Abstract
The mitochondrial integrated stress response (mitoISR) has emerged as a major adaptive pathway to respiratory chain deficiency, but both the tissue specificity of its regulation, and how mitoISR adapts to different levels of mitochondrial dysfunction are largely unknown. Here, we report that diverse levels of mitochondrial cardiomyopathy activate mitoISR, including high production of FGF21, a cytokine with both paracrine and endocrine function, shown to be induced by respiratory chain dysfunction. Although being fully dispensable for the cell-autonomous and systemic responses to severe mitochondrial cardiomyopathy, in the conditions of mild-to-moderate cardiac OXPHOS dysfunction, FGF21 regulates a portion of mitoISR. In the absence of FGF21, a large part of the metabolic adaptation to mitochondrial dysfunction (one-carbon metabolism, transsulfuration, and serine and proline biosynthesis) is strongly blunted, independent of the primary mitoISR activator ATF4. Collectively, our work highlights the complexity of mitochondrial stress responses by revealing the importance of the tissue specificity and dose dependency of mitoISR.
FGF21 regulates mitochondrial stress responses under mild OXPHOS dysfunction, thus preventing pathological cardiac remodeling.
INTRODUCTION
Mitochondria are organelles essential for energy production and numerous other cell processes, hence adapting to different exogenous and endogenous stresses to maintain cellular homeostasis. The ability of mitochondria to adapt to changing conditions by activating versatile signaling pathways is collectively known as the mitochondrial stress response, and recent studies have shown that it largely overlaps with the integrated stress response (ISR) (1–3). ISR is activated by various stress signals and metabolic cues, which lead to phosphorylation of eIF2α, resulting in the inhibition of global translation (3). In parallel, a subset of transcripts, including activating transcription factor 4 (Atf4), is preferentially translated to ensure effective responses to cellular stress (3). Recently, it was proposed that mitochondrial stress activates inner mitochondrial membrane (IMM) protease OMA1, which cleaves IMM protein DELE1. The cleaved DELE1 accumulates in the cytosol where it interacts and activates HRI, one of four eIF2α kinases, resulting in the activation of the mitochondrial (mito)ISR response (4, 5). Through ATF4 activation, mitoISR increases transcription of downstream target genes, including enzymes involved in 1C metabolism, serine biosynthesis, transsulfuration, and the glutamate-to-proline conversion pathway, as well as Gdf15 and Fgf21, the two cytokines shown to be excreted from tissues upon oxidative phosphorylation (OXPHOS) deficiency (1, 2, 6–8).
Fibroblast growth factor 21 (FGF21) is a cytokine released mainly from the liver and adipocytes during periods of increased feeding to regulate systemic glucose and lipid metabolism (9). Although FGF21 in mice is up-regulated after short-term starvation, in humans this seems to be more connected with pathophysiological response to prolonged starvation and accompanying tissue damage (10). FGF21 is also described as a mitochondrial cytokine or “mitokine,” which is expressed from noncanonical tissues, like skeletal muscles, heart, and brain, upon mitochondrial dysfunction (6, 11, 12). Early, high-regulation of Fgf21 expression was first described in Deletor mice (6), whose multiple mitochondrial DNA deletions serve as an animal model for mitochondrial dysfunction; since then however, upregulated Fgf21 expression has also been detected in many other models of mitochondrial dysfunction including the Dars2-heart and skeletal muscle–deficient mice (11), Clpp-deficient mice (13), mitochondrial DNA mutator mice (14), skeletal muscle–specific Opa1-deficient mice (15), and neuron-specific Drp1-deficient mice (12). While multiple roles have been proposed for FGF21 in different tissues, we still do not fully understand its role in cell-autonomous and systemic effects of mitochondrial dysfunction.
Although the heart produces negligible amounts of FGF21 under normal conditions, cardiac FGF21 secretion increases markedly in response to stress (11, 16). In the present study, we explored whether and how FGF21 mediates mitochondrial stress responses, primarily in the heart. Using a series of different models, we can demonstrate that FGF21 might be a modulator of stress signaling in mild-to-moderate mitochondrial dysfunction. However, the effects of FGF21 are dispensable or overtaken by other stress responses in severe mitochondrial dysfunction. Moreover, FGF21 has differential tissue-specific effects and its expression could be dose-dependent correlating with the severity of mitochondrial dysfunction.
RESULTS
Tissue-specific depletion of FGF21 does not influence the overall phenotype of the DARS2-deficient mice
To decipher the in vivo role of FGF21 in mitochondrial stress response, we selectively disrupted the Fgf21 gene in DARS2 (mitochondrial aspartyl-tRNA synthetase)–deficient heart and skeletal muscles, using a CRE recombinase expressed under muscle creatine kinase promoter (Ckmm-Cre) (11). These mice present severe cardiomyopathy accompanied by strong OXPHOS deficiency, resulting in an early death at 7 to 8 weeks of age (11). At 3 weeks of age, before OXPHOS deficiency is evident, several stress responses are activated in these animals, including a massive 250-fold up-regulation of Fgf21 levels exclusively in the heart (11). The eightfold Fgf21 up-regulation is one of the earliest changes observed already at 1 week of age in this model (11). To reduce the overall number of mattings and animals used in the experiments, triple heterozygous animals (Dars2+/L; Fgf21+/L; Ckmm-Cre+/T) were mated with Dars2L/L; Fgf21L/L animals to obtain control (all mice without Ckmm-Cre transgene), FGF21FL (Dars2+/L, Fgf21L/L, Ckmm-Cre+/T), DARS2FL (Dars2L/L, Fgf21+/L, Ckmm-Cre+/T), and DKOFL (Dars2L/L, Fgf21L/L, Ckmm-Cre+/T) mice (fig. S1A). Mice of all different genotypes were born at the expected Mendelian ratios. Control experiments were performed to ensure that FGF21FL and DARS2FL that were also heterozygous for Dars2 and Fgf21, respectively, did not differ from single mutants (Dars2L/L, Fgf21+/+, Ckmm-Cre+/T and Dars2+/+, Fgf21L/L, Ckmm-Cre+/T) in the observed phenotypes (fig. S1B).
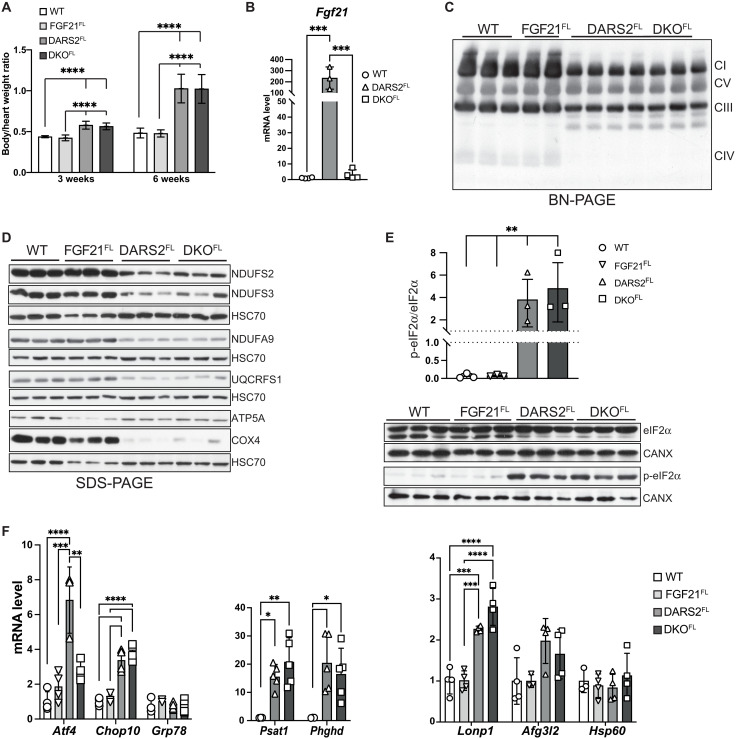
The severe shortening of life span to 7 to 8 weeks in DARS2FL mice was not affected by the loss of FGF21 in DKOFL hearts. The heart size and heart–to–body weight ratio at 3 and 6 weeks of age were not significantly different between DARS2FL and DKOFL mice (Fig. 1A). However, a strong up-regulation of Fgf21 expression previously observed in DARS2FL hearts was absent in DKOFL at 6 weeks of age (Fig. 1B).
Fig. 1. Tissue-specific depletion of FGF21 in DARS2-deficient hearts does not influence the levels of mitochondrial stress response, nor it changes the severity of OXPHOS deficiency.
(A) Relative heart–to–body weight ratio at 3 and 6 weeks of age (n = 4 to 20). (B) Relative Fgf21 transcript levels in wild-type (WT), FGF21FL, DARS2FL, and DKOFL animals (n = 3 to 4). (A and B) Bars represent means ± SD. One-way analysis of variance (ANOVA) and Tukey’s multiple comparisons test were used for significance (***P < 0.001 and ****P < 0.0001). (C) BN-PAGE analysis of the respiratory chain complexes in the presence of n-dodecyl-β-d-maltoside (DDM) followed by Western blot analysis using OXPHOS cocktail antibody (CI, NDUFB8; CII, SDHB; CIII, UQCRC2; CIV, COX1; CV, ATP5A). (D) Western blot analyses of different OXPHOS subunits. HSC70 was used as a loading control. Antibodies used were raised against proteins indicated in panels (n = 3). (E) Western blot (bottom) and quantification (top) of phosphorylated and nonphosphorylated levels of eIF2α. HSC70 and CANX were used as loading controls. Antibodies used were raised against proteins indicated in panels (n = 3). (B to E) The experiments are performed on heart lysates of 6-week-old animals. (F) Relative transcript levels of indicated genes in 6-week-old hearts (n = 3 to 5). (E and F) Bars represent means ± SD. One-way ANOVA and Tukey’s multiple comparisons test were used for significance (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
DARS2 deficiency severely affects levels of the mitochondrial respiratory chain (MRC), especially levels of complex I (CI) and IV (CIV) (11). To investigate whether FGF21 is needed for the maintenance of the respiratory chain, OXPHOS complexes were analyzed by blue native polyacrylamide gel electrophoresis (BN-PAGE) coupled with Western blot analysis. The levels of CI and CIV have equally decreased in DARS2FL and DKOFL mice at 6 weeks of age (Fig. 1C). The steady-state levels of individual OXPHOS subunits were evenly changed, confirming that FGF21 depletion does not affect the levels of MRC upon DARS2 loss (Fig. 1D). Strong OXPHOS deficiency in DARS2FL and DKOFL mice was confirmed by enzyme histochemical double staining for CIV (COX; cytochrome c oxidase) and CII [succinate dehydrogenase (SDH)] activities (fig. S1C). A high number of respiratory incompetent cells (COX−/SDH+) that stained purple/blue were detected in both DARS2FL and DKOFL mice, in contrast to control and FGF21FL hearts that showed uniform brown staining, indicative of OXPHOS competent (COX+) cells (fig. S1C). Diminished activity of respiratory chain complexes caused by the loss of DARS2 led to cardiomyopathy characterized by increased expression of cardiac hypertrophy marker Nppa regardless of the presence of FGF21 at 6 weeks of age (fig. S1D).
