOPMed Data Coordinating Center); NHLBI R01HL146860 (phenotype harmonization for coronary artery disease in TOPMed) (the funding supports for each study in TOPMed are listed in table S4); Biotechnology and Biological Sciences Research Council and British Heart Foundation (BHF) through UK Medical Research Council (MRC) grant MR/M012816/1 (UK Biobank telomere measurement); the Uehara Memorial Foundation Overseas Research Fellowship R-C12, 201740153 (to T.N.); British Heart Foundation (BHF) SP/16/4/32697 (to C.P.N.); National Institutes for Health Research (NIHR) Leicester Cardiovascular Biomedical Research Centre BRC-1215-20010 (to V.C., C.P.N., and N.J.S.); Hassenfeld Scholar Award from the Massachusetts General Hospital (to P.N.); NHLBI R01HL1427, R01HL148565, and R01HL148050 (to P.N.); Fondation Leducq TNE-18CVD04 (to T.N., P.N., and B.L.E.); NHLBI 1R01HL134892, the American Heart Association 18CSA34080399 (to P.L.); NHLBI R01HL046380, R01HL113338, and R35HL135818 (to S.R.); National Center for Advancing Translational Sciences KL2TR002490 (to L.M.R.); National Institute of Diabetes and Digestive and Kidney Disease R01DK110113, R01DK107786, R01DK124097, and R01HL142302 (to R.J.F.L.); and NHLBI PO1 HL132825 (to S.T.W.).
Author contributions: Conceptualization: T.N., A.G.B., A.P.R., R.A.M., and P.N. Methodology: T.N., J.L., N.P., R.D., and P.N. Investigation: T.N. and P.N. Visualization: T.N. Supervision: S.J., P.L., P.T.E., B.L.E., A.P.R., R.A.M., R.D., and P.N. Writing—original draft: T.N. and P.N. Writing—review and editing: A.G.B., C.L.C., M.C.H., J.S.W., J.C.B., D.D., L.M.R., S.S.R., M.H.C., B.M.P., M.F., C.K., J.D., N.J.S., P.L., A.P.R., R.A.M., and R.D. Data processing and analyses: T.N., A.G.B., M.A.T., S.M.Z., M.M.U., A.N., C.L.C., J.L., M.C.H., J.S.W., A.P., S.L.C., and R.B. Data acquisition: TOPMed Consortium (C.J.G., G.K.G., S.L.C., T.S.A., L.S.E., A.M.S., Q.W., J.B., C.A.L., A.T.K., A.V.S., T.W.B., Z.T.Y., J.M.P., D.W.B., M.R.I., M.B., W.Z., L.R.Y., K.L.W., J.E.H., C.C.G., G.M.P., D.M.R., M.S.R., C.-M.H., D.L.D., K.E.N., S.K., S.K.M., J.C.B., D.M.L.-J., J.M.J., M.P., R.P.T., P.A.P., D.Q., P.D., J.E.C., B.I.F., H.K.T., S.C., J.A.S., N.L.S., T.N.K., B.H., L.A.C., D.E.W., N.L.H., R.L.M., R.D., T.T.N., L.F., L.M.R., A.C.M., P.S.V., C.M.B., E.E.K., S.S.R., E.A.W., M.H.C., M.B.S., B.S.P., J.B., N.D.P., B.D.M., A.R.S., K.C.B., S.R., S.L.R.K., G.R.A., L.C.B., S.R.H., J.H., W.P., D.K.A., R.S.V., D.D., S.T.W., S.T.M., M.A., Y.-D.I.C., R.C.K., D.A.M., B.S.C., A.C., B.M.P., M.F., J.E.M., E.B., B.A.K., R.J.F.L., J.I.R., E.K.S., and C.K.). UK Biobank Telomere measurement (V.C., C.P.N., J.D., and N.J.S.). All authors approved the final submission.
