Abstract
Transient MyoD overexpression in concert with small molecule treatment reprograms mouse fibroblasts into induced myogenic progenitor cells (iMPCs). However, the molecular landscape and mechanisms orchestrating this cellular conversion remain unknown. Here, we undertook an integrative multiomics approach to delineate the process of iMPC reprogramming in comparison to myogenic transdifferentiation mediated solely by MyoD. Using transcriptomics, proteomics, and genome-wide chromatin accessibility assays, we unravel distinct molecular trajectories that govern the two processes. Notably, only iMPC reprogramming is characterized by gradual up-regulation of muscle stem cell markers, unique signaling pathways, and chromatin remodelers in conjunction with exclusive chromatin opening in core myogenic promoters. In addition, we determine that the Notch pathway is indispensable for iMPC formation and self-renewal and further use the Notch ligand Dll1 to homogeneously propagate iMPCs. Collectively, this study charts divergent molecular blueprints for myogenic transdifferentiation or reprogramming and underpins the heightened capacity of iMPCs for capturing myogenesis ex vivo.
An integrative multiomics analysis uncovers how fibroblasts convert into myocytes and satellite-like myogenic progenitors.
INTRODUCTION
Skeletal muscle is a soft tissue that governs voluntary movement and accounts for 30 to 40% of the normal human body mass. This tissue is predominantly composed of multinucleated muscle fibers that contract to generate movement and a variety of mononucleated resident cells that maintain tissue homeostasis (1). Satellite cells are resident skeletal muscle stem cells that can regenerate muscle fibers upon injury or disease and are characterized by high expression of the transcription factor paired box protein 7 (Pax7) (2–4). These cells reside in a unique anatomical location between the myofiber cell membrane and basal lamina and are quiescent during homeostasis, undergoing activation to repair the tissue following muscle damage (2, 3). During this repair process, activated satellite cells either divide symmetrically to increase the pool of muscle stem cells that regain quiescence or divide asymmetrically into myoblasts and fusion-competent myocytes that merge with damaged myofibers for tissue repair (5). Satellite cell activation is a stepwise process that commences by up-regulation of myogenic regulatory transcription factors including Myf5 and MyoD in myoblasts, Myogenin (Myog) and Myf6/MRF4 in myocytes, and myosin heavy chain (MyHC) isoforms in multinucleated myofibers (2, 3). Following isolation from skeletal muscles and in vitro propagation, satellite cells form a population of proliferative myoblasts that up-regulate MyoD and rapidly lose molecular attributes indicative of an in vivo activated satellite cell state (6). This loss of satellite cell attributes following in vitro expansion renders myoblasts cumbersome for regenerative medicine purposes and highlights the necessity to seek alternative methods to culture myogenic stem and progenitor cells (7, 8).
Direct lineage reprogramming denotes the conversion of one cell type into another. It is typically induced by forced overexpression of cell type–specific transcription factors or small molecule treatment (9). Manipulation of cell identity via this approach was first demonstrated in a milestone study that determined that overexpression of the transcription factor MyoD transdifferentiates fibroblasts into skeletal muscle cells (10). Since this seminal study, several works have reported on transdifferentiation of somatic cells into various cell types including neurons, cardiomyocytes, and hepatocytes (11–14). Moreover, further works have reported on direct reprogramming of somatic cells into tissue-specific multipotent stem and progenitor cells (15–20). One study recently reported on a method to directly reprogram mouse fibroblasts into “induced myogenic progenitor cells” (iMPCs) by transient MyoD overexpression in conjunction with three small molecules: the adenylate cyclase activator Forskolin (F), the transforming growth factor–β (TGF-β) receptor inhibitor RepSox (R), and the glycogen synthase kinase-3β (GSK-3β) inhibitor CHIR99021 (C) (abbreviated as F/R/C) (20). Reprogramming into iMPCs is markedly different from conventional transdifferentiation into myogenic cells solely by MyoD, which typically generates myocytes and multinucleated myotubes (10, 20). In contrast, MyoD overexpression in concert with F/R/C treatment gives rise to heterogeneous and expandable myogenic cultures consisting of skeletal muscle progenitor cells that express Pax7 and Myf5, in addition to a highly contractile myofiber network (20).
The divergent lineage conversion trajectories of MyoD and MyoD+F/R/C raise the question how small molecule administration endows a myogenic stem cell fate on fibroblasts in comparison to postmitotic myotubes via MyoD-mediated transdifferentiation, which has been extensively studied (21–23). Furthermore, it is of interest to investigate how molecularly akin Pax7+ iMPCs are to Pax7+ primary myoblasts, and whether Pax7+ iMPCs represent an improved capture of a myogenic stem cell state in vitro. To address these questions, here, we set out to delineate the molecular landscape of iMPCs using integrative multiomics approaches and further dissect the molecular trajectory guiding fibroblast conversion into iMPCs in comparison to transdifferentiation solely by MyoD. Collectively, we establish a body of knowledge in respect to the genes, proteins, and pathways that govern direct lineage conversion into myogenic progenitor cells and further establish their molecular similarity and disparity from primary myoblasts. Our results suggest that iMPCs capture a bona fide skeletal muscle differentiation program in vitro, thus establishing them as an exceptional method to culture and propagate skeletal muscle stem and progenitor cells.
RESULTS
An inducible reprogramming system to study fibroblast conversion into myogenic cells
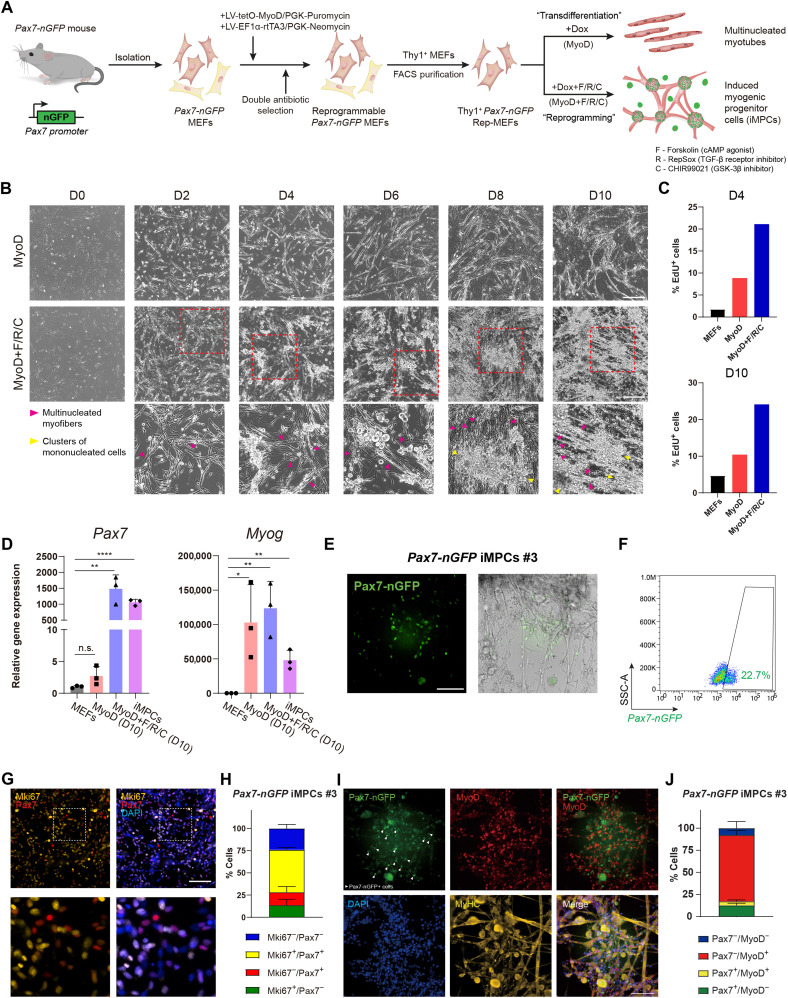