Stress responses are not affected by the loss of FGF21 in DARS2-deficient hearts
We previously reported that DARS2 deficiency in the heart, but not skeletal muscle, activates mitoISR (11, 17). Still, increased eIF2α phosphorylation in DARS2-deficient hearts was not affected by the FGF21 depletion (Fig. 1E). Consistently, Atf4 and Chop/Ddit3 were up-regulated upon DARS2 loss, but not Grp78/Bip, a marker of the canonical unfolded protein response (UPR; Fig. 1F). Although Atf4 transcripts were less up-regulated in the DKOFL than in DARS2FL hearts, we did not observe a difference in ATF4 protein level or the downstream targets of ATF4 such as 1C metabolism genes, which remained highly up-regulated at transcript (Psat1 and Phgdh) and protein level (SHMT2 and MTHFD2) upon FGF21 depletion in DARS2-deficient hearts (Fig. 1F and fig. S1, B and E).
In agreement with recent results showing that mitochondrial UPR is not the major stress response activated by the respiratory chain dysfunction in mammals, we observe major changes neither on the transcript levels of mitochondrial proteases Afg3l2 nor on the steady-state protein levels of mitochondrial chaperones HSP60 and GRP75 (mtHSP70; Fig. 1F and fig. S1F). The exception was LON protease whose transcript and protein levels were increased in DARS2FL and DKOFL hearts, which is not surprising considering that Lonp1 is also a direct ISR target (Fig. 1F and fig. S1F).
The mechanistic (mammalian) target of rapamycin (mTOR) complex I (mTORC1) is a major regulator of metabolic signaling that is activated by mitochondrial dysfunction (8). Phosphorylation of mTORC1 targets 4EBP1 and PRAS40, commonly used as a readout for its activation (18), was equally increased in DARS2FL and DKOFL hearts (fig. S1G). In agreement, the suppression of LC3B-I to LC3B-II conversion and accumulation of P62 and BECLIN were detected in both DARS2FL and DKOFL hearts, suggesting a reduction in autophagy caused by the mitochondrial dysfunction, which was again not affected by FGF21 depletion (fig. S1H). Together, these results indicate that the absence of FGF21 in the heart does not affect multiple stress responses activated by the strong mitochondrial dysfunction caused by DARS2 depletion.
The loss of FGF21 does not affect proteome remodeling dictated by the DARS2 depletion
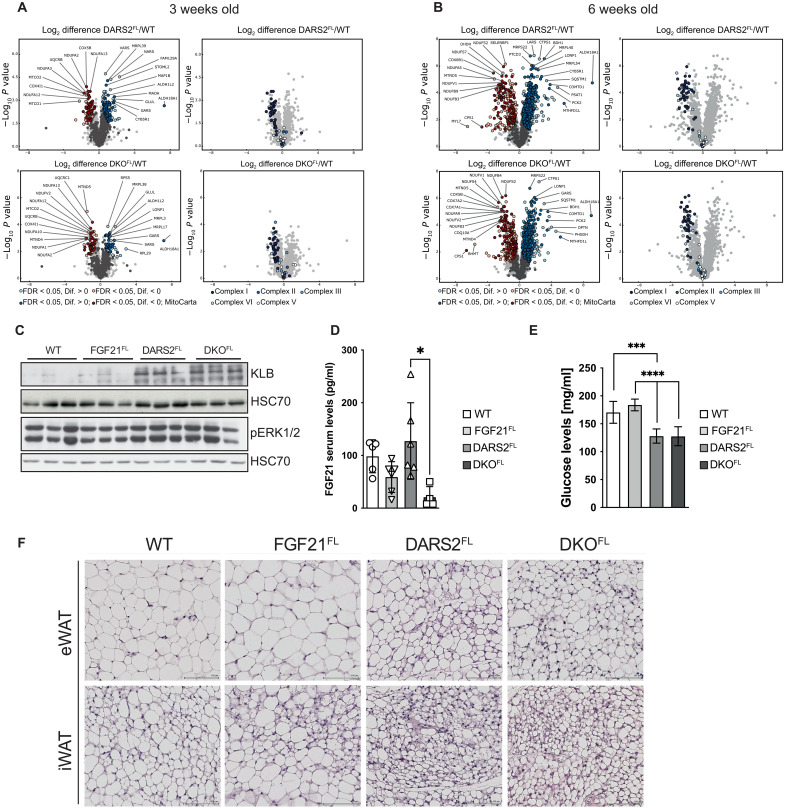
To exclude the possibility that the effects of FGF21 might be masked by a strong OXPHOS dysfunction observed in DARS2-deficient hearts at 6 weeks of age, we next measured the levels of OXPHOS subunits at 3 weeks of age. Milder changes in the steady-state levels of individual OXPHOS subunits were detected at this age, compared to 6-week-old animals (fig. S2A and Fig. 1D, respectively). Still, no difference between DARS2FL and DKOFL hearts was observed (fig. S2A). Further confirmation of these results came from the label-free whole proteome analysis of the 3-week-old animals that showed no significant changes between DARS2FL and DKOFL heart proteomes (Fig. 2A, fig. S2B, and table S1). Significant changes imposed by the loss of DARS2, compared to the wild-type (WT) hearts, could be identified, regardless of the FGF21 presence (Fig. 2A, fig. S2C, and table S1). Major down-regulated proteins and pathways enriched in both DARS2FL and DKOFL hearts included MRC complexes, in particular subunits of CI and CIV (Fig. 2A and fig. S2C). Three-quarters of all significantly down-regulated proteins were MRC subunits (table S1). Up-regulated proteins echoed the activation of mitoISR and included enzymes involved in the 1C and amino acid metabolism, as well as cytoplasmic amino acid tRNA synthases (Fig. 2A, fig. S2D, and table S1) (3, 11, 17). A significant increase in a large number of mitochondrial ribosomal subunits, likely as a compensatory response to disturbed protein synthesis, was also observed (fig. S2C and table S1).
Fig. 2. Whole-proteome changes in DARS2-deficient hearts are not dependent on tissue-specific FGF21 loss, as well as the browning of WAT.
(A and B) Volcano plots of whole-heart proteome changes in DARS2FL and DKOFL animals compared to WT at (A) 3 weeks and (B) 6 weeks of age [false discovery rate (FDR) < 0.05]. Dark blue and dark red dots represent MitoCarta proteins (left). Significant changes only in OXPHOS complexes (I to V) (right) (n = 4). (C) Western blot analysis of ERK1/2 phosphorylation levels. HSC70 was used as a loading control. Experiments were performed on cardiac lysates of mice at 6 weeks of age (n = 3). (D) Serum FGF21 (pg/ml) and (E) glucose levels (mg/ml) measured in nonfasted mice fed normal chow diet at 6 weeks of age (n = 4 to 8). Bars represent means ± SD (one-way ANOVA and Tukey’s multiple comparisons test; *P < 0.05, ***P < 0.001, and ****P < 0.0001). (F) Hematoxylin and eosin (H&E) staining of epidydimal WAT (eWAT) and inguinal WAT (iWAT) sections of mice at 6 weeks of age (n = 3). Scale bars, 100 μm.
Similar and further changes were detected when label-free proteomes of 6-week-old hearts were analyzed (table S2). Again, we did not observe significant changes (q < 0.05) between DARS2FL and DKOFL hearts (Fig. 2B, fig. S2B, and table S2). Still, the clear progression of the phenotype from 3 to 6 weeks of age could be observed (Fig. 2, A and B, and tables S1 and S2). In agreement with previous analyses, the large proportion of the most up-regulated proteins (10-fold to over hundred-fold increase) was involved in 1C metabolism (MTHFD2 and MTHFD1L) and amino acid synthesis (ALDH18A1, ASNS, PHGDH, PYCR2, PSAT1, and PCK1; table S2). We recently showed that already at 2 weeks of age, before the OXPHOS defect, the same proteins are the most up-regulated in DARS2-deficient hearts (17), suggesting that the primary stress response triggered by defective protein synthesis in mitochondria is directly related to the unbalanced amino acid metabolism. The most markedly decreased proteins in DARS2FL and DKOFL hearts were again OXPHOS subunits, mainly belonging to CI, CIII, and CIV subunits, reflecting a strong defect of mitochondrial translation (Fig. 2, A and B).
ERK1/2 signaling in the DARS2-deficient heart is still activated in the absence of FGF21
FGF21 produced in cardiomyocytes binds the FGFR1 receptor and its co-receptor β-KLOTHO (KLB) in an autocrine or paracrine fashion, thus activating the downstream signaling pathways (19). FGF21 signaling primarily leads to phosphorylation and consequent activation of extracellular signal–regulated kinases 1 and 2 [ERK1/2; also known as mitogen-activated protein kinase 3 (MAPK3)/MAPK1], which further regulate multiple metabolic processes (16, 20). Unexpectedly, we detected up-regulated protein levels of KLB and ERK1/2 upon DARS2 depletion in DARS2FL and DKOFL hearts (Fig. 2C). Klb transcript levels were also mildly up-regulated in DKOFL hearts, suggesting a compensatory up-regulation triggered by the lack of FGF21 (fig. S2E). The transcript levels of Pparα and Sirt1, involved in the propagation of Fgf21 expression in cardiomyocytes, were not affected by the FGF21 loss in DKOFL hearts (fig. S2E) (16). Last, we showed that levels of FGF19 and FGF23, two FGF factors of the same subfamily as FGF21, which could bind KLOTHO co-receptors and activate FGF receptor signaling, were not changed upon DARS2 depletion (fig. S2F). The activation of the heart FGF receptor (β-KLOTHO/FGFR1) by these cytokines is anyway highly unlikely as FGF19 binds primary β-KLOTHO/FGFR4 in the liver, while FGF23 specifically binds α-KLOTHO in combination with different FGFRs (FGFR1C/3C/4) in the kidney (19). Collectively, these results showed an unexpected activation of the FGF signaling cascade in DKOFL hearts, suggesting that endocrine FGF21 from liver or adipocyte tissues could activate the signaling cascade in the dysfunctional heart. The increased FGF21 serum levels were detected in DARS2-deficient mice, which is in line with previous findings (11). The levels of this cytokine were significantly down-regulated in DKOFL mice, but still present (Fig. 2D).
Besides mediating autocrine and paracrine signaling on the tissue of origin, FGF21 was proposed to have prominent effects on circulating glucose levels and the physiology of white (WAT) and brown (BAT) adipose tissues (10, 21). Still, even in the absence of FGF21 in the heart, DARS2-deficient animals had lower circulating glucose levels (Fig. 2E). Similarly, the loss of FGF21 did not affect prominent “browning” of the inguinal WAT (iWAT) and epididymal WAT (eWAT) or “whitening” of BAT in DKOFL animals (Fig. 2F and fig. S2G, respectively).