Competing interests: P.L. is an unpaid consultant to or involved in clinical trials for Amgen, AstraZeneca, Baim Institute, Beren Therapeutics, Esperion Therapeutics, Genentech, Kancera, Kowa Pharmaceuticals, Medimmune, Merck, Norvo Nordisk, Novartis, Pfizer, and Sanofi-Regeneron. P.L. is a member of the scientific advisory board for Amgen, Caristo, Cartesian, CSL Behring, DalCor Pharmaceuticals, Dewpoint, Kowa Pharmaceuticals, Olatec Therapeutics, Medimmune, Novartis, PlaqueTec, and XBiotech Inc. P.L.’s laboratory has received research funding in the past 2 years from Novartis. P.L. is on the Board of Directors of XBiotech Inc. P.L. has a financial interest in Xbiotech, a company developing therapeutic human antibodies. P.L.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. S.J. is a paid consultant for Novartis, Genentech, AVRO Bio, and Foresite Labs. L.M.R. is a consultant for the TOPMed Administrative Coordinating Center (through Westat). M.H.C. has received grant funding from GSK and Bayer and speaking or consulting fees from AstraZeneca, Illumina, and Genentech. P.L. is an inventor on a patent (“Use of canakinumab”) related to this work filed by Brigham and Women’s Hospital (U.S. patent application no. 20200239564, filed 18 August 2020). J.S.W., S.J., and A.G.B. are inventors on a patent related to this work filed by Stanford University (U.S. application serial no. 63/274,331, filed on 21 January 2022). P.N. is a cofounder and A.B., P.L., S.J., and B.L.E. are founding scientific advisors of TenSixteen Bio Inc., which had no role in this manuscript. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S16
Tables S1 to S3, S7 to S9
Other Supplementary Material for this manuscript includes the following:
Tables S4 to S6
REFERENCES AND NOTES
- 1.Sfeir A., de Lange T., Removal of shelterin reveals the telomere end-protection problem. Science 336, 593–597 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackburn E. H., Epel E. S., Lin J., Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., Futcher A. B., Greider C. W., Harley C. B., Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 89, 10114–10118 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brouilette S., Singh R. K., Thompson J. R., Goodall A. H., Samani N. J., White cell telomere length and risk of premature myocardial infarction. Arterioscler. Thromb. Vasc. Biol. 23, 842–846 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Vasa-Nicotera M., Brouilette S., Mangino M., Thompson J. R., Braund P., Clemitson J.-R., Mason A., Bodycote C. L., Raleigh S. M., Louis E., Samani N. J., Mapping of a major locus that determines telomere length in humans. Am. J. Human Genet. 76, 147–151 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhan Y., Hägg S., Telomere length and cardiovascular disease risk. Curr. Opin. Cardiol. 34, 270–274 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Hastings R., Qureshi M., Verma R., Lacy P. S., Williams B., Telomere attrition and accumulation of senescent cells in cultured human endothelial cells. Cell Prolif. 37, 317–324 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Childs B. G., Li H., van Deursen J. M., Senescent cells: A therapeutic target for cardiovascular disease. J. Clin. Invest. 