We commenced our investigation by establishing an inducible cellular conversion system in fibroblasts that enables comparison of reprogramming via MyoD+F/R/C to transdifferentiation solely by MyoD. To this end, we derived mouse embryonic fibroblasts (MEFs) from a transgenic mouse strain that carries a Pax7-nuclear GFP (Pax7-nGFP) reporter, which allows prospective skeletal muscle stem cell purification using fluorescence-activated cell sorting (FACS) (24). To initiate controlled MyoD overexpression in Pax7-nGFP MEFs, we engineered a doxycycline (dox)–inducible Tet-On lentiviral dual-plasmid system harboring a MyoD coding sequence under a TRE3G promoter (tetO-MyoD/PGK-Puromycin) and a Tet3G activator under the control of a constitutive EF1-α promoter (EF1α-rtTA3/PGK-Neomycin) (Fig. 1A). Each respective plasmid also contained an antibiotic resistance gene, thus enabling selection for transduced reprogrammable MEFs (Rep-MEFs) that carry both lentiviral constructs (Fig. 1A). Since MEF cultures may contain different cell types, we FACS-purified fibroblasts from MEFs using Thy1, a fibroblast-specific marker (fig. S1A) (25). We then subjected Thy1+ Pax7-nGFP Rep-MEFs to either dox (MyoD) or dox + F/R/C (MyoD+F/R/C) treatment. Following 1 to 3 days of MyoD overexpression with or without F/R/C treatment, we documented the formation of multinucleated myotubes; however, from days 6 to 8, Rep-MEFs treated with MyoD+F/R/C formed a highly contractile network of myofibers in conjunction with the appearance of proliferative mononucleated cells (Fig. 1, B and C). To assess the expression of myogenic markers upon either MyoD or MyoD+F/R/C treatment, we performed quantitative real-time polymerase chain reaction (qRT-PCR) at day 10 of reprogramming and documented robust up-regulation of the myogenic stem cell markers Pax7 and Myf5 only in MyoD+F/R/C treatment, whereas the differentiation marker Myog was up-regulated under both conditions (Fig. 1D and fig. S1B). Substantial down-regulation of the fibroblast marker Col1a1 was observed only under the MyoD+F/R/C condition (fig. S1B).
Fig. 1. A reprogrammable system to study lineage conversion into myogenic cells.
(A) Schematic of experimental design denoting the conversion of Thy1+ Pax7-nGFP Rep-MEFs into multinucleated myotubes solely by MyoD overexpression or iMPCs by MyoD+F/R/C treatment. (B) Representative bright-field images of Thy1+ Rep-MEFs subjected to either MyoD or MyoD+F/R/C treatment for the indicated days. Scale bars, 400 μm. (C) Bar graphs based on flow cytometry analysis showing the percentage of EdU+ cells in Thy1+ Rep-MEFs subjected to either the MyoD or MyoD+F/R/C condition for the indicated days. (D) qRT-PCR for the indicated myogenic genes at day 10. Data are shown as means ± SD. N = 3 cell lines per group. Statistical significance was determined by a two-tailed unpaired t test (*P < 0.05; **P < 0.01; ****P < 0.0001; n.s., nonsignificant). (E) Representative images of a stable Pax7-nGFP iMPC clone at passage 1. Scale bar, 100 μm. (F) Flow cytometry analysis of a Pax7-nGFP iMPC clone at P1. (G) Representative immunofluorescence images of iMPCs immunostained for Mki67 and Pax7. Nuclei were counterstained with DAPI. Scale bar, 100 μm. (H) Quantification of (G). Three different images were quantified from the respective iMPC clone. (I) Representative immunofluorescence images of Pax7-nGFP iMPCs immunostained for MyoD and MyHC. Nuclei were counterstained with DAPI. White arrowheads point to mononucleated Pax7-nGFP+ cells. Scale bar, 100 μm. (J) Quantification of (I). Four different images were quantified from the respective iMPC clone.
We next wished to determine whether F/R/C supplementation without ectopic MyoD expression can dedifferentiate myocytes that have been produced solely by MyoD overexpression into Pax7+ progenitor cells. To investigate this question, we used Rep-MEFs harboring Pax7-CreERT2; R26-LoxSTOPLox-ntdTomato reporter alleles, which allows labeling of Pax7+ cells in the presence of 4-hydroxytamoxifen (fig. S1C) (26). We then solely overexpressed MyoD for 2 or 4 days via dox administration followed by dox withdrawal and addition of F/R/C supplementation for up to 10 days. We observed that MEFs subjected to MyoD overexpression for only 2 or 4 days followed by F/R/C administration successfully produced Pax7+ iMPC–like clusters, whereas F/R/C treatment alone did not (fig. S1, D to F). Notably, continuous overexpression of MyoD with F/R/C treatment throughout the entire reprogramming process yielded a higher number of Pax7+ cells (fig. S1, D to F). We conclude that exogenous MyoD overexpression is essential for the production of iMPCs; however, it may only be necessary during the initial phase of reprogramming to generate myocytes, which can further dedifferentiate into iMPCs in the presence of F/R/C supplementation.
As the next step, we reprogrammed several Rep-MEF lines using MyoD+F/R/C treatment for 10 days and then withdrew dox and expanded the cells in medium containing F/R/C for several passages. This enabled us to establish dox-independent iMPC clones that proliferated robustly and expressed an array of myogenic progenitor and differentiation genes (Fig. 1, D and E, and fig. S1B). Noticeably, these iMPC clones were composed of contractile myofiber network together with mononucleated Pax7-nGFP+ cells, ranging between 3.6 and 22.7% (Fig. 1, E and F, and fig. S1G). A Pax7 immunofluorescence analysis of a stable iMPC clone revealed Pax7+ cells that were predominately positive for Mki67+, unveiling a highly proliferative cell state of Pax7+ iMPCs (Fig. 1, G and H). We also documented the presence of Pax7-nGFP+/MyoD−/MyHC− mononucleated cells in a stable clone, suggesting that iMPCs contain more immature myogenic stem/progenitor cells as previously reported (Fig. 1I) (20). Quantification of the various cell populations comprising a stable iMPC clone determined the presence of Pax7+/MyoD− (12.6 ± 2.5%), Pax7+/MyoD+ (4.7 ± 1.5%), Pax7−/MyoD+ (74.9 ± 5.3%), and Pax7−/MyoD− (7.9 ± 7.4%) cells, highlighting the heterogeneous cell composition of iMPCs (Fig. 1J). Collectively, we successfully established an improved reprogramming system to interrogate transdifferentiation by MyoD or reprogramming by MyoD+F/R/C. Furthermore, we demonstrate via this system that subjecting fibroblasts to MyoD+F/R/C treatment forms heterogeneous iMPC cultures consisting of proliferative Pax7+ cells that can propagate without ectopic MyoD overexpression.