Endocrine FGF21 does not regulate adaptive responses upon DARS2 depletion
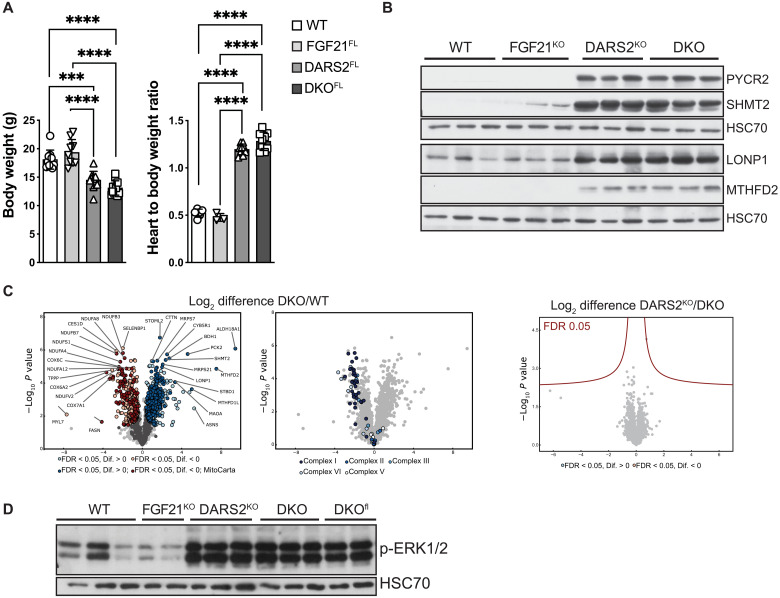
To investigate whether the circulating FGF21 could still activate the signaling pathways in DARS2-deficient hearts and mediate the cell nonautonomous effects, a new mouse model lacking FGF21 in the whole body was created [double-knockout (DKO)—Dars2L/L, Ckmm-Cre+/T; Fgf21−/−]. The new DKO mice had comparable body weight, heart–to–body weight ratio, and life span to DARS2KO mice (Fig. 3A). The complete loss of FGF21 did not affect respiratory chain deficiency in DARS2-deficient hearts (fig. S3A). The analysis of mitoISR in DARS2KO and DKO hearts revealed a uniform increase in the level of phosphorylated eIF2α and corresponding downstream targets (MTHFD2, SHMT2, PYCR1, and LONP1; Fig. 3B and fig. S3B). Furthermore, a comparable level of mTOR activation was detected in DARS2 single- or double-deficient mice (DKO; fig. S3B). Label-free quantitative profiling of 6-week-old mice mirrored previous results and showed no significant changes between DARS2KO and DKO heart proteome (Fig. 3C). When compared to WT hearts, the most up-regulated pathways in DKO animals reflected robust mitoISR activation and strong down-regulation of respiratory chain complexes (table S3). The most up-regulated Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways included cytoplasmic and mitochondrial translation and ribosomes, and processes involved in Ca2+ handling in the cell and mitochondria (fig. S3C). Besides OXPHOS complexes, down-regulated KEGG pathways suggested problems with cardiomyocyte organization and lipoprotein transport (fig. S3C). Almost identical changes were observed in DKOFL animals when compared to WT mice (fig. S3C).
Fig. 3. Overall phenotype and stress response of DARS2-deficient mice is not changed with the FGF21 depletion in the whole body.
(A) Body weight (left; n = 9 to 12) and heart–to–body weight ratio (right; n = 3 to 12) of WT, FGF21FL, DARS2FL, and DKOFL animals at 6 weeks of age. (B) Western blot analyses of cardiac lysates of mice at 6 weeks of age. Antibodies used were raised against proteins indicated in panels. HSC70 was used as a loading control. (C) Volcano plots of whole cardiac proteome changes in DKOKO compared to WT mice (FDR < 0.05). Dark blue and dark red dots represent MitoCarta proteins (left). Significant changes only in OXPHOS complexes (I to V) between DKO and WT hearts (middle). Volcano plot showing no significant (FDR < 0.05) changes between DKOKO and DARS2KO hearts at 6 weeks of age (right). (D) Western blot analysis of ERK1/2 phosphorylation levels. HSC70 was used as a loading control. Experiments were performed on cardiac lysates of mice at 6 weeks of age (n = 3).
Last, phosphorylated ERK1/2 levels were analyzed in DARS2KO and DKO heart lysates. ERK1/2 was phosphorylated, hence activated to the same level in DARS2KO, DKOFL, and DKO mice, independently of the autocrine/paracrine or endocrine origin of FGF21 (Fig. 3D). These data suggest that severe mitochondrial dysfunction in the heart does not rely (just) on FGF21 for the activation of ERK1/2-mediated pathway but triggers additional compensatory pathways in the absence of this cytokine. Therefore, loss of FGF21 does not influence the signaling that leads to the stress responses activated upon DARS2 deficiency, either in mitochondrially deficient heart or in a cell nonautonomous manner in the rest of the body.
FGF21 regulates mitoISR in the heart upon mild mitochondrial dysfunction caused by CLPP depletion
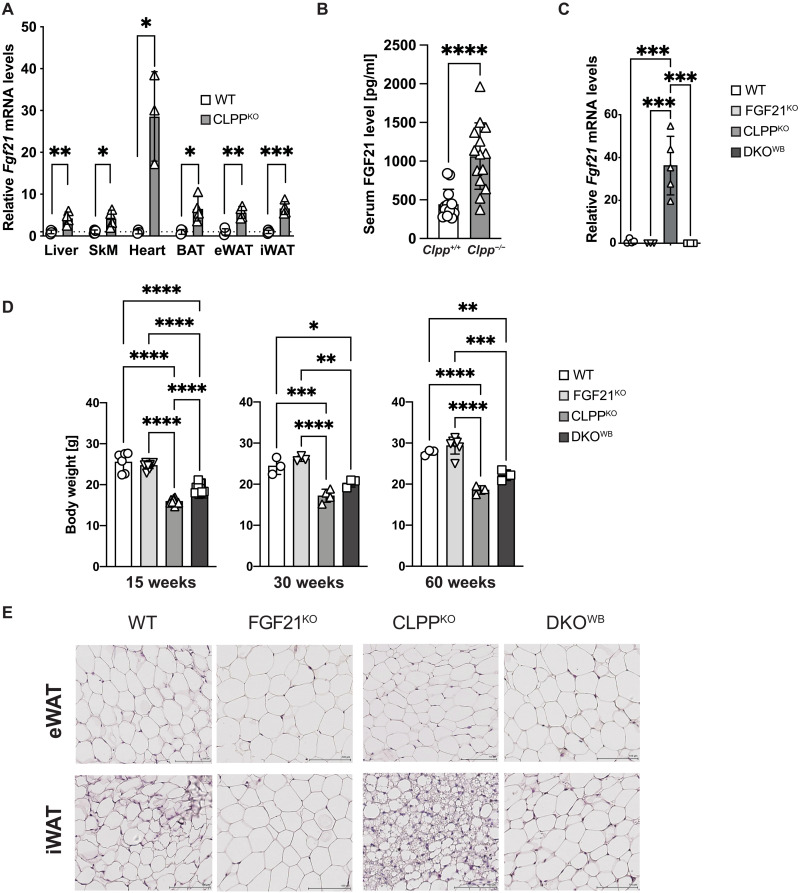
To test whether, in the context of milder mitochondrial deficiency in the heart, FGF21 up-regulation could have a more defined role, FGF21 was depleted in mice deficient for the mitochondrial matrix protease CLPP (DKOWB—Clpp−/−; Fgf21−/−) (22). Previously, we have shown that in the heart, CLPP protease regulates the mitoribosome assembly and the rate of mitochondrial protein synthesis through timely removal of its substrate ERAL1 (22). Our further studies demonstrated that CLPP promotes the well-being of the MRC by promoting the CI salvage pathway (23). Therefore, in the absence of CLPP, animals develop mild-to-moderate respiratory chain deficiency that does not affect life span but renders animals resistant to diet-induced obesity and resulting in intolerance to cold (13). In agreement with the OXPHOS defect, CLPP KO mice showed consistent up-regulation of Fgf21 levels in multiple tissues, which led to an increase in FGF21 circulating levels (Fig. 4, A and B). The observed increase in Fgf21 transcripts was an order-of-magnitude higher in the heart than in other tissues and was fully suppressed in DKOWB animals (Fig. 4, A and C). Depletion of FGF21 in DKOWB mice did not influence the level of OXPHOS dysfunction resulting from the CLPP deficiency (fig. S4, A to C). OXPHOS supercomplex levels and activity were equally affected in CLPP KO and DKOWB hearts (fig. S4, A and B).
Fig. 4. Phenotypical and metabolic characterization of CLPP- and FGF21-deficient mice.
(A) Relative Fgf21 transcript levels in different tissues of CLPPKO mice (n = 3). SkM, skeletal muscle. (B) Serum FGF21 (pg/ml) levels measured in nonfasted WT and CLPPKO mice fed normal chow diet at 17 (±2) weeks of age (n = 15). (A and B) Bars represent means ± SD (unpaired two-tailed Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). (C) Relative Fgf21 transcript levels in WT, FGF21KO, CLPPKO, and DKOKO mice at 17 (±2) weeks of age (n = 3 to 5). (D) Body weight of male mice at different ages. (A to D) One-way ANOVA and Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (E) H&E staining of eWAT and iWAT tissue sections of mice at 17 (±2) weeks of age (n = 3). Scale bars, 100 μm.
FGF21-deficient mice exhibit a mild weight gain, increased adipocyte size, and slightly impaired glucose homeostasis (24). The loss of FGF21 in the whole body of DKOWB animals led to small but significant increase in the body weight compared to CLPP KO mice throughout the life span (Fig. 4D). These mice, however, never reached the weight of the WT animals (Fig. 4D). During the same period, FGF21 KO animals started from a similar size as WT, only to become heavier as they grew older (Fig. 4D). The weight gain was dependent on the increasing WAT depos caused by the loss of FGF21 in both WT and CLPP-deficient mice (Fig. 4E). A clear trend toward less efficient clearance of glucose was observed in DKOWB mice when compared to CLPP KO animals (fig. S4D).