128, 1217–1228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Childs B. G., Baker D. J., Wijshake T., Conover C. A., Campisi J., van Deursen J. M., Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science 354, 472–477 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Telomeres Mendelian Randomization Collaboration, Haycock P. C., Burgess S., Nounu A., Zheng J., Okoli G. N., Bowden J., Wade K. H., Timpson N. J., Evans D. M., Willeit P., Aviv A., Gaunt T. R., Hemani G., Mangino M., Ellis H. P., Kurian K. M., Pooley K. A., Eeles R. A., Lee J. E., Fang S., Chen W. V., Law M. H., Bowdler L. M., Iles M. M., Yang Q., Worrall B. B., Markus H. S., Hung R. J., Amos C. I., Spurdle A. B., Thompson D. J., O’Mara T. A., Wolpin B., Amundadottir L., Stolzenberg-Solomon R., Trichopoulou A., Onland-Moret N. C., Lund E., Duell E. J., Canzian F., Severi G., Overvad K., Gunter M. J., Tumino R., Svenson U., van Rij A., Baas A. F., Bown M. J., Samani N. J., van t’Hof F. N. G., Tromp G., Jones G. T., Kuivaniemi H., Elmore J. R., Johansson M., Mckay J., Scelo G., Carreras-Torres R., Gaborieau V., Brennan P., Bracci P. M., Neale R. E., Olson S. H., Gallinger S., Li D., Petersen G. M., Risch H. A., Klein A. P., Han J., Abnet C. C., Freedman N. D., Taylor P. R., Maris J. M., Aben K. K., Kiemeney L. A., Vermeulen S. H., Wiencke J. K., Walsh K. M., Wrensch M., Rice T., Turnbull C., Litchfield K., Paternoster L., Standl M., Abecasis G. R., SanGiovanni J. P., Li Y., Mijatovic V., Sapkota Y., Low S.-K., Zondervan K. T., Montgomery G. W., Nyholt D. R., van Heel D. A., Hunt K., Arking D. E., Ashar F. N., Sotoodehnia N., Woo D., Rosand J., Comeau M. E., Brown W. M., Silverman E. K., Hokanson J. E., Cho M. H., Hui J., Ferreira M. A., Thompson P. J., Morrison A. C., Felix J. F., Smith N. L., Christiano A. M., Petukhova L., Betz R. C., Fan X., Zhang X., Zhu C., Langefeld C. D., Thompson S. D., Wang F., Lin X., Schwartz D. A., Fingerlin T., Rotter J. I., Cotch M. F., Jensen R. A., Munz M., Dommisch H., Schaefer A. S., Han F., Ollila H. M., Hillary R. P., Albagha O., Ralston S. H., Zeng C., Zheng W., Shu X.-O., Reis A., Uebe S., Hüffmeier U., Kawamura Y., Otowa T., Sasaki T., Hibberd M. L., Davila S., Xie G., Siminovitch K., Bei J.-X., Zeng Y.-X., Försti A., Chen B., Landi S., Franke A., Fischer A., Ellinghaus D., Flores C., Noth I., Ma S.-F., Foo J. N., Liu J., Kim J.-W., Cox D. G., Delattre O., Mirabeau O., Skibola C. F., Tang C. S., Garcia-Barcelo M., Chang K.-P., Su W.-H., Chang Y.-S., Martin N. G., Gordon S., Wade T. D., Lee C., Kubo M., Cha P.-C., Nakamura Y., Levy D., Kimura M., Hwang S.-J., Hunt S., Spector T., Soranzo N., Manichaikul A. W., Barr R. G., Kahali B., Speliotes E., Yerges-Armstrong L. M., Cheng C.-Y., Jonas J. B., Wong T. Y., Fogh I., Lin K., Powell J. F., Rice K., Relton C. L., Martin R. M., Smith G. D., Association between telomere length and risk of cancer and non-neoplastic diseases: A Mendelian randomization study. JAMA Oncol. 3, 636–651 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li C., Stoma S., Lotta L. A., Warner S., Albrecht E., Allione A., Arp P. P., Broer L., Buxton J. L., Alves A. D. S. C., Deelen J., Fedko I. O., Gordon S. D., Jiang T., Karlsson R., Kerrison N., Loe T. K., Mangino M., Milaneschi Y., Miraglio B., Pervjakova N., Russo A., Surakka I., van der Spek A., Verhoeven J. E., Amin N., Beekman M., Blakemore A. I., Canzian F., Hamby S. E., Hottenga J. J., Jones P. D., Jousilahti P., Mägi R., Medland S. E., Montgomery G. W., Nyholt D. R., Perola M., Pietiläinen K. H., Salomaa V., Sillanpää E., Suchiman H. E., van Heemst D., Willemsen G., Agudo A., Boeing H., Boomsma D. I., Chirlaque M.-D., Fagherazzi G., Ferrari P., Franks P., Gieger C., Eriksson J. G., Gunter M., Hägg S., Hovatta I., Imaz L., Kaprio J., Kaaks R., Key T., Krogh V., Martin N. G., Melander O., Metspalu A., Moreno C., Onland-Moret N. C., Nilsson P., Ong K. K., Overvad K., Palli D., Panico S., Pedersen N. L., Penninx B. W. J. H., Quirós J. R., Järvelin M.-R., Rodríguez-Barranco M., Scott R. A., Severi G., Slagboom P. E., Spector T. D., Tjonneland A., Trichopoulou A., Tumino R., Uitterlinden A. G., van der Schouw Y. T., van Duijn C. M., Weiderpass E., Denchi E. L., Matullo G., Butterworth A. S., Danesh J., Samani N. J., Wareham N. J., Nelson C. P., Langenberg C., Codd V., Genome-wide association analysis in humans links nucleotide metabolism to leukocyte telomere length. Am. J. Hum. Genet. 