Transcriptional dynamics during fibroblast conversion into skeletal muscle cells
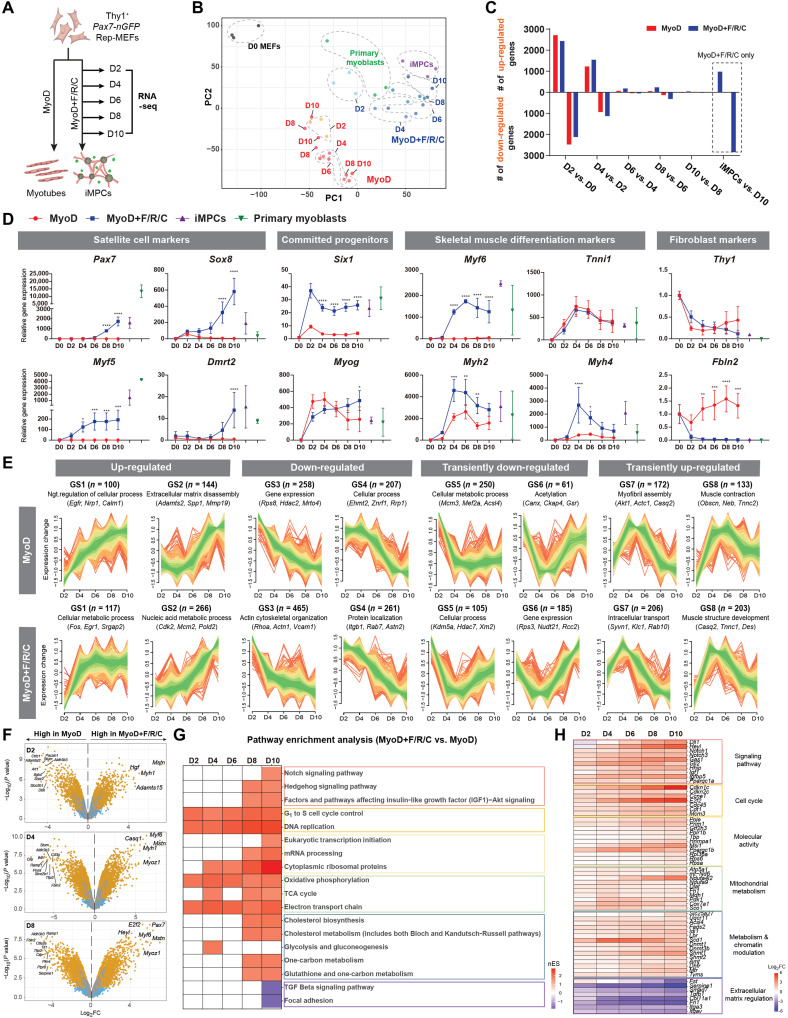
To gain molecular insights into the transcriptional changes that occur during iMPC formation, we performed bulk RNA sequencing (RNA-seq) and dissected transcriptional dynamics following either MyoD or MyoD+F/R/C treatment of fibroblasts. To this end, we subjected Thy1+ Rep-MEFs to the MyoD or MyoD+F/R/C condition, followed by RNA-seq at days 2, 4, 6, 8, and 10 of the conversion process (Fig. 2A). As positive controls, we used stable iMPC clones and satellite cell–derived Pax7-nGFP primary myoblasts. Principal component analysis (PCA) and dendrogram clustering separated the MyoD- and MyoD+F/R/C-treated cells into two distinct groups, which were both transcriptionally divergent from parental MEFs (Fig. 2B and fig. S2A). For the PCA, a stepwise temporal trajectory was documented during the MyoD+F/R/C reprogramming course, as cells gradually clustered further away from parental MEFs and became more akin to stable iMPCs (Fig. 2B). In contrast, MEFs subjected only to MyoD overexpression exhibited a haphazard temporal trajectory with no clear separation between time points (Fig. 2B). We next documented the number of differentially expressed genes (DEGs) during either the MyoD or MyoD+F/R/C condition. This analysis revealed a prominent transcriptional wave of up-regulated and down-regulated genes under both conditions at days 2 and 4, followed by reduced transcriptional activity during days 4 to 6, and an additional transcriptional wave predominantly under the MyoD+F/R/C condition at day 8 (Fig. 2C). We then investigated individual gene expression dynamics during the reprogramming course and noted an up-regulation of the differentiation markers Myog, Tnni1, Actn2, and Myh2 under both MyoD and MyoD+F/R/C conditions (Fig. 2D and fig. S2B). However, prominent expression of other key myogenic progenitor and differentiation markers including Myf5, Myf6, Six1, Six4, Myh1, and Myh4 was significantly or solely up-regulated under the MyoD+F/R/C condition (Fig. 2D and fig. S2B). Notably, significant up-regulation of myogenic stem cell markers including Pax7 and Dmrt2 was only observed under the MyoD+F/R/C condition starting at days 6 to 8 of the reprogramming process (Fig. 2D and fig. S2C). In addition, pronounced down-regulation of fibroblast-specific genes such as Fbln2, Col1a1, and Col5a1 was most notably observed in the presence of F/R/C treatment, whereas Thy1 expression was significantly down-regulated under both conditions (Fig. 2D and fig. S2, B and C). Last, we wished to compare our transcriptome analysis to a recently published work that dissected iMPC formation via a transgenic MyoD-reprogrammable mouse strain (27). To this end, we compared their RNA-seq dataset for MyoD+F/R/C reprogramming during defined time points to our own. Overall, we documented remarkably similar transcriptional patterns between the two reprogramming systems, albeit a few differentiation genes were up-regulated at different time points (fig. S2, D and E).
Fig. 2. Global transcriptome dynamics during myogenic conversions.
(A) Experimental design. (B) PCA of global RNA-seq using all gene read counts. N = 3 cell lines per group. (C) Bar graphs showing the number of DEGs between each indicated comparison. DEGs were calculated using |log2FC| > 0.5 (P < 0.01). (D) Gene expression dynamics for the indicated genes and conditions based on bulk RNA-seq. The data are shown as means ± SD. N = 3 cell lines per group. Statistical significance was determined by two-way analysis of variance (ANOVA) between samples at each time point (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). (E) Fuzzy clustering based on bulk RNA-seq for gene expression dynamics during the indicated days and conditions. Expression changes are represented as log2FC for each time point versus parental MEFs. N = 3 cell lines per group. (F) Volcano plots showing DEGs between the indicated conditions and days. Significant DEGs are shown as yellow dots (|log2FC| > 0.5, P < 0.05). N = 3 cell lines per group. (G) Heatmap of enriched pathways under MyoD+F/R/C versus MyoD conditions at the indicated time points. Normalized enrichment score (nES) obtained from GSEA is displayed. (H) Heatmap of relative gene expression for the indicated conditions and time points. Each gene group is associated with a pathway represented in (G). Log2FC is presented as a color gradient. N = 3 cell lines per group.
We next analyzed broad transcriptional trends in the two cellular conversions via fuzzy clustering. In total, eight gene subsets that exhibited a similar expression pattern during either the MyoD or MyoD+F/R/C condition were defined (Fig. 2E). The first group involved two gene sets (GS1 and GS2) that were up-regulated under both conditions, albeit for the MyoD condition, these genes annotated with negative regulation of cellular processes, whereas metabolic processes were defined for the MyoD+F/R/C condition (Fig. 2E). Specifically, 266 genes in GS2 of MyoD+F/R/C treatment exhibited a pattern of gradual gene up-regulation and contained genes that annotated with regulation of cell cycle (Cdk2, Cdkn1c, and Smc3) and striated muscle tissue development (Pax7, Heyl, and Tbx3) (fig. S2F). The second gene sets (GS3 and GS4) involved down-regulated genes for both conditions (Fig. 2E). Last, gene subsets showing transient down-regulation (GS5 and GS6) or up-regulation (GS7 and GS8) patterns were similarly defined for both conditions (Fig. 2E). Notably, the MyoD condition showed transient down-regulation of genes associated with metabolic processes (GS5 under the MyoD condition), whereas the MyoD+F/R/C condition exhibited gradual up-regulation of metabolic genes (GS1 and GS2 under the MyoD+F/R/C condition). This observation suggests that MyoD+F/R/C treatment of fibroblasts may lead to a more metabolically active cell state, encompassing biochemical reactions in the form of oxidative phosphorylation and tricarboxylic acid (TCA) cycle (GS1) in addition to nucleic acid metabolic processing involved in DNA binding, mRNA splicing, and epigenetic regulation (GS2).