FGF21 modulates mitoISR response in CLPP-deficient hearts
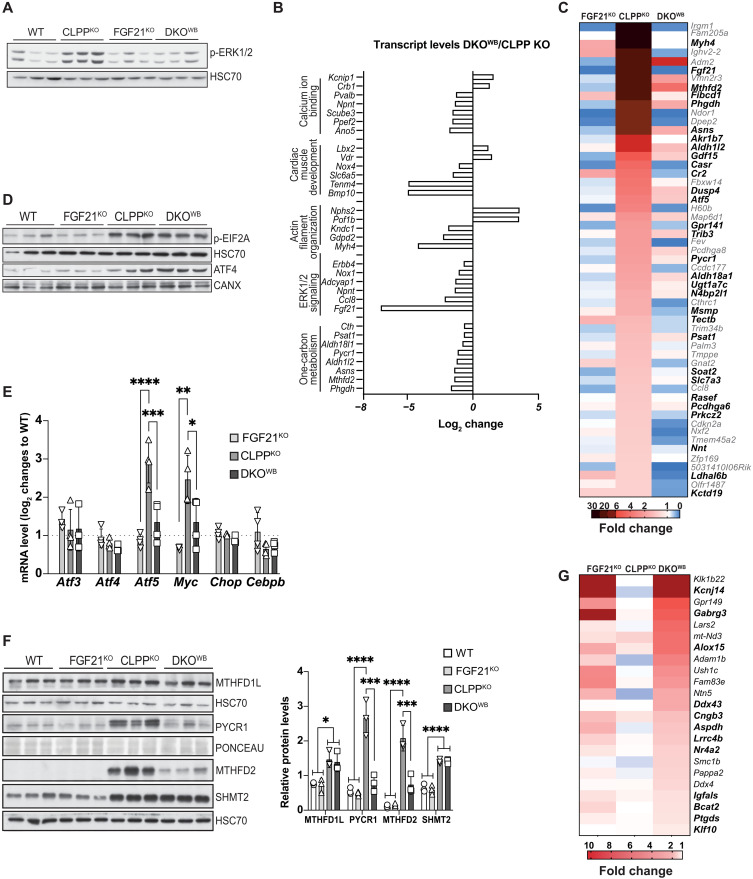
We next looked at whether the loss of FGF21 affected ERK1/2 phosphorylation in the CLPP-deficient cardiomyocytes. Given the previous findings in the DARS2-deficient background, it was startling to observe that the ERK1/2 activation in CLPP KO cardiomyocytes was fully suppressed by the loss of FGF21 (Fig. 5A). We next sought to identify pathways that are affected by FGF21 deficiency in CLPP KO animals by comparing global changes in mRNA levels in FGF21 KO, CLPP KO, and DKOWB versus control hearts. Although we detected around 1500 up- or down-regulated genes in the CLPP KO compared to control hearts, only a small proportion of transcripts was differentially regulated in DKOWB animals (table S4). Out of these, many were up-regulated in CLPP KO animals and down-regulated in DKOWB (Fig. 5B). Gene Ontology (GO) enrichment analysis using DAVID (Database for Annotation, Visualization and Integrated Discovery) (25) not only confirmed the down-regulation of ERK1/2 pathway and response to FGF signaling but also identified changes in cell adhesion, actin cytoskeleton, Ca2+ signaling, and cardiac development (Fig. 5B and table S5). The transcription factor (TF) enrichment analyses of binding motifs in genes down-regulated in DKOWB hearts (in bold) identified the CARE/AARE motif as the most prominent one found in half of the transcripts that are up-regulated in CLPP KO hearts and down-regulated more than twofold in DKOWB animals (Fig. 5C). These include mitoISR genes encoding for proteins involved in 1C metabolism, de novo serine and proline biosynthesis, and transsulfuration, previously shown to be regulated by ATF4 (1, 2, 8). As mentioned before, ATF4 is primarily regulated on the level of protein synthesis as it is preferentially translated when eIF2α phosphorylation is increased (3). The presence of FGF21 did not affect the up-regulation of eIF2α phosphorylation and ATF levels in CLPP-deficient mice (Fig. 5D). ATF4 is not the only TF able to bind to AARE elements, as the same was shown for the other members of the ATF/CEBP family (ATF3, ATF5, CHOP, and CEBP/β) that are direct ATF4 targets and are involved in the fine-tuning of mitoISR (17, 26–29). Out of these TFs, only Atf5 transcripts were highly up-regulated in the CLPP KO animals and strongly down-regulated upon FGF21 depletion in DKOWB hearts (Fig. 5E). Mirroring the changes in transcript levels (fig. S5A), we detected lower levels of proteins involved in the 1C metabolism in the DKOWB hearts, suggesting a blunted mitoISR response (Fig. 5F). The effect was less pronounced in skeletal muscle, possibly because myocytes do not express β-KLOTHO; hence, FGF21 is unable to act in an autocrine/paracrine manner in this tissue (fig. S5, B to E). Even more surprising was to see that FGF21 loss in adipose tissues (BAT and iWAT) does not affect the 1C metabolism enzymes, particularly in brown adipocytes that show a prominent OXPHOS-deficient phenotype in CLPP-deficient animals (fig. S5F) (13). Besides ATF/CEBP TFs, c-MYC was also proposed to be involved in the regulation of mitoISR in hearts in vivo (30), and we observed its negative regulation upon FGF21 depletion in DKOWB animals (Fig. 5D).
Fig. 5. FGF21 loss in CLPP-deficient mice leads to suppression of different constituents of one-carbon metabolism through ERK1/2 pathway and up-regulation of genes involved in cardiomyopathy.
(A) Western blot analysis of ERK1/2 phosphorylation levels. HSC70 was used as a loading control. Experiments were performed on cardiac lysates of WT, FGF21KO, CLPPKO, and DKOKO mice at 17 weeks of age (±2) (n = 3). (B) Log2 transcript changes in DKOWB hearts compared to CLPPKO, extracted from the RNA sequencing (RNA-seq) data (n = 4). (C) Heatmap of total mRNA fold changes of significantly changed genes significantly up-regulated in CLPPKO animals and more than twofold down-regulated in DKOWB (n = 4). Transcripts with CARE/AARE motifs in their promoters [as predicted by Cytoscape plug-in iRegulon (58)] are in bold. (D) Western blot analyses of ATF4 and p-eIF2α levels in cardiac lysates of mice at 17 weeks of age (±2). HSC70 and CANX were used as loading controls (n = 3). (E) mRNA levels (log2 change) of mitoISR TFs relative to WT level extracted from RNA-seq data (n = 4). (F) Western blot analyses of different proteins involved in 1C metabolism (left) and corresponding quantification in cardiac lysates of mice at 17 weeks of age (±2) (right). Antibodies used were raised against proteins indicated in panels (n = 3). (E and F) Bars represent means ± SD (one-way ANOVA and Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). (G) Heatmap of total mRNA fold changes representing transcripts significantly up-regulated in DKOWB mice but unchanged or down-regulated in CLPPKO animals (n = 4). Genes involved in the development of cardiomyopathies are in bold.
Only a handful of transcripts were up-regulated in DKOWB animals when compared to WT and CLPP KO (Fig. 5G). Although GO term analysis did not show any meaningful pathways in which these genes are involved, many of them (in bold) code for the proteins identified as important for cardiac homeostasis (Fig. 5G).
FGF21 protects from the pathological cardiac remodeling
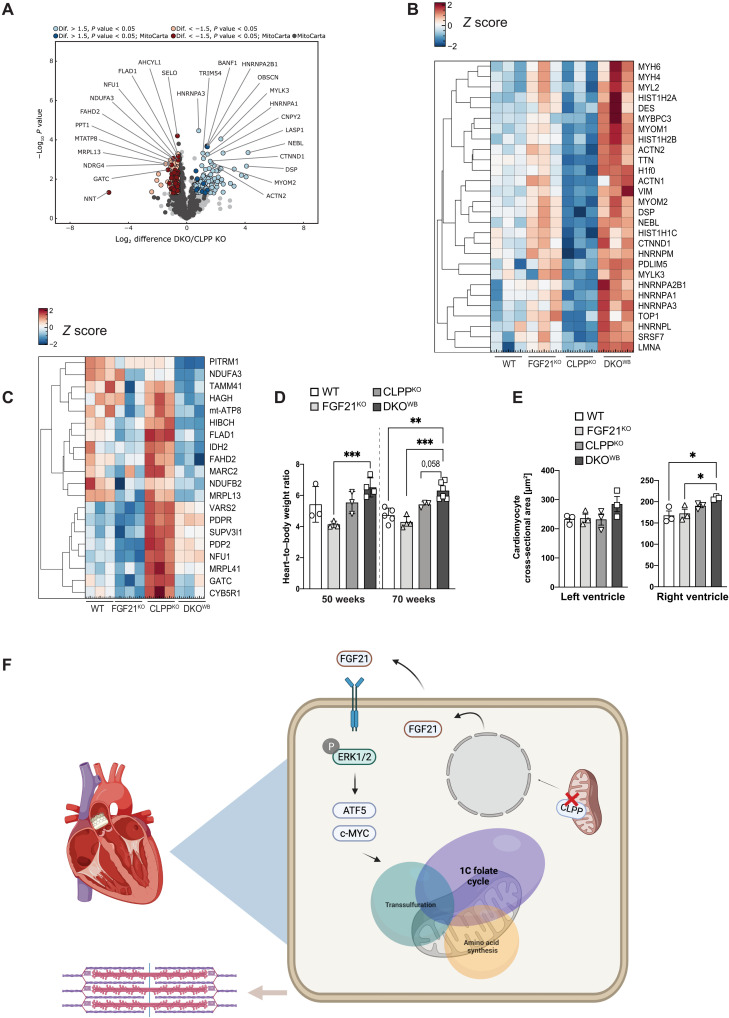
To get a broader aspect of the changes in DKOWB hearts, label-free quantitative profiling of proteome was performed. The data showed 51 significantly up-regulated and 39 down-regulated proteins in DKOWB compared to CLPP KO hearts (Fig. 6A and table S6). KEGG and GO enrichment analyses of up-regulated proteins revealed pathways related to cardiac muscle contraction, organization, and development to be the most prevalent ones (table S7). The most prominent change on the proteome level was the up-regulation of different sarcomeric proteins (α-actinin, myosin, vimentin, titin, and desmin), the structural building blocks of the cardiomyocytes that are vital for cardiac muscle contractility and relaxation (Fig. 6B). Several subunits of the cytoplasmic ribosome and spliceosome machinery were also up-regulated in DKOWB hearts, indicating additional stress response up-regulated by the lack of FGF21. Most of these proteins were significantly down-regulated in CLPP KO hearts (table S6). The most prominent KEGG pathways included different types of cardiomyopathies (dilated, hypertrophied, and right ventricular arrhythmogenic; table S7). In agreement, more than half of up-regulated proteins (28 of 53) were identified as related to heart diseases (Fig. 6B) through the publicly available database for functional annotations Hamonizome (31) using GWASdb (32) and DISEASES datasets (33). In contrast, down-regulated proteins are primarily located in the mitochondria (Fig. 6C and tables S6 and S7). They are involved in different mitochondrial pathways, most notably OXPHOS synthesis and assembly (mt-ATP8, MRPL13, NDUFB2, NDUFA3, MRPL41, VARS2, SUP3VL1, and GATC), but also pyruvate metabolism (IDH2, PDPR, and PDP2), phospholipid synthesis (TAMM41 and CDS2), and mitochondrial import (PITRM1). This suggests that loss of FGF21 might interfere with the mitochondrial biogenesis that seems to be increased upon CLPP depletion, likely as a compensatory mechanism for the respiratory chain deficiency.