106, 389–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ojha J., Codd V., Nelson C. P., Samani N. J., Smirnov I. V., Madsen N. R., Hansen H. M., de Smith A. J., Bracci P. M., Wiencke J. K., Wrensch M. R., Wiemels J. L., Walsh K. M., Genetic variation associated with longer telomere length increases risk of chronic lymphocytic leukemia. Cancer Epidemiol. Biomarkers Prev. 25, 1043–1049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murnane J. P., Telomeres and chromosome instability. DNA Repair 5, 1082–1092 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Chiba K., Lorbeer F. K., Shain A. H., McSwiggen D. T., Schruf E., Oh A., Ryu J., Darzacq X., Bastian B. C., Hockemeyer D., Mutations in the promoter of the telomerase gene TERT contribute to tumorigenesis by a two-step mechanism. Science 357, 1416–1420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hastie N. D., Dempster M., Dunlop M. G., Thompson A. M., Green D. K., Allshire R. C., Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Armanios M., Blakburn E. H., The telomere syndromes. Nat. Rev. Genet. 13, 693–704 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mangaonkar A. A., Patnaik M. M., Short telomere syndromes in clinical practice: Bridging bench and bedside. Mayo Clin. Proc. 93, 904–916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNally E. J., Luncsford P. J., Armanios M., Long telomeres and cancer risk: The price of cellular immortality. J. Clin. Invest. 129, 3474–3481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Codd V., Wang Q., Allara E., Musicha C., Kaptoge S., Stoma S., Jiang T., Hamby S. E., Braund P. S., Bountziouka V., Budgeon C. A., Denniff M., Swinfield C., Papakonstantinou M., Sheth S., Nanus D. E., Warner S. C., Wang M., Khera A. V., Eales J., Ouwehand W. H., Thompson J. R., Angelantonio E. D., Wood A. M., Butterworth A. S., Danesh J. N., Nelson C. P., Samani N. J., Polygenic basis and biomedical consequences of telomere length variation. Nat. Genet. 53, 1425–1433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steensma D. P., Bejar R., Jaiswal S., Lindsley R. C., Sekeres M. A., Hasserjian R. P., Ebert B. L., Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie M., Lu C., Wang J., McLellan M. D., Johnson K. J., Wendl M. C., McMichael J. F., Schmidt H. K., Yellapantula V., Miller C. A., Ozenberger B. A., Welch J. S., Link D. C., Walter M. J., Mardis E. R., DiPersio J. F., Chen F., Wilson R. K., Ley T. J., Ding L., Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 20, 1472–1478 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genovese G., Kähler A. K., Handsaker R. E., Lindberg J., Rose S. A., Bakhoum S. F., Chambert K., Mick E., Neale B. M., Fromer M., Purcell S. M., Svantesson O., Landén M., Höglund M., Lehmann S., Gabriel S. B., Moran J. L., Lander E. S., Sullivan P. F., Sklar P., Grönberg H., Hultman C. M., McCarroll S. A., Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P. V., Mar B. G., Lindsley R. C., Mermel C. H., Burtt N., Chavez A., Higgins J. M., Moltchanov V., Kuo F. C., Kluk M. J., Henderson B., Kinnunen L., Koistinen H. A., Ladenvall C., Getz G., Correa A., Banahan B. F., Gabriel S., Kathiresan S., Stringham H. M., McCarthy M. I., Boehnke M., Tuomilehto J., Haiman C., Groop L., Atzmon G., Wilson J. G., Neuberg D., Altshuler D., Ebert B. L., Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaiswal S., Natarajan P., Silver A. J., Gibson C. J., Bick A. G., Shvartz E., McConkey M., Gupta N., Gabriel S., Ardissino D., Baber U., Mehran R., Fuster V., Danesh J., Frossard P., Saleheen D., Melander O., Sukhova G. K., Neuberg D., Libby P., Kathiresan S., Ebert B. L., Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuster J. J., MacLauchlan S., Zuriaga M. A., Polackal M. N., Ostriker A. C., Chakraborty R., Wu C.