The transcriptome analysis revealed prominent gene expression differences between the MyoD and MyoD+F/R/C conditions, prompting us to perform analysis between the two cellular conversions to uncover key DEGs. The top 30 up-regulated genes for both MyoD and MyoD+F/R/C conditions (day 10) versus MEFs were associated with skeletal muscle differentiation markers including Trim72, Tnnt1, Tnni1, Tnnc2, Neb, Myh7, Myh8, and Actn2 (fig. S3A). However, the top 30 up-regulated genes for MyoD+F/R/C versus MyoD conditions at day 10 were mainly myogenic stem and progenitor cell markers including Pax7, Sox8, Msc, Lgr5, Fgfr4, Dbx1, and Heyl (fig. S3A). Given the elevated expression of myogenic stem cell markers in MyoD+F/R/C in comparison to MyoD at day 10, we next performed sequential analysis of DEGs under MyoD+F/R/C versus MyoD conditions. The most up-regulated genes at days 2, 4, and 6 for MyoD+F/R/C were predominantly skeletal muscle differentiation markers including Mstn, Myoz1, Myh1, Myf6, and Casq1 (Fig. 2F and fig. S3, B and C). In addition, the most differentially up-regulated genes at a late reprogramming stage of the MyoD+/F/R/C condition (days 8 and 10) consisted of stem cell markers including Pax7, Sox8, and Dbx1 as well as the differentiation markers Myf6, Mstn, and Myoz1 (Fig. 2F and fig. S3B). To further validate the up-regulation of muscle stem cell genes during reprogramming via MyoD+F/R/C, we performed a meta comparison of the transcriptomes of Pax7-nGFP primary myoblasts with MEFs subjected to either the MyoD or MyoD+F/R/C condition. Notably, we documented that satellite cell–associated genes are only shared between primary myoblasts and MyoD+F/R/C-treated cells during days 6 to 10, whereas several differentiation markers were detected under all conditions for all inspected time points (fig. S3D). To acquire additional insights into the signaling pathways that are enriched during MyoD+F/R/C treatment, we performed pathway enrichment analysis of DEGs between MyoD+F/R/C and MyoD conditions using gene set enrichment analysis (GSEA) for each reprogramming time point. Pathways enriched for the MyoD+F/R/C condition across all time points were associated with “cell cycle” and “mitochondrial metabolism,” whereas “metabolism/chromatin modulation” and “Notch and Hedgehog signaling pathways” were enriched under the MyoD+F/R/C condition only at days 8 and 10, thus corroborating our fuzzy clustering analysis (Fig. 2, G and H).
In summary, using bulk RNA-seq, we delineated transcriptional dynamics during cellular reprogramming manifested by either the MyoD or MyoD+F/R/C condition and demonstrated divergent transcriptional changes. Solely overexpressing MyoD in fibroblasts induces fast and direct formation of multinucleated myotubes that expressed only a partial cohort of myogenic differentiation genes. In contrast, MyoD+F/R/C treatment entails a stepwise reprogramming process that commences with an early transcriptional wave that is characterized by up-regulation of a plethora of myogenic differentiation genes before a second transcriptional wave that is characterized by up-regulation of myogenic stem cell markers. Furthermore, direct transcriptional comparison between MyoD+F/R/C and MyoD conditions at defined time points revealed expression of canonical myogenic stem cell and differentiation markers in addition to signaling pathways that are unique to iMPC reprogramming.
Proteome dynamics during direct conversion of fibroblasts into myogenic cells
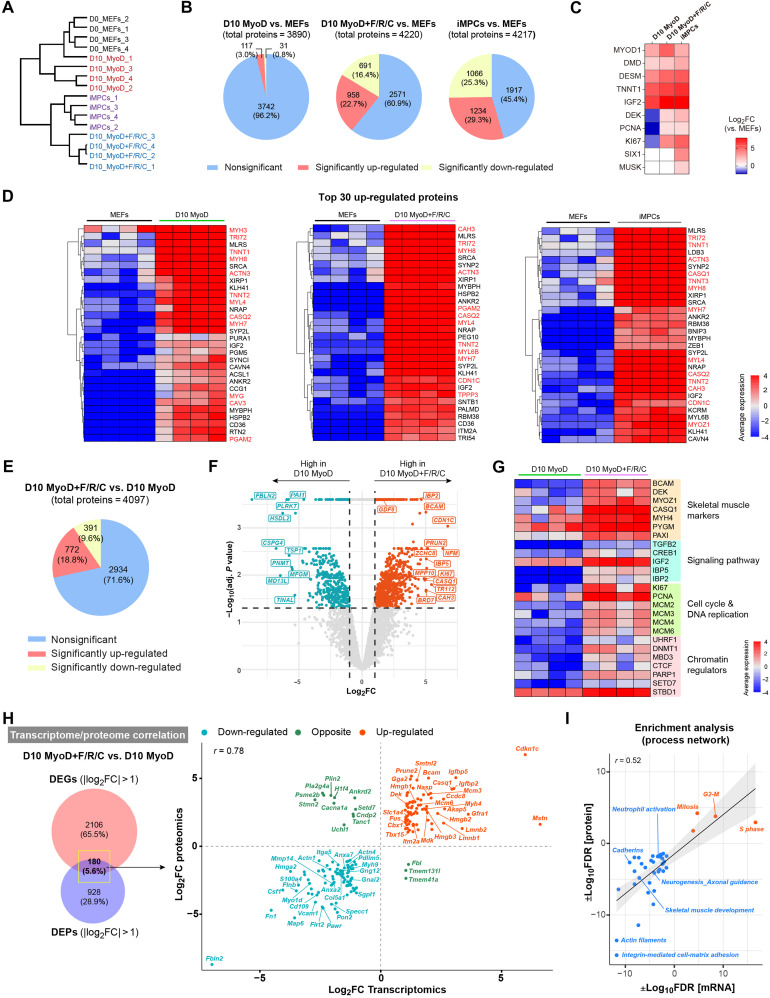
The RNA-seq analysis revealed pronounced transcriptional differences between MEFs subjected to the MyoD condition and those subjected to the MyoD+F/R/C condition. Given this observation, we next wished to assess whether these differences manifest at the protein level as well. To explore this possibility, we performed liquid chromatography–mass spectrometry (LC-MS) analysis of multiple MEFs, iMPC clones, and MEFs subjected to either the MyoD or MyoD+F/R/C condition at day 10. Dendrogram clustering and PCA separated MEFs and MyoD-treated MEFs into one group and iMPCs and MyoD+F/R/C-treated MEFs into another group, with further subdivision in each respective group (Fig. 3A and fig. S4A). To corroborate this observation further, we identified the number of differentially expressed proteins (DEPs) between the various groups and MEFs. In accordance with the hierarchical clustering, the percentage of DEPs was higher for MEFs subjected to MyoD+F/R/C treatment or iMPCs than MEFs subjected to the MyoD condition alone, suggesting that MyoD+F/R/C manifests in more pronounced proteome changes (Fig. 3B). However, in comparison to parental MEFs, we documented in all groups an up-regulation of several skeletal muscle–associated proteins such as DESM, DMD, and TNNT1, albeit the proliferation markers PCNA and KI67 and the myogenic protein DEK were significantly up-regulated only under the MyoD+F/R/C condition and stable iMPCs (Fig. 3C). We then analyzed the top 30 up-regulated DEPs under each condition versus MEFs and documented the up-regulation of typical skeletal muscle proteins in all groups including MYH7, MYH8, TNNT2, ACTN3, and CASQ2 (Fig. 3D). Furthermore, a functional enrichment analysis of DEPs highlighted the up-regulation of several skeletal muscle gene networks in all groups (fig. S4B). We then performed a direct comparison between MyoD+F/R/C and MyoD conditions at day 10 and documented 772 up-regulated and 391 down-regulated proteins of 4097 total detected proteins (Fig. 3E). Notably, the most up-regulated proteins under the MyoD+F/R/C versus MyoD conditions were associated with skeletal muscle differentiation markers and signaling proteins such as CASQ1, MYOZ1, GDF8 (Mstn), IBP2, and IBP5 and cell proliferation and chromatin regulators including KI67, CDN1C, DNMT1, and BRD7 (Fig. 3, F and G). Following MyoD+F/R/C treatment, we further observed significant enrichment for cell proliferation and DNA replication proteins (MCM2, MCM3, MCM4, MCM6, PCNA, and KI67), chromatin regulators (DNMT1, UHRF1, MBD3, PARP1, and SETD7), signaling proteins (IGF2, TGFB2, IBP2, IBP5, and CREB1), and, most notably, skeletal muscle–associated proteins (PAXI, PYGM, MYH4, DEK, BCAM, CASQ1, and MYOZ1) (Fig. 3G).