Fig. 6. Depletion of FGF21 in CLPP-deficient mice leads to cardiac remodeling and onset of mild cardiomyopathy.
(A) Volcano plots of whole cardiac proteome changes in DKOWB compared to CLPPKO mice (FDR < 0.05). Dark blue and dark red dots represent MitoCarta proteins (left) (n = 3 to 4). (B) Heatmap of whole proteome fold changes (log2) of significantly altered proteins involved in cardiomyopathy development, which are down-regulated in CLPPKO and up-regulated in DKOWB hearts (n = 3). (C) Heatmap of whole proteome fold changes (log2) of significantly altered proteins, which are down-regulated in DKOWB hearts (n = 3). (D) Relative heart–to–body weight ratio at 50 and 70 weeks of age in WT, FGF21KO, CLPPKO, and DKOKO mice (n = 3 to 5). (E) Measurements of the cardiomyocyte cross-sectional area after H&E staining in both left and right ventricles. For the left ventricle, 60 to 70 cross-sectional cardiomyocytes could be identified, and for the right ventricle, 20 to 30 cross-sectional cardiomyocytes could be identified (n = 3). (D and E) Bars represent means ± SD (one-way ANOVA and Tukey’s multiple comparisons test, *P < 0.05, **P < 0.01, and ***P < 0.001). (F) CLPP depletion leads to increased FGF21 levels, which further induce phosphorylation of ERK1/2 in cardiomyocytes and consequent activation of ATF5 and c-MYC TFs, which are involved in the regulation of 1C folate cycle, transsulfuration, and amino acid synthesis pathways in a protective manner.
The changes in the proteome of DKOWB cardiomyocytes suggest heart remodeling consistent with impairment in muscle contraction that precedes cardiomyopathy. This was further supported by the heart–to–body weight ratio that was found to be increased in the older DKOWB animals (Fig. 6D). Cross sections of right and left ventricles additionally confirmed cardiac hypertrophy, as we observed increased size of cardiomyocytes in DKOWB hearts, as an early sign of cardiac impairment (Fig. 6, C and D).
DISCUSSION
Understanding the role of FGF21 in the context of mitochondrial dysfunction, especially when it comes to tissue specificity, remains incomplete. Here, we demonstrate that upon strong OXPHOS deficiency caused by DARS2 depletion, loss of either paracrine or endocrine FGF21 does not affect ERK1/2 signaling and overall phenotypes. Conversely, we present evidence that in the context of milder mitochondrial dysfunction, caused by the loss of CLPP protease, FGF21 protects the heart from early pathological changes that precede cardiomyopathy by activating the ERK1/2 signaling pathway. We further show that FGF21 controls Atf5 and c-Myc levels in the CLPP-deficient hearts, which, in turn, regulate enzymes of the 1C metabolism that are part of mitoISR (Fig. 6E). This selective blunting of mitoISR results in the changes characteristic for the early stages of the heart remodeling during cardiomyopathy (Fig. 6E). This seems to be specific to cardiomyocytes, as a similar effect was not observed in skeletal muscle, WAT, or BAT of CLPP KO mice upon FGF21 depletion. This is particularly surprising for the BAT, one of the primary FGF21 target tissues, which in CLPP-deficient animals displays OXPHOS deficiency that corresponds to the one observed in the heart (13, 22).
Besides presenting different levels of OXPHOS dysfunction, these two models also differ in the type of defect they induce. In the absence of DARS2, mitochondrial protein synthesis is strongly dysregulated; hence, the OXPHOS complexes are not made and assembled at normal levels (11). In contrast, the loss of CLPP results in the overall moderate decrease in mitochondrial translation combined with a defect in the maintenance of OXPHOS complexes (22, 23). These are different molecular pathologies that might activate distinctive types of mitoISR, regardless of the severity of OXPHOS dysfunction.
The MAPK signaling cascade, of which the ERK1/2 branch is probably the most meticulously studied one, is initiated by a two-step protein kinase cascade starting with the RAF family [MAPK kinase kinase (MAPKKK)] and the MEK1/MEK2 isoforms [MAPK kinase (MAPKK)] (34). This signaling pathway can be activated by several stimuli in the dysfunctional heart including G protein–coupled receptors (angiotensin II, endothelin-1, and adrenergic receptors), receptor tyrosine kinases (insulin-like growth factor, transforming growth factor–β, and FGF receptors), cardiotrophin-1 (gp130 receptor), and stress stimuli, including mechanical stress arising from hypertrophic enlargement of individual myofibers (35). Our results argue that severe dysregulation of mitochondrial translation, stemming from DARS2 loss, and the resulting strong OXPHOS deficiency in the heart induce specific stress pathways; hence, even in the absence of FGF21, ERK1/2 signaling is activated. Similarly, although inducing WAT lipolysis is one of the main functions of endocrine FGF21, in the context of severe mitochondrial cardiomyopathy, this is obviously not the only pathway that ensures a constant influx of triacylglycerides from circulation to the failing heart. Other types of heart failure were shown to induce WAT lipolysis through at least two other pathways: activation of β-adrenergic receptors through the action of norepinephrine and epinephrine, or signaling cascade dependent on natriuretic peptides (NPPA and NPPB) and their receptor NPRA (36). Although similar changes in white adipocyte morphology were observed when milder OXPHOS dysfunction was induced by CLPP deficiency, these phenotypes were largely reverted by the loss of FGF21. It is possible that different levels of OXPHOS deficiency subsequently induce compensatory mechanisms, with FGF21 activation being the predominant initial one, while the others follow just when dysfunction becomes more severe. Alternatively, CLPP deficiency, as in the case of Deletor mice, is present in all cell types and tissues, and therefore, it is possible that changes to FGF21 signaling or the absence of it originates from the underlying perturbations of mitochondrial function in white adipocytes (6, 13).
There is compelling evidence that Fgf21 expression can be stimulated by ISR through the eIF2α/ATF4 axis to counteract cellular stress, including mitochondrial dysfunction, endoplasmic reticulum stress, or amino acid deprivation (37, 38). Our data demonstrate that FGF21 itself regulates multiple mitoISR genes containing CARE/AARE motifs, likely by controlling ATF5 and c-MYC, both of which are proposed to play important roles in the mitoISR activation (27, 30, 39). The down-regulation of ATF5 and c-MYC in DKOWB hearts leads to a decrease of a subset of transcript involved in the remodeling of metabolism but does not affect the expression of other genes typically up-regulated by (mito)ISR through the p-eIF2α/ATF4 axis, i.e., cytosolic amino acid–tRNA synthetases (17, 30, 40). In the skeletal muscles of Deletor mice, ATF5 was shown to drive the second phase of mitoISR by controlling transcript levels of Atf3, serine de novo biosynthesis, and transsulfuration pathway, but not MTHFD2, a rate-limiting enzyme of the mitochondrial 1C folate cycle (27). These differences suggest a tissue-specific dependency on the FGF21 action upon mitochondrial dysfunction that may stem from the fact that cardiomyocytes express receptor and co-receptor (Klb and Fgfr1) for FGF21 autocrine/paracrine action, while Klb is absent in skeletal muscle (19). Therefore, the transcriptional control of Atf5 expression might differ in the two tissues, resulting in a diverse output when it comes to the regulation of 1C metabolism. Curiously, some of the previously described c-MYC target genes in the 1C metabolism, e.g., Shmt1/2 (serine hydroxymethyltransferase 1 and 2) (41), were not changed upon FGF21 loss. Together, these results imply that the individual mitoISR genes that are important for the metabolic adaptation to mitochondrial dysfunction are differentially regulated depending on the tissue specificity and/or the type of mitochondrial dysfunction.
Together with ATF4, c-MYC was found to be the prevalent TF regulating mitoISR in hearts of five different models of mitochondrial dysfunction caused by the loss of essential enzymes involved in various steps of mitochondrial gene expression (30). FGF21-mediated regulation of Atf5 and c-Myc levels appears to require ERK1/2 signaling, as the down-regulation was observed only in CLPP-deficient animals when FGF21 is depleted and ERK1/2 signaling is abrogated. This is reminiscent of the recently described pathway by which KRAS and the ERK1/2 signaling activate c-MYC, which, in turn, promotes Mthfd2 expression in colorectal cancer, thus stimulating malignancy (42).
The mTOR signaling cascade has been shown to also play an important role in the regulation of mitoISR (8). Various types of mitochondrial dysfunction have been shown to activate mTOR signaling, and its blunting by rapamycin had generally positive effect on observed phenotypes (8, 43, 44), although the deleterious tissue-specific effects were also observed (45). mTOR function was also shown to be decreased in liver, but not heart of mice haplodeficient for PTCD1, a protein required for mitoribosome assembly (46), that presents an overall mild phenotype (47). In contrast, mTOR function was increased in PTCD1-deficient heart that develops severe mitochondrial cardiomyopathy comparable to DARS2 KO mice (46). While we showed that FGF21 depletion does not affect strong mTOR activation in DARS2 KO mice upon tissue-specific or systemic FGF21 depletion, our data (e.g., significant increase in ribosomal proteins) in CLPP KO mice suggest the activation of mTOR pathway only upon FGF21 depletion. Therefore, it would be very important to further investigate the role of mTOR in different tissue-specific mitochondrial dysfunction settings, particularly as mTOR is considered as one of the potential therapeutic targets in disease patients.
The blunting of the part of mitoISR in CLPP-deficient mice depleted of FGF21 results in the remodeling of various sarcomeric proteins, a hallmark of a cardiac disease characterized by reduced ventricular function (48). The initial phase of cardiac hypertrophy, which precedes heart failure, is characterized by the accumulation of the sarcomeric proteins to counteract the increased strain imposed on the myocardium. Other genes previously related to pathological changes in heart physiology were also up-regulated in DKOWB hearts, including Kcnj14 (>75-fold increase), which encodes for a potassium channel shown to be low in the healthy ventricles but highly increased in dilated cardiomyopathy (49). Similarly, Alox15, which encodes 12/15-lipoxygenase, was more than sevenfold up-regulated in DKOWB cardiomyocytes and was found to be highly expressed in human ischemic heart tissue (50).
Some histone-H1 family members or so-called “linker-histones” (H1F0, HIST1H1B, HIST1H1C, HIST1H2B; HIST1H2A) were also up-regulated in DKOWB, suggesting changes in chromatin structure. Changes in chromatin structure alter cellular transcriptional programs, as in adult-onset cardiomyopathies where transcriptional reprogramming led to initial hypercontractility followed by heart failure (51, 52). Nevertheless, very little is known about how different “core” and “linker” histones change during cardiomyopathy, and to what extent this contributes to pathological heart remodeling.