-L., Sano S., Muralidharan S., Rius C., Vuong J., Jacob S., Muralidhar V., Robertson A. A. B., Cooper M. A., Andrés V., Hirschi K. K., Martin K. A., Walsh K., Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bick A. G., Pirruccello J. P., Griffin G. K., Gupta N., Gabriel S., Saleheen D., Libby P., Kathiresan S., Natarajan P., Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation 141, 124–131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zink F., Stacey S. N., Norddahl G. L., Frigge M. L., Magnusson O. T., Jonsdottir I., Thorgeirsson T. E., Sigurdsson A., Gudjonsson S. A., Gudmundsson J., Jonasson J. G., Tryggvadottir L., Jonsson T., Helgason A., Gylfason A., Sulem P., Rafnar T., Thorsteinsdottir U., Gudbjartsson D. F., Masson G., Kong A., Stefansson K., Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bick A. G., Weinstock J. S., Nandakumar S. K., Fulco C. P., Bao E. L., Zekavat S. M., Szeto M. D., Liao X., Leventhal M. J., Nasser J., Chang K., Laurie C., Burugula B. B., Gibson C. J., Lin A. E., Taub M. A., Aguet F., Ardlie K., Mitchell B. D., Barnes K. C., Moscati A., Fornage M., Redline S., Psaty B. M., Silverman E. K., Weiss S. T., Palmer N. D., Vasan R. S., Burchard E. G., Kardia S. L. R., He J., Kaplan R. C., Smith N. L., Arnett D. K., Schwartz D. A., Correa A., Andrade M., Guo X., Konkle B. A., Custer B., Peralta J. M., Gui H., Meyers D. A., McGarvey S. T., Chen I. Y.-D., Shoemaker M. B., Peyser P. A., Broome J. G., Gogarten S. M., Wang F. F., Wong Q., Montasser M. E., Daya M., Kenny E. E., North K. E., Launer L. J., Cade B. E., Bis J. C., Cho M. H., Lasky-Su J., Bowden D. W., Cupples L. A., Mak A. C. Y., Becker L. C., Smith J. A., Kelly T. N., Heckbert S. R., Tiwari H. K., Yang I. V., Heit J. A., Lubitz S. A., Johnsen J. M., Curran J. E., Wenzel S. E., Weeks D. E., Rao D. C., Darbar D., Moon J.-Y., Tracy R. P., Buth E. J., Rafaels N., Loos R. J. F., Durda P., Liu Y., Hou L., Lee J., Kachroo P., Freedman B. I., Levy D., Bielak L. F., Hixson J. E., Floyd J. S., Whitsel E. A., Ellinor P. T., Irvin M. R., Fingerlin T. E., Raffield L. M., Armasu S. M., Wheeler M. M., Sabino E. C., Blangero J., Williams L. K., Levy B. D., Sheu W. H.-H., Roden D. M., Boerwinkle E., Manson J. E., Mathias R. A., Desai P., Taylor K. D., Johnson A. D.; NHLBI Trans-Omics for Precision Medicine Consortium, Auer P. L., Kooperberg C., Laurie C. C., Blackwell T. W., Smith A. V., Zhao H., Lange E., Lange L., Rich S. S., Rotter J. I., Wilson J. G., Scheet P., Kitzman J. O., Lander E. S., Engreitz J. M., Ebert B. L., Reiner A. P., Jaiswal S., Abecasis G., Sankaran V. G., Kathiresan S., Natarajan P., Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taliun D., Harris D. N., Kessler M. D., Carlson J., Szpiech Z. A., Torres R., Taliun S. A. G., Corvelo A., Gogarten S. M., Kang H. M., Pitsillides A. N., LeFaive J., Lee S., Tian X., Browning B. L., Das S., Emde A.-K., Clarke W. E., Loesch D. P., Shetty A. C., Blackwell T. W., Smith A. V., Wong Q., Liu X., Conomos M. P., Bobo D. M., Aguet F., Albert C., Alonso A., Ardlie K. G., Arking D. E., Aslibekyan S., Auer P. L., Barnard J., Barr R. G., Barwick L., Becker L. C., Beer R. L., Benjamin E. J., Bielak L. F., Blangero J., Boehnke M., Bowden D. W., Brody J. A., Burchard E. G., Cade B. E., Casella J. F., Chalazan B., Chasman D. I., Chen Y.-D. I., Cho M. H., Choi S. H., Chung M. K., Clish C. B., Correa A., Curran J. E., Custer B., Darbar D., Daya M., de Andrade M., DeMeo D. L., Dutcher S. K., Ellinor P. T., Emery L. S., Eng C., Fatkin D., Fingerlin T., Forer L., Fornage M., Franceschini N., Fuchsberger C., Fullerton S. M., Germer S., Gladwin M. T., Gottlieb D. J., Guo X., Hall M. E., He J., Heard-Costa N. L., Heckbert S. R., Irvin M. R., Johnsen J. M., Johnson A. D., Kaplan R., Kardia S. L. R., Kelly T., Kelly S., Kenny E. E., Kiel D. P., Klemmer R., Konkle B. A., Kooperberg C., Köttgen A., Lange L. A., Lasky-Su J., Levy D., Lin X., Lin K.