Fig. 3. Proteome dynamics during myocyte and iMPC formation.
(A) Hierarchical clustering based on total proteome data. N = 4 cell lines per group. (B) Pie charts showing quantification of DEPs for the indicated comparisons. The significance threshold was set as |log2FC| > 1 and adjusted P < 0.05. (C) Heatmap showing relative protein expression for canonical myogenic markers. The calculated log2FC in each cell line versus MEFs is shown. Nondetected proteins are represented by a white box. N = 4 cell lines per group. (D) Heatmaps based on proteome data. The average protein expression is presented as gradient. N = 4 cell lines per group. Myogenic differentiation–related proteins are highlighted in red. (E) Pie chart showing quantification of DEPs between the indicated conditions. The significance threshold was set as |log2FC| > 1 and adjusted P < 0.05. (F) Volcano plot for DEPs between the indicated conditions. Significant DEPs (|log2FC| > 1, adjusted P < 0.05) are shown as blue and red dots. N = 4 cell lines per group. (G) Heatmap based on protein expression for the indicated markers together with a manual annotation. The average protein expression is presented as gradient. N = 4 cell lines per group. (H) Left: Venn diagram for DEGs and DEPs between the indicated conditions (|log2FC| > 1, P < 0.05). Right: Scatterplot showing the correlation between the transcriptome and proteome datasets. (I) Scatterplot showing correlated process networks that are enriched under MyoD+F/R/C versus MyoD conditions as determined by Metacore. Up-regulated (red) and down-regulated (blue) process networks are shown.
Next, we performed an integrative analysis of the RNA-seq and proteome datasets. We first inspected significant DEGs and DEPs in iMPCs compared to MEFs and recorded 443 shared genes/proteins with a high correlation (r = 0.81) (fig. S4C). Notably, enriched genes/proteins in iMPCs included skeletal muscle–related markers, chromatin regulators, and signaling pathway factors, whereas MEFs were enriched for fibroblast-specific genes and proteins (fig. S4C, right). In addition, by comparing iMPCs with MEFs, we determined that several actin cytoskeleton–related markers such as Sorbs2, Bcl6, and Fmnl1 demonstrated significant up-regulation at the mRNA but not at the protein level, suggesting potential posttranscriptional regulation in iMPCs (fig. S4D). We then performed a similar integrative analysis of MyoD+F/R/C versus MyoD conditions at day 10 of reprogramming and identified a group of 180 shared DEGs/DEPs (Fig. 3H, left). We noted a high correlation between the mRNA and protein levels (r = 0.78) and detected, following F/R/C treatment, a unique up-regulation of skeletal muscle markers such as Dek, Igfbp2, Igfbp5, Mstn, Bcam, Myh4, and Casq1, in addition to cell proliferation and DNA replication markers such as Mcm3, Mcm6, and Nasp (Fig. 3H, right). Last, we performed enrichment analysis for each category of up-regulated and down-regulated markers, revealing up-regulation of “cell proliferation” and down-regulation of “developmental processes”– and “cell adhesion”–associated networks following F/R/C treatment (Fig. 3I).
Collectively, the LC-MS analysis uncovered a unique expression of proteins that are substantially up-regulated in iMPCs or under the MyoD+F/R/C condition. These proteins include skeletal muscle differentiation proteins, signaling factors, chromatin regulators, and proliferation markers. The high expression of proteins such as MYOZ1, PYGM, MYH4, and DEK predominantly in the presence of F/R/C treatment suggests that iMPCs preferentially express mature muscle markers in comparison to myotubes produced via transdifferentiation by MyoD.
Unique chromatin accessibility changes during formation of iMPCs
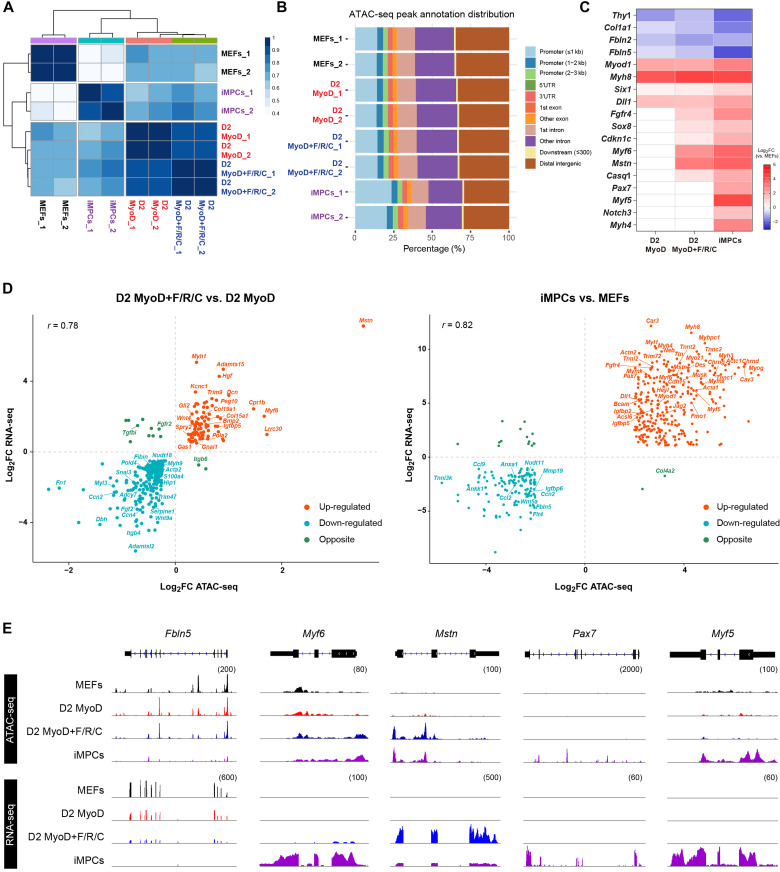
Enhanced chromatin accessibility in gene promoter regions is well known to be associated with elevated gene expression. Given the prominent differences at the mRNA and protein levels between MyoD and MyoD+F/R/C conditions, we reasoned that divergent chromatin accessibility changes in promoter regions of key myogenic genes may accompany the observed differences in the transcriptome and proteome during transdifferentiation and reprogramming. To address this question, we performed an assay for transposase-accessible chromatin using sequencing (ATAC-seq) of MEFs, MEFs subjected to either the MyoD or MyoD+F/R/C condition for 2 days, and established iMPC clones and established iMPC clones. Similar to the previous multiomics analyses, iMPCs were characterized by a distinct chromatin accessibility profile in comparison to MEFs and unexpectedly demonstrated a high percentage of annotated peaks around promoter regions (Fig. 4, A and B). Given this observation, we performed differential accessible region analysis in gene promoter regions of canonical myogenic and fibroblast genes. In comparison to MEFs, we detected a decrease in chromatin accessibility in fibroblast-specific gene promoters such as Thy1, Fbln2, Fbln5, and Col1a1 and increased accessibility in myogenic gene promoters including Myh8, Six1, and Myod1 for all myogenic cell lines (Fig. 4C). In accordance with the transcriptome analysis, we also documented a preferential increase in chromatin accessibility in MyoD+F/R/C-treated MEFs and iMPCs in multiple myogenic differentiation genes such as Myf6, Mstn, and Casq1 as well as progenitor genes such as Sox8 and Fgfr4 (Fig. 4C). Notably, open chromatin configuration in the promoters of muscle stem cell markers such as Pax7, Myf5, and Notch3 was solely documented in established iMPCs versus MEFs (Fig. 4C).