Adult-onset mitochondrial dysfunction–triggered cardiomyopathies induce compensatory mitochondrial biogenesis most commonly through the activity of PGC-1α, the master regulator of mitochondrial on-demand proliferation (53). In the heart, c-MYC contributes to cardiac mitochondrial biogenesis during times of growth or ischemia, as a possible adaptive response to stress independent of the canonical PGC-1α pathway (54). This phenomenon seems also to occur in DKOWB hearts; although only a subset of proteins was down-regulated in CLPP KO hearts upon FGF21 depletion, that subset consisted mostly of MRC subunits or proteins involved in expression of those subunits, as well as enzymes involved in different mitochondrial energy-production pathways, likely regulated by c-MYC levels.
In conclusion, we have defined a mechanism by which FGF21 fine-tunes activation of cardiac mitoISR upon mitochondrial dysfunction through regulation of the ERK1/2 pathway and ATF5 and c-MYC levels, thus preventing cardiac remodeling and safeguarding against cardiomyopathy. Our results not only dismiss the idea of a universal path that regulates mitoISR but also highlight the need to further understand adaptive changes to mitochondrial deficiency in different contexts and levels of dysfunction that might help us understand the tissue specificity of mitochondrial diseases.
MATERIALS AND METHODS
Mouse experiments and animal care
All mice procedures were conducted in compliance with protocols and approved by local government authorities (Bezirksregierung Köln, Cologne, Germany) and were in accordance with National Institutes of Health guidelines and following procedures previously described in (55). The mice (Mus musculus, C57B1/6) were housed in groups of three to five mice per cage at an ambient temperature of 22° to 24°C and kept at a 12-hour light/12-hour dark cycle in the pathogen-free animal facility of the Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD) Institute. Mice were sacrificed by cervical dislocation (55).
For conducting this research, different mouse lines were used for the creation of DKO models. Previously described (11) mitochondrial aspartyl-tRNA synthetase (Dars2) gene targeting was carried out as part of the International Knockout Mouse Consortium (KOMP) (information available at www.mousephenotype.org/data/genes/MGI:2442510). The mice lacking Dars2 in both heart and skeletal muscle were generated by mating Dars2loxP/loxP animals with transgenic mice expressing cre recombinase under the control of muscle creatine kinase promoter (Ckmm-cre) (56). Conditional targeting of Fgf21loxP mice was carried out in the laboratory of D. Mangelsdorf (57). Fgf21loxP mice were ordered from The Jackson Laboratory [strain: B6.129S6(SJL)-Fgf21tm1.2Djm/J]. DKO mice, in heart and skeletal muscle, were generated by mating homozygous Dars2loxP/loxP; Fgf21oxP/loxP mice with triple heterozygous ones—Dars2loxP/wt; Fgf21loxP/wt, Ckmm-Cretg/wt. To generate full-body Clpp knockout mice, Clppfl/fl were mated with transgenic mice ubiquitously expressing Cre recombinase under the control of the β-actin promoter, resulting in Clpp+/− mice. Heterozygous Clpp+/− mice were further intercrossed to obtain homozygous. For the generation of double-deficient Clpp and Fgf21 knockout animals, Clppfl/fl Fgf21fl/fl mice were mated with transgenic mice ubiquitously expressing Cre recombinase (β-actin Cre), as described in (55).
Blood and serum analyses
Blood glucose levels were determined from whole venous tail blood using an automatic glucose monitor (Contour Next, Bayer) from either random fed or 6-hour fasted animals. Glucose concentrations in the fed state were measured after submandibular bleeding using glucose strips, which were read for absorbance in a reflectance meter (ACCU-CHEK AVIVA, Roche, Mannheim, Germany). Serum was obtained by collecting blood from the submandibular vein. Blood samples were then incubated at room temperature for 45 min and centrifuged at 1500g for 15 min. The resulting serum was stored at −80°C for subsequent analysis. The levels of FGF21 in serum were measured with the Mouse/Rat FGF-21 Quantikine ELISA Kit (R&D Systems, Minneapolis, USA), using the provided instructions, as previously described (55).
Glucose tolerance test, insulin tolerance test, and insulin signaling
Glucose tolerance tests were carried out in 15-week-old animals following a 6-hour fast and procedures previously described in (55). After measuring fasted blood glucose levels, mice were injected intraperitoneally with glucose (2 mg/g body weight, 20% glucose solution; Sigma-Aldrich). Blood glucose levels were determined at 15, 30, 60, and 90 min after glucose injection. Insulin tolerance tests were performed with ad libitum–fed 16-week-old animals. After recording baseline glucose levels, each animal was administered intraperitoneally with insulin (0.6 U/kg body weight) (Insuman Rapid, Sanofi-Aventis), and blood glucose levels were monitored at 15, 30, 60, and 90 min after injection (55). For insulin signaling analysis, mice were injected with insulin (0.6 U/kg body weight) (Insuman Rapid, Sanofi-Aventis) or saline following a 2-hour fast. Mice were then sacrificed 30 min after injection, and tissues were collected and frozen in liquid nitrogen (55).
Extraction of the total RNA from tissues
Total RNA was isolated from mouse tissues using TRIzol reagent (Life Technologies) and following previously described procedures (55). Initially, 50 to 100 mg of fresh or snap-frozen tissue were placed in 2-ml tubes (Peqlab) together with ceramic beads (Mobio, Dianova GmbH) and 1 ml of TRIzol. The tissue was homogenized using a Precellys 24 fast-prep machine (Bertin) at 5500 rpm for 2 × 20 s. For tissues with a high-fat content (eWAT and iWAT), an additional centrifugation step following homogenization was performed at 12,000g for 10 min at 4°C. Homogenates were then transferred to a new tube, and it proceeded with the addition of chloroform and subsequent phase separation according to the manufacturer’s instruction. The RNA pellet was resuspended in ribonuclease (RNase)/deoxyribonuclease (DNase)–free H2O (Gibco) (55).
Reverse transcription PCR and quantitative real-time PCR
The total RNA that was previously isolated was treated with DNase. Digested RNA (100 ng μl−1 per sample) was reverse-transcribed using the High-Capacity Reverse Transcription Kit (Applied Biosystems, Life Technologies). So generated complementary DNA (cDNA) was amplified with Brilliant III Ultra-Fast SYBR QPCR Master Mix (Agilent Technologies) using different primers (see the Supplementary Materials) (55).
Real-time polymerase chain reaction (PCR) was performed on QuantStudio K FlexSystem (Applied Biosystems, Life Technologies). Samples were adjusted for total RNA content by hypoxanthine-guanine phosphoribosyltransferase (HPRT). Relative expression of mRNAs was determined using a comparative method (2−δδCT) according to the ABI Relative Quantification Method (55).
Mitochondria isolation from heart
The freshly dissected tissues for mitochondria isolation were transferred with mitochondria isolation buffer [MIB; 100 mM sucrose, 50 mM KCl, 1 mM EDTA, 20 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES), 0.2% bovine serum albumin (BSA) free from fatty acids, pH 7.2], as previously described (55). Tissues were homogenized with a Potter S homogenizer (Sartorius) at 1200 rpm with 10 to 15 strokes [subtilisin A (1 mg/ml) was added]. The homogenate was transferred and centrifuged at 8500g for 5 min at 4°C. After discarding the supernatant, the pellet was resuspended by shaking in 30 ml of MIB. Another centrifugation step (at 800g for 5 min at 4°C) was performed, and this time, the supernatant, consisting of mitochondria, was transferred into a new 50-ml tube and centrifuged at 8500g for 5 min at 4°C to pellet mitochondria. The remaining pellet is resuspended in 50 μl of MIB without BSA. The concentrations were measured by Bradford reagent (Sigma-Aldrich, Seelze, Germany) according to the manufacturer’s instructions (55).
BN-PAGE Western blot analysis
BN-PAGE was conducted using the Novex Bis-Tris system (Life Technologies GmbH, Darmstadt, Germany) following the manufacturer’s instructions, as previously described (55). For the mitochondrial complex analysis, approximately 20 to 40 μg were lysed using 1% n-dodecyl-β-d-maltoside (DDM). The lysed mitochondria were run on 4 to 16% bis-tris gradient gels. The transfer onto polyvinylidene difluoride membranes was followed by immunodetection using antibodies for mitochondrial complexes (55).
CI in-gel activity measurements
CI in-gel activity was measured by incubating BN-PAGE gel in NADH (reduced form of nicotinamide adenine dinucleotide) (0.1 mg/ml) and nitro blue tetrazolium (2.5 mg/ml) in 5 mM tris (pH 7.4) for 1 hour.
SDS-PAGE Western blot analysis
Tissue protein lysates
Homogenization of 25 mg of cardiac tissue samples in 400 μl of cold organ lysis buffer [50 mM Hepes (pH 7.4), 50 mM NaCl, 1% (v/v) Triton X-100, 0.1 M NaF, 10 mM EDTA, 0.1% SDS (w/v), 10 mM Na-orthovanadate, 2 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail (Sigma-Aldrich), and 1× PhosSTOP phosphatase inhibitor cocktail (Roche)] was performed with Precellys CK 14 (Bertin Technologies) (5000 rpm, 30 s). Cleared protein lysates (45 min, 20,000g, 4°C) were transferred into fresh tubes. Determination of protein concentration was performed with Bradford reagent (Sigma-Aldrich) according to the manufacturer’s instructions. Protein lysates were stored at 80°C.
SDS–polyacrylamide gel electrophoresis
Protein samples were dissolved in SDS-PAGE loading buffer [50 mM tris-HCl (pH 6.8), 2% (w/v) SDS, 10% (v/v) glycerol, 1% β-mercaptoethanol, 12.5 mM EDTA, and 0.02% bromophenol blue] before denaturation. Depending on the required range of protein sizes, the proteins were separated on 8 to 15% acrylamide gels [stacking gel: 5% acrylamide-bisacrylamide (37.5:1), 12.5 mM tris-HCl, 0.1% (w/v) SDS, 0.25% ammonium persulfate (APS), 0.25% tetramethylethylenediamine (TEMED), pH 6.8; separating gel: 8 to 15% acrylamide-bisacrylamide (37.5:1), 37.5 mM tris-HCl, 0.1% (w/v) SDS, 0.1% APS, 0.1% TEMED, pH 8.8] in running buffer [25 mM tris-HCl, 250 mM glycine, 0.1% SDS (w/v), pH 8.3].
Western blot
Transfer of proteins on a nitrocellulose membrane by Western blot was conducted in transfer buffer (30 mM tris-HCl, 240 mM glycine, 0.037% SDS, 20% methanol) at 400 mA for 2 hours at 4°C. For a first evaluation of the transfer, shortly washed membranes (dH2O) were stained with Ponceau S solution (Sigma-Aldrich). Depending on the antibody requirements, destaining and blocking of membranes were performed for 1 hour in either 5% milk-PBST (phosphate-buffered saline–Tween 20) or 3% BSA-TBST (tris-buffered saline–Tween 20) on a gently shaking platform before subsequent immunodecoration with the indicated antibodies according to the manufacturer’s instructions. Secondary horseradish peroxidase–coupled antibodies (1:5000) were incubated for 1 hour before detection by Pierce ECL Western blotting substrate (Thermo Fisher Scientific). Densitometry-based quantification of Western blots was performed with ImageJ and Image Studio Lite Software.