-H., Liu C., Loos R. J. F., Garman L., Gerszten R., Lubitz S. A., Lunetta K. L., Mak A. C. Y., Manichaikul A., Manning A. K., Mathias R. A., McManus D. D., McGarvey S. T., Meigs J. B., Meyers D. A., Mikulla J. L., Minear M. A., Mitchell B. D., Mohanty S., Montasser M. E., Montgomery C., Morrison A. C., Murabito J. M., Natale A., Natarajan P., Nelson S. C., North K. E., O’Connell J. R., Palmer N. D., Pankratz N., Peloso G. M., Peyser P. A., Pleiness J., Post W. S., Psaty B. M., Rao D. C., Redline S., Reiner A. P., Roden D., Rotter J. I., Ruczinski I., Sarnowski C., Schoenherr S., Schwartz D. A., Seo J., Seshadri S., Sheehan V. A., Sheu W. H., Shoemaker M. B., Smith N. L., Smith J. A., Sotoodehnia N., Stilp A. M., Tang W., Taylor K. D., Telen M., Thornton T. A., Tracy R. P., Berg D. J. V. D., Vasan R. S., Viaud-Martinez K. A., Vrieze S., Weeks D. E., Weir B. S., Weiss S. T., Weng L.-C., Willer C. J., Zhang Y., Zhao X., Arnett D. K., Ashley-Koch A. E., Barnes K. C., Boerwinkle E., Gabriel S., Gibbs R., Rice K. M., Rich S. S., Silverman E. K., Qasba P., Gan W.; NHLBI Trans-Omic for Precision Medicine (TOPMed) Consortium, Papanicolaou G. J., Nickerson D. A., Browning S. R., Zody M. C., Zöllner S., Wilson J. G., Cupples L. A., Laurie C. C., Jaquish C. E., Hernandez R. D., O’Connor T. D., Abecasis G. R., Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hout C. V. V., Tachmazidou I., Backman J. D., Hoffman J. D., Liu D., Pandey A. K., Gonzaga-Jauregui C., Khalid S., Ye B., Banerjee N., Li A. H., O’Dushlaine C., Marcketta A., Staples J., Schurmann C., Hawes A., Maxwell E., Barnard L., Lopez A., Penn J., Habegger L., Blumenfeld A. L., Bai X., O’Keeffe S., Yadav A., Praveen K., Jones M., Salerno W. J., Chung W. K., Surakka I., Willer C. J., Hveem K., Leader J. B., Carey D. J., Ledbetter D. H.; Geisinger-Regeneron DiscovEHR Collaboration, Cardon L., Yancopoulos G. D., Economides A., Coppola G., Shuldiner A. R., Balasubramanian S., Cantor M.; Regeneron Genetics Center, Nelson M. R., Whittaker J., Reid J. G., Marchini J., Overton J. D., Scott R. A., Abecasis G. R., Yerges-Armstrong L., Baras A., Exome sequencing and characterization of 49,960 individuals in the UK Biobank. Nature 586, 749–756 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taub M. A., Conomos M. P., Keener R., Iyer K. R., Weinstock J. S., Yanek L. R., Lane J., Miller-Fleming T. W., Brody J. A., Raffield L. M., McHugh C. P., Jain D., Gogarten S. M., Laurie C. A., Keramati A., Arvanitis M., Smith A. V., Heavner B., Barwick L., Becker L. C., Bis J. C., Blangero J., Bleecker E. R., Burchard E. G., Celedón J. C., Chang Y. P. C., Custer B., Darbar D., de las Fuentes L., DeMeo D. L., Freedman B. I., Garrett M. E., Gladwin M. T., Heckbert S. R., Hidalgo B. A., Irvin M. R., Islam T., Johnson W. C., Kaab S., Launer L., Lee J., Liu S., Moscati A., North K. E., Peyser P. A., Rafaels N., Seidman C., Weeks D. E., Wen F., Wheeler M. M., Williams L. K., Yang I. V., Zhao W., Aslibekyan S., Auer P. L., Bowden D. W., Cade B. E., Chen Z., Cho M. H., Cupples L. A., Curran J. E., Daya M., Deka R., Eng C., Fingerlin T. E., Guo X., Hou L., Hwang S.-J., Johnsen J. M., Kenny E. E., Levin A. M., Liu C., Minster R. L., Naseri T., Nouraie M., Reupena M. S., Sabino E. C., Smith J. A., Smith N. L., Lasky-Su J., Taylor J. G., Telen M. J., Tiwari H. K., Tracy R. P., White M. J., Zhang Y., Wiggins K. L., Weiss S. T., Vasan R. S., Taylor K. D., Sinner M. F., Silverman E. K., Shoemaker M. B., Sheu W. H.