Fig. 4. Chromatin accessibility changes unique to iMPC reprogramming.
(A) Correlation matrix for the indicated samples based on ATAC-seq peaks for global chromatin accessibility. Pearson’s correlation coefficient r is displayed as a color gradient. (B) Distribution of annotated ATAC-seq peaks across the respective genomic regions. 3′UTR, 3′ untranslated region; 5′UTR, 5′ untranslated region. (C) Heatmap showing relative chromatin accessibility levels in gene promoter regions (±1 kb of TSS) for the indicated samples. Log2FC is shown as a color gradient. Nonsignificant log2FC values (P > 0.01) are shown in white. N = 2 cell lines per group. TSS, transcription start site. (D) Scatterplot for overlapping genes showing the correlation between chromatin accessibility in promoter regions (±1 kb of TSS) and gene expression at day 2 of MyoD+F/R/C- versus MyoD-treated MEFs (left) and iMPCs versus MEFs (right). The significant genes (P < 0.01) with |log2FC| > 0.5 (left) and |log2FC| > 2 (right) are shown. (E) IGV tracks for ATAC-seq peaks and mRNA coverage based on bulk RNA-seq data for the indicated genes.
To investigate further whether chromatin accessibility in promoter regions is associated with differential gene expression, we performed an integrative analysis of ATAC-seq and bulk RNA-seq datasets between MyoD- and MyoD+F/R/C-treated MEFs at day 2, as well as established iMPCs and MEFs. For both comparisons, we noted a positive and high correlation between increased gene expression and an accessible, more “open” chromatin configuration (Fig. 4D). Notably, we detected an increase in both gene expression and chromatin accessibility in the promoter regions of myogenic genes that were preferentially expressed following F/R/C treatment at day 2 including Mstn, Myf6, Myh1, and Gas1 (Fig. 4, D and E). Concomitantly, we noticed a similar trend for myogenic stem and differentiated cell markers when comparing iMPCs to parental MEFs, including Pax7, Myf5, Bcam, Heyl, Mymk, Myh1, and Myh4 (Fig. 4, D and E). Notably, a high concordance of gene expression and chromatin accessibility was detected between the MyoD+F/R/C condition at day 2 and MEFs; however, very few genes exhibited an anticorrelation trend (fig. S5). Together, our results demonstrate that as early as 2 days following either MyoD or MyoD+F/R/C treatment of MEFs, prominent chromatin accessibility changes can be detected at myogenic promoters; however, F/R/C treatment further elicits preferential open chromatin in multiple key myogenic gene promoters, in agreement with their unique transcriptomic and proteomic profiles.
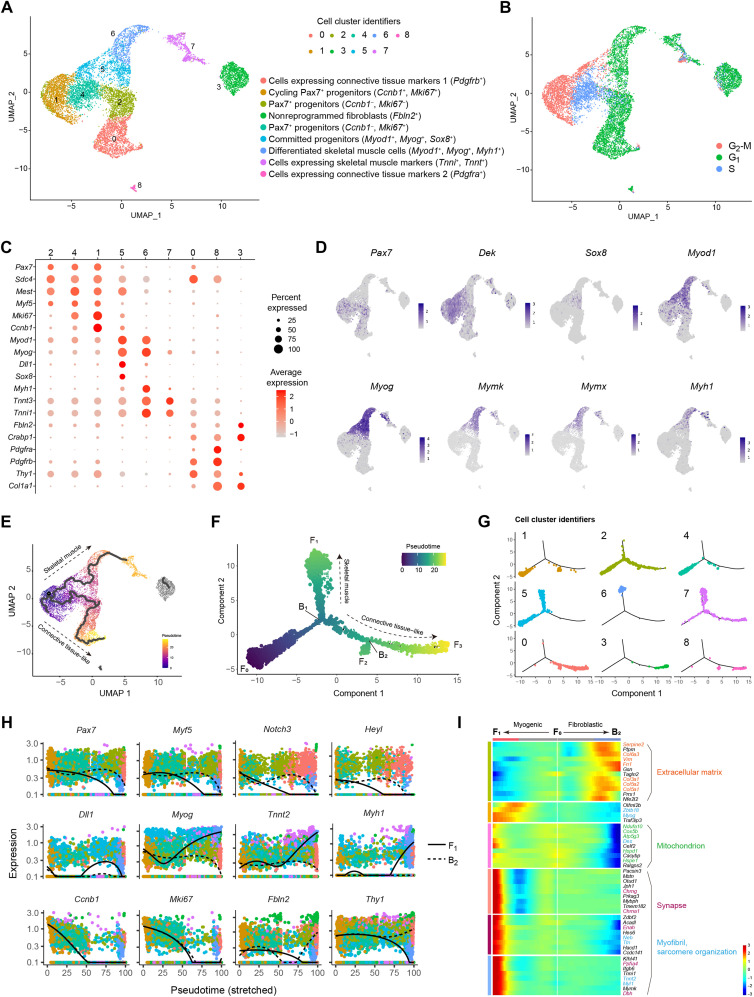
Cell types and differentiation trajectories in iMPCs uncovered by single-cell RNA-seq
The heterogeneity of iMPCs renders their molecular characterization challenging using bulk multiomics tools. To address this challenge and to identify the underlying cell types that comprise the heterogeneous iMPC cultures, we performed single-cell RNA-seq (scRNA-seq) of a stable iMPC clone, which showed a strong correlation (r = 0.7) with the bulk RNA-seq dataset of iMPCs (fig. S6A). Using unsupervised clustering (28), we then identified nine distinct cell clusters present in iMPCs, characterized by three divergent cell cycle states (Fig. 5, A and B). Three cell clusters (1, 2, and 4) represented the Pax7+ progenitor cell population and could be further separated into cycling (C1, 16.25%) or less-cycling cell populations (C2, 14.88% or C4, 12.58%) (Fig. 5, A to C). These clusters were also highly enriched for the myogenic stem and progenitor cell markers Myf5, Dek, Dbx1, and Mest (Fig. 5, C and D, and fig. S6B). Notably, cells in cluster 5 (C5, 10.38%) expressed high levels of Sox8, Myod1, and Myog and the canonical Notch ligand Dll1, thus representing committed progenitors, whereas late-stage differentiation markers including Myf6, Mstn, Mymk, Mymx, and Myh1 were present in cluster 6, thus representing differentiated skeletal muscle cells (C6, 7.64%) (Fig. 5, C and D, and fig. S6B). Cluster 7 (C7, 6.62%) may correspond to cells expressing skeletal muscle markers given the high expression of Tnnt3 and Tnni1 but the absence of MyoD or Myog expression (Fig. 5, C and D). Moreover, three clusters (0, 3, and 8) highly expressed Col1a1, Thy1, Pdgfra, and Pdgfrb, which are markers of connective tissue cells (Fig. 5, A and C). To identify the origin of these connective tissue–like cells, we performed scRNA-seq of parental MEFs and observed that these markers are highly expressed in fibroblasts (fig. S6C). In particular, pseudo average gene expression of C3 in iMPCs showed a very high concordance with MEFs (r = 0.98), thus suggesting that C3 may represent a population of nonreprogrammed fibroblasts in iMPC cultures (fig. S6D). Clusters C0 and C8 also demonstrated high correlation with MEFs (r = 0.89), suggesting that these are fibroblasts or other connective tissue cell types (fig. S6D).