Histological analyses
BAT, eWAT, and iWAT were embedded in paraffin and sectioned at 5 μm. Deparaffinized and rehydrated tissue sections were stained with Mayer’s hematoxylin and counterstained with eosin according to procedures previously described in (55).
For fresh-frozen sections, tissues were directly isolated after sacrificing the animals and directly embedded in Tissue-Tek (Sakura, Alphen aan den Rijn, The Netherlands). Frozen tissues were cut on a Leica CM1850 cryostat with a thickness of 7 μm. COX-SDH staining was performed on fresh-frozen sections, which were air-dried for half an hour after removal from −80°C. First, COX staining solution (0.8 ml of 3,3′-diaminobenzidine tetrahydrochloride, 0.2 ml of 500 μM cytochrome c, a few grains of catalase) was applied in amount to cover the sectioned tissue and incubated in a humid chamber at 37°C for 40 min. After the washing step with PBS, sections were further incubated in SDH staining solution (0.8 ml of 1.875 mM nitro blue tetrazolium, 0.1 ml of 1.3 M sodium succinate, 0.1 ml of 2 mM phenazine methosulfate, 0.01 ml of 100 mM sodium azide) for half an hour at 37°C and consequentially washed in PBS, dehydrated with increasing ethanol concentration (75% for 2 min, 95% for 2 min, 100% for 10 min), air-dried, and mounted in D.P.X. (VWR, Darmstadt, Germany). The stained sections were visualized at ×20 magnification (Leica SCN400 slide scanner and Client software) (55).
Cardiomyocyte cross-sectional area measurements
Cardiomyocyte size was determined separately for the left and right ventricles. Paraffin-embedded heart tissue was hematoxylin and eosin–stained and subsequently analyzed using QuPath software. Only cross-sectional, almost circular shaped cardiomyocytes were included in the analysis. For this purpose, the QuPath polygon tool was used to encircle the cells and determine the area. For the left ventricle, 60 to 70 cross-sectional cardiomyocytes could be identified, and for the right ventricle, 20 to 30 cross-sectional cardiomyocytes could be identified.
Label-free quantification of the cardiac proteome
Heart tissue was used for label-free quantification of the proteome. First, the tissues were disrupted using liquid nitrogen and dissolved in 8 M urea buffer, with the addition of a 1× proteinase inhibitor. After incubation (30 min, 4°C, on a rotating wheel), lysates were cleared (15 min, 4°C, full speed) and protein concentration was determined with Bradford reagent (Sigma-Aldrich) according to the manufacturer’s instructions. Next, 50 μg of protein per sample was precipitated with the fourfold volume of cold 100% acetone (60 min, −80°C), pelleted (15 min, 4°C, full speed), and washed twice with cold 90% acetone (5 min, 4°C, full speed). The air-dried pellet was subsequently resuspended in 300 μl of 8 M urea buffer. In solution, digest was performed by addition of dithiothreitol to a final concentration of 5 mM (1 hour, 37°C), addition of chloroacetamide to a final concentration of 40 mM (20 min, protect from light), the addition of endoproteinase Lys-C at an enzyme-substrate ratio of 1:75 (4 hours, 37°C), and dilution with 50 mM triethylammonium bicarbonate (TEAB) to a urea concentration of 2 M before adding trypsin at an enzyme-substrate ratio of 1:75 (overnight, 37°C, protect from light). The digest was stopped with formic acid (1% final concentration). Digested protein lysates were loaded on SDB-RPS stage tips and submitted to the CECAD proteomic facility for further processing.
RNA sequencing of cardiac total mRNA
RNA isolation for RNA-seq was performed using the “Direct-zol RNA MiniPrep Kit” (Zymo Research) according to the manufacturer’s instruction. Purified RNA was submitted to the Cologne Centre for Genomics for further processing. Libraries were prepared using the “TruSeq mRNA Stranded Sample Preparation Kit” (Illumina) and validated on a tape station (Agilent). Equimolar amounts of libraries were pooled, quantified (Peqlab KAPA Library Quantification Kit, Applied Biosystems 7900HT Sequence Detection System), and finally sequenced on a “HiSeq4000 sequencer” (Illumina).
Statistical analyses
Statistical significance of datasets with two independent groups was analyzed using two-tailed unpaired Student’s t test (55). Datasets with more than two groups were analyzed using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. All P values below 0.05 were considered statistically significant; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. All data are presented as means ± SD (55). Statistical analysis and data visualization were managed using Microsoft Excel, GraphPad Prism 6 (GraphPad Software), and Instant Clue (55).
Acknowledgments
We thank the CECAD Proteomics Core Facility and Cologne Center for Genomics (CCG) for their excellent support. We also thank A. Kukat for critically reading the manuscript.
Funding: This work was supported by German Research Foundation [Deutsche Forschungsgemeinschaft (DFG)] SFB 1218 Projektnummer 269925409, German Research Foundation (DFG) TR 1018/8-1, and Centre for Molecular Medicine Cologne, University of Cologne C15 (to A.T.). M.C. received a scholarship from Marie Curie ITN—Marriage (Grant Agreement ID:316964). M.P and L.G.-F. were supported by the German Research Foundation (DFG, GRK-2407). S.R. is supported by the German Research Foundation (DFG, GRK-2407 and SFB-TRR259).
Author contributions: Conceptualization: A.T., M.C., K.Sz., M.K., and S.R. Data curation: M.C., A.T., K.Sz., M.P., C.L., T.B., L.G.-F., R.J.A., H.K., and C.P. Formal analysis: M.C., A.T., K.Sz., K.Se., M.P., C.L., T.B., L.G.-F., R.J.A., H.K., and C.P. Funding acquisition: A.T. Investigation: M.C., K.Sz., K.Se., M.P., C.L., T.B., L.G.-F., R.J.A., H.K., C.P., C.P.B., and A.A. Visualization: M.C., A.T., K.Sz., M.P., T.B., L.G.-F., and R.J.A. Writing: A.T. and M.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The mouse and cell lines can be provided by A.T. pending scientific review and a completed material transfer agreement. The raw mass spectrometry proteomic data have been deposited to the Proteome Exchange Consortium via the “PRIDE” partner repository with the dataset identifier PXD031324, username: reviewer_pxd031324@ebi.ac.uk; password: edsaq2JQ (www.ebi.ac.uk/pride/profile/%20reviewer_pxd031324). The RNA-seq raw data have been deposited to Gene Expression Omnibus (GEO) Submission (GSE195947) (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE195947).
Supplementary Materials
This PDF file includes:
Supplementary Material and Methods
Figs. S1 to S5
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S7
REFERENCES AND NOTES
- 1.Bao X. R., Ong S. E., Goldberger O., Peng J., Sharma R., Thompson D. A., Vafai S. B., Cox A. G., Marutani E., Ichinose F., Goessling W., Regev A., Carr S. A., Clish C. B., Mootha V. K., Mitochondrial dysfunction remodels one-carbon metabolism in human cells. eLife 5, e10575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quiros P. M., Prado M. A., Zamboni N., D’Amico D., Williams R. W., Finley D., Gygi S. P., Auwerx J., Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216, 2027–2045 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa-Mattioli M., Walter P., The integrated stress response: From mechanism to disease. Science 368, eaat5314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fessler E., Eckl E. M., Schmitt S., Mancilla I. A., Meyer-Bender M. F., Hanf M., Philippou-Massier J., Krebs S., Zischka H., Jae L. T., A pathway coordinated by DELE1 relays mitochondrial stress to the cytosol. Nature 579, 433–437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo X., Aviles G., Liu Y., Tian R., Unger B. A., Lin Y. T., Wiita A. P., Xu K., Correia M. A., Kampmann M., Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 579, 427–432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyynismaa H., Carroll C. J., Raimundo N., Ahola-Erkkila S., Wenz T., Ruhanen H., Guse K., Hemminki A., Peltola-Mjosund K. E., Tulkki V., Oresic M., Moraes C. T., Pietilainen K., Hovatta I., Suomalainen A., Mitochondrial myopathy induces a starvation-like response. Hum. Mol. Genet. 19, 3948–3958 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Yatsuga S., Fujita Y., Ishii A., Fukumoto Y., Arahata H., Kakuma T., Kojima T., Ito M., Tanaka M., Saiki R., Koga Y., Growth differentiation factor 15 as a useful biomarker for mitochondrial disorders. Ann. Neurol. 78, 814–823 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan N. A., Nikkanen J., Yatsuga S., Jackson C., Wang L., Pradhan S., Kivela R., Pessia A., Velagapudi V., Suomalainen A., mTORC1 regulates mitochondrial integrated stress response and mitochondrial myopathy progression. Cell Metab. 26, 419–428.e415 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Potthoff M. J., Kliewer S. A., Mangelsdorf D. J., Endocrine fibroblast growth factors 15/19 and 21: From feast to famine. Genes Dev. 26, 312–324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazeli P. K., Lun M., Kim S. M., Bredella M. A., Wright S., Zhang Y., Lee H., Catana C., Klibanski A., Patwari P., Steinhauser M. L., FGF21 and the late adaptive response to starvation in humans. J. Clin. Invest. 125, 4601–4611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dogan S. A., Pujol C., Maiti P., Kukat A., Wang S., Hermans S., Senft K., Wibom R., Rugarli E. I., Trifunovic A., Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 19, 458–469 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Restelli L. M., Oettinghaus B., Halliday M., Agca C., Licci M., Sironi L., Savoia C., Hench J., Tolnay M., Neutzner A., Schmidt A., Eckert A., Mallucci G., Scorrano L., Frank S., Neuronal mitochondrial dysfunction activates the integrated stress response to induce fibroblast growth factor 21. Cell Rep. 24, 1407–1414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker C., Kukat A., Szczepanowska K., Hermans S., Senft K., Brandscheid C. P., Maiti P., Trifunovic A., CLPP deficiency protects against metabolic syndrome but hinders adaptive thermogenesis. EMBO Rep. 19, e45126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kukat A., Dogan S. A., Edgar D., Mourier A., Jacoby C., Maiti P., Mauer J., Becker C., Senft K., Wibom R., Kudin A. P., Hultenby K., Flogel U., Rosenkranz S., Ricquier D., Kunz W. S., Trifunovic A., Loss of UCP2 attenuates mitochondrial dysfunction without altering ROS production and uncoupling activity. PLOS Genet. 10, e1004385 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira R. O., Tadinada S. M., Zasadny F. M., Oliveira K. J., Pires K. M. P., Olvera A., Jeffers J., Souvenir R., McGlauflin R., Seei A., Funari T., Sesaki H., Potthoff M. J., Adams C. M., Anderson E. J., Abel E. D., OPA1 deficiency promotes secretion of FGF21 from muscle that prevents obesity and insulin resistance. EMBO J. 36, 2126–2145 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Planavila A., Redondo I., Hondares E., Vinciguerra M., Munts C., Iglesias R., Gabrielli L. A., Sitges M., Giralt M., van Bilsen M., Villarroya F., Fibroblast growth factor 21 protects against cardiac hypertrophy in mice. Nat. Commun. 