-H., Sciurba F., Schwartz D. A., Rotter J. I., Roden D., Redline S., Raby B. A., Psaty B. M., Peralta J. M., Palmer N. D., Nekhai S., Montgomery C. G., Mitchell B. D., Meyers D. A., McGarvey S. T.; Fernando D. Martinez on behalf of the NHLBI CARE Network, Mak A. C. Y., Loos R. J. F., Kumar R., Kooperberg C., Konkle B. A., Kelly S., Kardia S. L. R., Kaplan R., He J., Gui H., Gilliland F. D., Gelb B. D., Fornage M., Ellinor P. T., de Andrade M., Correa A., Chen Y.-D. I., Boerwinkle E., Barnes K. C., Ashley-Koch A. E., Arnett D. K., Albert C.; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; TOPMed Hematology and Hemostasis Working Group; TOPMed Structural Variation Working Group, Laurie C. C., Abecasis G., Nickerson D. A., Wilson J. G., Rich S. S., Levy D., Ruczinski I., Aviv A., Blackwell T. W., Thornton T., O’Connell J., Cox N. J., Perry J. A., Armanios M., Battle A., Pankratz N., Reiner A. P., Mathias R. A., Genetic determinants of telomere length from 109,122 ancestrally diverse whole-genome sequences in TOPMed. Cell Genom. 2, 100084 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Z., Mangino M., Aviv A.; UK10K Consortium, Spector T., Durbin R., Estimating telomere length from whole genome sequence data. Nucleic Acids Res. 42, e75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schratz K. E., Haley L., Danoff S. K., Blackford A. L., DeZern A. E., Gocke C. D., Duffield A. S., Armanios M., Cancer spectrum and outcomes in the Mendelian short telomere syndromes. Blood 135, 1946–1956 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gonzalo S., Jaco I., Fraga M. F., Chen T., Li E., Esteller M., Blasco M. A., DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat. Cell Biol. 8, 416–424 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Yang J., Guo R., Wang H., Ye X., Zhou Z., Dan J., Wang H., Gong P., Deng W., Yin Y., Mao S., Wang L., Ding J., Li J., Keefe D. L., Dawlaty M. M., Wang J., Xu G., Liu L., Tet enzymes regulate telomere maintenance and chromosomal stability of mouse ESCs. Cell Rep. 15, 1809–1821 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Tutton S., Lieberman P. M., A role for p53 in telomere protection. Mol. Cell. Oncol. 4, e1143078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haycock P. C., Heydon E. E., Kaptoge S., Butterworth A. S., Thompson A., Willeit P., Leucocyte telomere length and risk of cardiovascular disease: Systematic review and meta-analysis. BMJ 349, g4227 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunt S. C., Kark J. D., Aviv A., Association between shortened leukocyte telomere length and cardio-metabolic outcomes. Circ. Cardiovasc. Genet. 8, 4–7 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Verbanck M., Chen C.-Y., Neale B., Do R., Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tingley D., Yamamoto T., Hirose K., Keele L., Imai K., mediation: R Package for causal mediation analysis. J. Stat. Soft. 59, 1–38 (2014). [Google Scholar]
- 41.Giaccherini M., Macauda A., Sgherza N., Sainz J., Gemignani F., Maldonado J. M. S., Jurado M., Tavano F., Mazur G., Jerez A., Góra-Tybor J., Gołos A., Mohedo F. H., Lopez J. M., Várkonyi J., Spadano R., Butrym A., Canzian F., Campa D., Genetic polymorphisms associated with telomere length and risk of developing myeloproliferative neoplasms. Blood Cancer J. 10, 89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernard L., Belisle C., Mollica L., Provost S., Roy D.-C., Gilliland D. G., Levine R. L., Busque L., Telomere length is severely and similarly reduced in JAK2V617F-positive and -negative myeloproliferative neoplasms. Leukemia 23, 287–291 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aviv A., Anderson J. J., Shay J. W., Mutations, cancer and the telomere length paradox. Trends Can. 3, 253–258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watson C. J., Papula A. L., Poon G. Y. P., Wong W. H., Young A. L., Druley T. E., Fisher D. S., Blundell J. R., The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 367, 1449–1454 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Blasco M. A., Lee H.