Fig. 5. scRNA-seq uncovers cell types and differentiation trajectories in iMPCs.
(A) UMAP projection based on scRNA-seq data of iMPCs representing 9184 cells. (B) UMAP projection colored by cell cycle states. (C) Dot plot for individual gene expression based on scRNA-seq of iMPCs. (D) UMAP projection showing all cells colored by the indicated myogenic markers. (E) UMAP projection showing all cells colored by pseudotime as calculated from unsupervised single-cell trajectory using Monocle3. (F) Minimum spanning tree showing ordered cells based on semi-supervised single-cell trajectory analysis reconstructed by Monocle2 and colored by pseudotime. The pseudotime initiates from F0 and bifurcates at the branch point B1 toward two main cell fates (denoted as F1 and F3). Note that C3 and C8 are part of the trajectory due to a different algorithm used by Monocle2 versus Monocle3. (G) Minimum spanning tree corresponding to (F) and showing the cells for each cell cluster identifier. (H) Plots showing the expression kinetics of the indicated genes as a function of pseudotime, which emanates from F0 and proceeds toward F1 (solid line) or B2 (dashed line). The data correspond to (F), and the dots indicate cells colored by cell cluster identifiers. (I) Heatmap for DEGs regulated at B1 in (F). Color-coded gradient represents normalized gene expression level for each gene across all cells. The GO terms were annotated using DAVID v6.8 for each respective gene cluster and shown using the same color coding.
To dissect how iMPCs recapitulate a myogenic differentiation program in vitro, we performed a pseudotime lineage trajectory analysis using the Monocle package (29). As a first step, we performed an unsupervised trajectory analysis, which identified two major cellular progressions, both emanating from the cycling Pax7+ progenitors (C1) (Fig. 5E). One branch further gave rise to several myogenic differentiation cell types (C5 and C6), whereas another branch emanated toward a less-proliferative population of Pax7+ cells, which gave rise to a connective tissue–like cell fate (C0). Aside from these two main trajectories, C3 was not associated with an active lineage progression, further supporting the notion that these cells may indeed correspond to nonreprogrammed MEFs (Fig. 5E). To validate the reproducibility of the trajectory analysis, we reconstructed a cellular progression in a semi-supervised manner. By way of this analysis, we detected two distinct differentiation branch points (denoted as B1 and B2) and three different cell fates (denoted as F1, F2, and F3) emanating from the root of trajectory F0, which was mainly composed of Pax7+ progenitors (Fig. 5, F and G, and fig. S7A). The B1 branching point represents a clear bifurcation into two distinct reprogramming routes, F1 and F3 (Fig. 5F). Notably, the progression from B1 to F1 mainly includes cells that express committed and differentiation myogenic genes (C5, C6, and C7), whereas the progression from B1 to F3 consists of connective tissue–like cells (C0 and C8) (Fig. 5, F and G). Notably, the small bifurcation leading from B2 to F2 corresponds to cells that up-regulated several stem cell and metabolic-related genes (fig. S7, B and C). This unexpected observation cautiously suggests that these cells may have reverted back into a cycling Pax7+ cell state; however, further experimental work is needed to characterize this small cell population.
As the next step, we set out to dissect individual gene expression in the two differentiation routes emanating from B1. First, stem and progenitor cell markers such as Pax7 and Myf5 were highly expressed at the start of the pseudotime and were rapidly down-regulated as they progressed into F1 (Fig. 5H and fig. S7D). In addition, the committed progenitor genes Myod1 and Myog were only up-regulated from B1 to F1, in conjunction with the appearance of differentiation markers such as Myh1 and Tnni1 (Fig. 5H and fig. S7D). Dll1- and Sox8-expressing cells were detected in the middle of the branch leading to F1, further suggesting that these are committed myogenic progenitors before their differentiation (Fig. 5H and fig. S7D). Furthermore, the cell cycle proliferation markers Ccnb1 and Mki67 were substantially down-regulated in both routes, whereas the connective tissue cell markers Fbln2, Thy1, Col1a1, Pdgfra, and Pdgfrb were only up-regulated in the path leading to B2. A differential gene expression analysis between these two paths further corroborated the clear bifurcation into either skeletal muscle or connective tissue–like cell fate (Fig. 5I).
The intriguing observation that iMPCs may give rise to a connective tissue–like cell fate prompted us to investigate whether purified iMPCs can similarly give rise to this cell population in the absence of nonreprogrammed fibroblasts. To this end, we FACS-purified Pax7-nGFP+ cells from iMPCs and reestablished a heterogeneous iMPC clone consisting of Pax7-nGFP+ cells, fibers, and about 1% Pdgfrb+ cells at passage 1 (fig. S8, A to C). Following seven passages, the number of Pdgfrb+ cells increased to 11.8%, suggesting that Pdgfrb+ cells are highly proliferative or alternatively derived from Pax7+ cells as suggested by the pseudotime analysis (fig. S8C). In addition, we FACS-purified Thy1+ and Thy1− cells from an iMPC clone and observed that the Thy1+ cell population gave rise to fibroblast-like cells, whereas the Thy1− cell population generated iMPC-like clusters (fig. S8D). These observations support the notion that progenitor cells present in iMPCs can also give rise to another cell type or alternatively revert back into a connective tissue–like cell fate.
In summary, using scRNA-seq, we identified the various cell populations that comprise the heterogeneous iMPC cultures, demonstrating the presence of cycling Pax7+/Myf5+ stem cells, Sox8+/Myod1+/Myog+ committed progenitors, and Myf6+/Myh1+ differentiated cells, in addition to connective tissue–like cells. Moreover, we reconstructed the myogenic program in an iMPC clone using a pseudotime trajectory analysis and delineated the differentiation route from activated satellite–like cells into committed progenitors and differentiated skeletal muscle cells. This analysis also unveiled an alternative differentiation route into a connective tissue–like cell fate, cautiously suggesting that iMPCs may harbor a bipotential differentiation propensity.
The molecular landscape of purified Pax7-nGFP+ iMPCs in comparison to myoblasts
The scRNA-seq analysis uncovered a unique cell population consisting of Pax7+ stem cells; however, using bulk RNA-seq, we could only detect a few myogenic stem cell–associated genes because of the heterogeneity of iMPC cultures. Furthermore, using MS, we did not detect satellite cell– and myoblast-associated proteins in bulk iMPCs. This could be due to technical limitations in using LC-MS to detect lowly expressed proteins in the form of stem cell–specific transcription factors due to the high expression of structural and signaling proteins emanating from the multinucleated myofibers of iMPCs.