4, 2019 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Kaspar S., Oertlin C., Szczepanowska K., Kukat A., Senft K., Lucas C., Brodesser S., Hatzoglou M., Larsson O., Topisirovic I., Trifunovic A., Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR. Sci. Adv. 7, eabf0971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degirolamo C., Sabba C., Moschetta A., Therapeutic potential of the endocrine fibroblast growth factors FGF19, FGF21 and FGF23. Nat. Rev. Drug Discov. 15, 51–69 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Planavila A., Redondo-Angulo I., Villarroya F., FGF21 and cardiac physiopathology. Front. Endocrinol. 6, 133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher F. M., Kleiner S., Douris N., Fox E. C., Mepani R. J., Verdeguer F., Wu J., Kharitonenkov A., Flier J. S., Maratos-Flier E., Spiegelman B. M., FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev. 26, 271–281 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szczepanowska K., Maiti P., Kukat A., Hofsetz E., Nolte H., Senft K., Becker C., Ruzzenente B., Hornig-Do H. T., Wibom R., Wiesner R. J., Kruger M., Trifunovic A., CLPP coordinates mitoribosomal assembly through the regulation of ERAL1 levels. EMBO J. 35, 2566–2583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szczepanowska K., Senft K., Heidler J., Herholz M., Kukat A., Hohne M. N., Hofsetz E., Becker C., Kaspar S., Giese H., Zwicker K., Guerrero-Castillo S., Baumann L., Kauppila J., Rumyantseva A., Muller S., Frese C. K., Brandt U., Riemer J., Wittig I., Trifunovic A., A salvage pathway maintains highly functional respiratory complex I. Nat. Commun. 11, 1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badman M. K., Koester A., Flier J. S., Kharitonenkov A., Maratos-Flier E., Fibroblast growth factor 21-deficient mice demonstrate impaired adaptation to ketosis. Endocrinology 150, 4931–4940 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Bruhat A., Averous J., Carraro V., Zhong C., Reimold A. M., Kilberg M. S., Fafournoux P., Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 277, 48107–48114 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Forsstrom S., Jackson C. B., Carroll C. J., Kuronen M., Pirinen E., Pradhan S., Marmyleva A., Auranen M., Kleine I. M., Khan N. A., Roivainen A., Marjamaki P., Liljenback H., Wang L., Battersby B. J., Richter U., Velagapudi V., Nikkanen J., Euro L., Suomalainen A., Fibroblast growth factor 21 drives dynamics of local and systemic stress responses in mitochondrial myopathy with mtDNA deletions. Cell Metab. 30, 1040–1054.e1047 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S., ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 277, 24120–24127 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Siu F., Chen C., Zhong C., Kilberg M. S., CCAAT/enhancer-binding protein-beta is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 276, 48100–48107 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Kuhl I., Miranda M., Atanassov I., Kuznetsova I., Hinze Y., Mourier A., Filipovska A., Larsson N. G., Transcriptomic and proteomic landscape of mitochondrial dysfunction reveals secondary coenzyme Q deficiency in mammals. eLife 6, e30952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouillard A. D., Gundersen G. W., Fernandez N. F., Wang Z., Monteiro C. D., McDermott M. G., Ma’ayan A., The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, baw100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M. J., Liu Z., Wang P., Wong M. P., Nelson M. R., Kocher J. P., Yeager M., Sham P. C., Chanock S. J., Xia Z., Wang J., GWASdb v2: An update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 44, D869–D876 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pletscher-Frankild S., Palleja A., Tsafou K., Binder J. X., Jensen L. J., DISEASES: Text mining and data integration of disease-gene associations. Methods 74, 83–89 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Sugden P. H., Clerk A., Regulation of the ERK subgroup of MAP kinase cascades through G protein-coupled receptors. Cell. Signal. 9, 337–351 (1997). [DOI] [PubMed] [Google Scholar]
- 35.Bueno O. F., Molkentin J. D., Involvement of extracellular signal-regulated kinases 1/2 in cardiac hypertrophy and cell death. Circ. Res. 91, 776–781 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Kintscher U., Foryst-Ludwig A., Haemmerle G., Zechner R., The role of adipose triglyceride lipase and cytosolic lipolysis in cardiac function and heart failure. Cell Rep. Med. 1, 100001 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim K. H., Jeong Y. T., Oh H., Kim S. H., Cho J. M., Kim Y. N., Kim S. S., Kim D. H., Hur K. Y., Kim H. K., Ko T., Han J., Kim H. L., Kim J., Back S. H., Komatsu M., Chen H., Chan D. C., Konishi M., Itoh N., Choi C. S., Lee M. S., Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat. Med. 19, 83–92 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Salminen A., Kaarniranta K., Kauppinen A., Integrated stress response stimulates FGF21 expression: Systemic enhancer of longevity. Cell. Signal. 40, 10–21 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Nikkanen J., Forsstrom S., Euro L., Paetau I., Kohnz R. A., Wang L., Chilov D., Viinamaki J., Roivainen A., Marjamaki P., Liljenback H., Ahola S., Buzkova J., Terzioglu M., Khan N. A., Pirnes-Karhu S., Paetau A., Lonnqvist T., Sajantila A., Isohanni P., Tyynismaa H., Nomura D. K., Battersby B. J., Velagapudi V., Carroll C. J., Suomalainen A., Mitochondrial DNA replication defects disturb cellular dntp pools and remodel one-carbon metabolism. Cell Metab. 23, 635–648 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Shan J., Zhang F., Sharkey J., Tang T. A., Ord T., Kilberg M. S., The C/ebp-Atf response element (CARE) location reveals two distinct Atf4-dependent, elongation-mediated mechanisms for transcriptional induction of aminoacyl-tRNA synthetase genes in response to amino acid limitation. Nucleic Acids Res. 44, 9719–9732 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikiforov M. A., Chandriani S., O’Connell B., Petrenko O., Kotenko I., Beavis A., Sedivy J. M., Cole M. D., A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol. Cell. Biol. 22, 5793–5800 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ju H. Q., Lu Y. X., Chen D. L., Zuo Z. X., Liu Z. X., Wu Q. N., Mo H. Y., Wang Z. X., Wang D. S., Pu H. Y., Zeng Z. L., Li B., Xie D., Huang P., Hung M. C., Chiao P. J., Xu R. H., Modulation of redox homeostasis by inhibition of MTHFD2 in colorectal cancer: Mechanisms and therapeutic implications. J. Natl. Cancer Inst. 111, 584–596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson S. C., Yanos M. E., Kayser E. B., Quintana A., Sangesland M., Castanza A., Uhde L., Hui J., Wall V. Z., Gagnidze A., Oh K., Wasko B. M., Ramos F. J., Palmiter R. D., Rabinovitch P. S., Morgan P. G., Sedensky M. M., Kaeberlein M., mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Civiletto G., Dogan S. A., Cerutti R., Fagiolari G., Moggio M., Lamperti C., Beninca C., Viscomi C., Zeviani M., Rapamycin rescues mitochondrial myopathy via coordinated activation of autophagy and lysosomal biogenesis. EMBO Mol. Med. 10, e8799 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ignatenko O., Nikkanen J., Kononov A., Zamboni N., Ince-Dunn G., Suomalainen A., Mitochondrial spongiotic brain disease: Astrocytic stress and harmful rapamycin and ketosis effect. Life Sci. Alliance 3, e202000797 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perks K. L., Rossetti G., Kuznetsova I., Hughes L. A., Ermer J. A., Ferreira N., Busch J. D., Rudler D. L., Spahr H., Schondorf T., Shearwood A. J., Viola H. M., Siira S. J., Hool L. C., Milenkovic D., Larsson N. G., Rackham O., Filipovska A., PTCD1 is required for 16S rRNA maturation complex stability and mitochondrial ribosome assembly. Cell Rep. 23, 127–142 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Perks K. L., Ferreira N., Richman T. R., Ermer J. A., Kuznetsova I., Shearwood A. J., Lee R. G., Viola H. M., Johnstone V. P. A., Matthews V., Hool L. C., Rackham O., Filipovska A., Adult-onset obesity is triggered by impaired mitochondrial gene expression. Sci. Adv. 3, e1700677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Velden J., Stienen G. J. M., Cardiac disorders and pathophysiology of sarcomeric proteins. Physiol. Rev. 99, 381–426 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Szuts V., Menesi D., Varga-Orvos Z., Zvara A., Houshmand N., Bitay M., Bogats G., Virag L., Baczko I., Szalontai B., Geramipoor A., Cotella D., Wettwer E., Ravens U., Deak F., Puskas L. G., Papp J. G., Kiss I., Varro A., Jost N., Altered expression of genes for Kir ion channels in dilated cardiomyopathy. Can. J. Physiol. Pharmacol. 91, 648–656 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Magnusson L. U., Lundqvist A., Asp J., Synnergren J., Johansson C. T., Palmqvist L., Jeppsson A., Hulten L. M., High expression of arachidonate 15-lipoxygenase and proinflammatory markers in human ischemic heart tissue. Biochem. Biophys. Res. Commun. 424, 327–330 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Mahmoud S. A., Poizat C., Epigenetics and chromatin remodeling in adult cardiomyopathy. J. Pathol. 231, 147–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang H., Yu Z., He J., Hua B., Zhang G., Identification of the molecular mechanisms underlying dilated cardiomyopathy via bioinformatic analysis of gene expression profiles. Exp. Ther. Med. 13, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.G. W. Dorn II, Vega R. B., Kelly D. P., Mitochondrial biogenesis and dynamics in the developing and diseased heart. Genes Dev. 29, 1981–1991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahuja P., Zhao P., Angelis E., Ruan H., Korge P., Olson A., Wang Y., Jin E. S., Jeffrey F. M., Portman M., Maclellan W. R., Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J. Clin. Invest. 120, 1494–1505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.M. Aradjanski, “The role of fibroblast growth factor 21 in different mouse models of mitochondrial dysfunction,” thesis, University of Cologne, Cologne, Germany (2018). [Google Scholar]
- 56.Larsson N. G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G. S., Clayton D. A., Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18, 231–236 (1998). [DOI] [PubMed] [Google Scholar]
- 57.Potthoff M. J., Inagaki T., Satapati S., Ding X., He T., Goetz R., Mohammadi M., Finck B. N., Mangelsdorf D. J., Kliewer S. A., Burgess S. C., FGF21 induces PGC-1α and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc. Natl. Acad. Sci. U.S.A. 106, 10853–10858 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janky R., Verfaillie A., Imrichova H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D., Svetlichnyy D., Kalender Atak Z., Fiers M., Marine J. C., Aerts S., iRegulon: From a gene list to a gene regulatory network using large motif and track collections. PLOS Comput. Biol. 10, e1003731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material and Methods
Figs. S1 to S5
References
Tables S1 to S7