-W., Hande M. P., Samper E., Lansdorp P. M., DePinho R. A., Greider C. W., Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 91, 25–34 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Jeong M., Park H. J., Celik H., Ostrander E. L., Reyes J. M., Guzman A., Rodriguez B., Lei Y., Lee Y., Ding L., Guryanova O. A., Li W., Goodell M. A., Challen G. A., Loss of Dnmt3a immortalizes hematopoietic stem cells in vivo. Cell Rep. 23, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stefanick M. L., Cochrane B. B., Hsia J., Barad D. H., Liu J. H., Johnson S. R., The women’s health initiative postmenopausal hormone trials: Overview and baseline characteristics of participants. Ann. Epidemiol. 13, S78–S86 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Honigberg M. C., Zekavat S. M., Niroula A., Griffin G. K., Bick A. G., Pirruccello J. P., Nakao T., Whitsel E. A., Farland L. V., Laurie C., Kooperberg C., Manson J. E., Gabriel S., Libby P., Reiner A. P., Ebert B. L.; NHLBI Trans-Omics for Precision Medicine Program, Natarajan P., Premature menopause, clonal hematopoiesis, and coronary artery disease in postmenopausal women. Circulation 143, 410–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cibulskis K., Lawrence M. S., Carter S. L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E. S., Getz G., Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pedersen B. S., Quinlan A. R., Mosdepth: Quick coverage calculation for genomes and exomes. Bioinformatics 34, 867–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura M., Stone R. C., Hunt S. C., Skurnick J., Lu X., Cao X., Harley C. B., Aviv A., Measurement of telomere length by the Southern blot analysis of terminal restriction fragment lengths. Nat. Protoc. 5, 1596–1607 (2010). [DOI] [PubMed] [Google Scholar]
- 52.Balduzzi S., Rücker G., Schwarzer G., How to perform a meta-analysis with R: A practical tutorial. Evid. Based Mental Health 22, 153–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burgess S., Smith G. D., Davies N. M., Dudbridge F., Gill D., Glymour M. M., Hartwig F. P., Holmes M. V., Minelli C., Relton C. L., Theodoratou E., Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 4, 186 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemani G., Zheng J., Elsworth B., Wade K. H., Haberland V., Baird D., Laurin C., Burgess S., Bowden J., Langdon R., Tan V. Y., Yarmolinsky J., Shihab H. A., Timpson N. J., Evans D. M., Relton C., Martin R. M., Smith G. D., Gaunt T. R., Haycock P. C., The MR-Base platform supports systematic causal inference across the human phenome. eLife 7, e34408 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.W. Revelle, Procedures for Psychological, Psychometric, and Personality Research [R package psych version 2.0.9]. Comprehensive R Archive Network (CRAN) (2020); https://cran.r-project.org/web/packages/psych/index.html.
- 56.Yavorska O. O., Burgess S., MendelianRandomization: An R package for performing Mendelian randomization analyses using summarized data. Int. J. Epidemiol. 46, 1734–1739 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Imai K., Keele L., Tingley D., A general approach to causal mediation analysis. Psychol. Methods 15, 309–334 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Bycroft C., Freeman C., Petkova D., Band G., Elliott L. T., Sharp K., Motyer A., Vukcevic D., Delaneau O., O’Connell J., Cortes A., Welsh S., Young A., Effingham M., McVean G., Leslie S., Allen N., Donnelly P., Marchini J., The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S16
Tables S1 to S3, S7 to S9
Tables S4 to S6