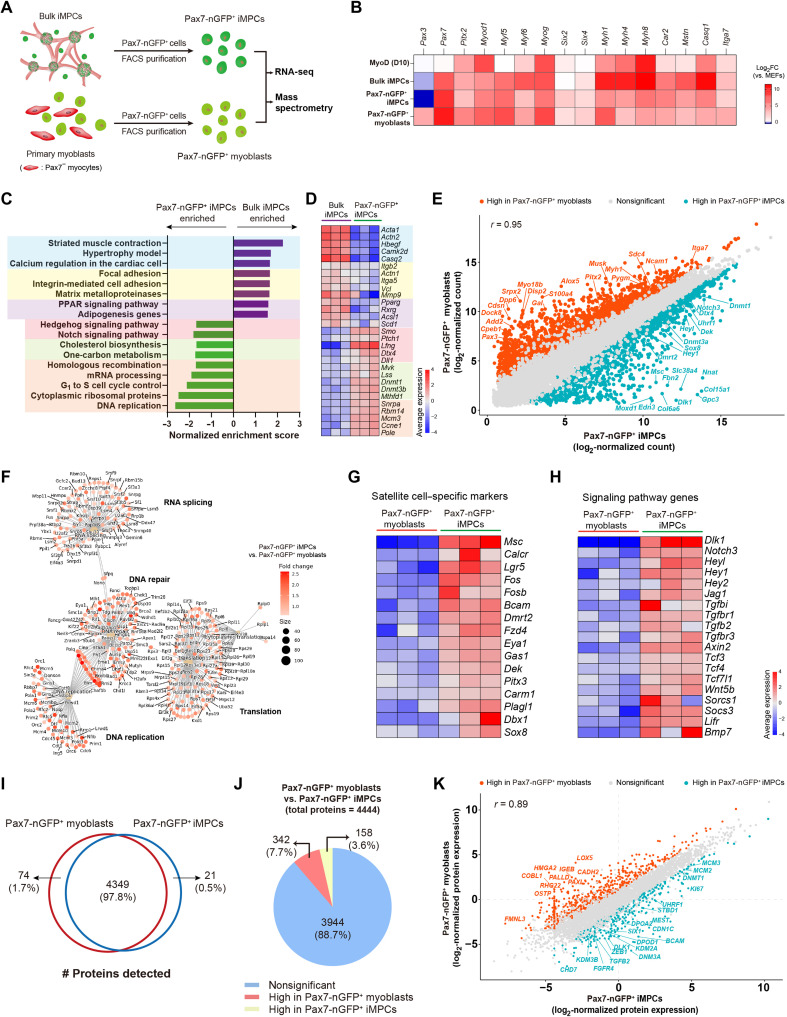
To address this limitation and characterize the Pax7+ stem cell population in depth, we opted to FACS-purify Pax7-nGFP+ cells from iMPC clones and molecularly compare them to FACS-purified Pax7-nGFP+ myoblasts by bulk RNA-seq and LC-MS (Fig. 6A). The expression levels of canonical satellite cell markers such as Pax7 and Six2 were higher in Pax7-nGFP+ iMPCs and Pax7-nGFP+ myoblasts, whereas the expression of skeletal muscle differentiation genes such as Myh1, Myh4, Myh8, and Casq1 was higher in bulk iMPCs, indicating that the stem cell purification strategy from bulk iMPCs was successful (Fig. 6B). Next, to identify pathways unique to the stem cell subsets of iMPCs, we performed a pathway enrichment analysis between bulk iMPCs and Pax7-nGFP+ iMPCs, thus revealing gene categories that were highly enriched in bulk iMPCs associated with differentiated muscle cells (Fig. 6, C and D). In contrast, cell proliferation– and metabolism/chromatin modulation–associated pathways were highly enriched in Pax7-nGFP+ iMPCs in addition to the Notch and Hedgehog signaling pathways (Fig. 6, C and D).
Fig. 6. Molecular characterization of FACS-purified Pax7-nGFP+ iMPCs and myoblasts.
(A) Experimental design. (B) Heatmap based on bulk RNA-seq. Log2FC for the respective cell types versus MEFs is displayed as a color gradient. N = 3 cell lines per group. (C) Pathway enrichment analysis using GSEA. (D) Heatmap based on bulk RNA-seq corresponding to (C). The average expression is presented as gradient. N = 3 cell lines per group. (E) Scatterplot showing gene correlation between the two cell types based on log2-normalized gene counts from bulk RNA-seq. Red and blue dots denote statistically significant up-regulated or down-regulated genes, respectively. N = 3 cell lines per group. (F) ORA of statistically significant gene sets (log2FC > 0.5, P < 0.01) in the indicated comparison. (G) Heatmap based on bulk RNA-seq. The average gene expression is presented as gradient. N = 3 cell lines per group. (H) Heatmap based on bulk RNA-seq. The average gene expression is presented as gradient. N = 3 cell lines per group. (I) Venn diagram showing the number of proteins detected in the indicated cell types. (J) Pie chart showing the number of DEPs for the indicated comparison. The significance threshold was set as |log2FC| > 1 and adjusted P < 0.1. N = 4 cell lines per group. (K) Scatterplot based on log2-normalized protein expression. Red and blue dots denote statistically significant up-regulated or down-regulated genes, respectively. N = 4 cell lines per group.
The FACS purification strategy further allowed us to directly compare Pax7-nGFP+ iMPCs to Pax7-nGFP+ myoblasts. As expected, the two cell types were transcriptionally similar (r = 0.95) (Fig. 6E). However, we also documented statistically significant DEGs that included satellite cell markers in addition to proliferation and chromatin regulators in Pax7-nGFP+ iMPCs whereas other genes were uniquely expressed in Pax7-nGFP+ myoblasts (Fig. 6E). Furthermore, an overrepresentation analysis (ORA) showed divergent transcriptional categories unique to each respective cell type (Fig. 6F and fig. S9A). Notably, we detected elevated expression of known activated satellite cell markers in Pax7-nGFP+ iMPCs that were not detectable or very lowly expressed in Pax7-nGFP+ myoblasts (Fig. 6G). These included Calcr, Musculin (Msc), Lgr5, Fos, Dmrt2, Fzd4, Gas1, Dek, Pitx3, Carm1, Sox8, Dbx1, and Plagl, many of which have been reported to regulate in vivo muscle regeneration and satellite cell activation (Fig. 6G) (30–38). Moreover, gene groups associated with critical pathways for satellite cell activation and proliferation including Notch, TGF-β, Janus kinase (JAK)–signal transducer and activator of transcription (STAT), and WNT were significantly up-regulated in Pax7-nGFP+ iMPCs (Fig. 6H). Last, in comparison to myoblasts, we documented in Pax7-nGFP+ iMPCs enrichment for chromatin remodelers, including Tet1, Tet3, Dnmt1, Dnmt3a, Dnmt3b, and Uhrf1; members of the transcription factor families Myc, KLF, and BEX; and a plethora of cell proliferation and DNA replication markers (fig. S9B).
To examine whether the transcriptional differences could be further detected at the protein level, we opted to perform LC-MS on FACS-purified Pax7-nGFP+ iMPCs and Pax7-nGFP+ myoblasts. Using this approach, we detected 4444 proteins, of which 4349 were detected in both cell types (Fig. 6I). From this protein group, 342 and 158 proteins were differentially expressed in Pax7-nGFP+ myoblasts and Pax7-nGFP+ iMPCs, respectively (Fig. 6J). A comparison of DEPs between the two cell types documented transcription factors, signaling molecules, and chromatin regulators that were significantly more expressed in Pax7-nGFP+ iMPCs in comparison to Pax7-nGFP+ myoblasts including BCAM, CHD7, SIX1, DNM3A, ZEB1, DEK, FGFB2, FGFR4, and KDM2A and the cell proliferation markers KI67, MCM2, and MCM3 (Fig. 6K). In summary, using a transgenic Pax7 reporter, we successfully purified and characterized the transcriptome and a portion of the proteome of Pax7-nGFP+ iMPCs and myoblasts. This analysis established a unique expression signature in Pax7-nGFP+ iMPCs, which is reminiscent of activated satellite cells and distinct from primary myoblasts.
The Notch pathway is critical for iMPC formation and maintenance
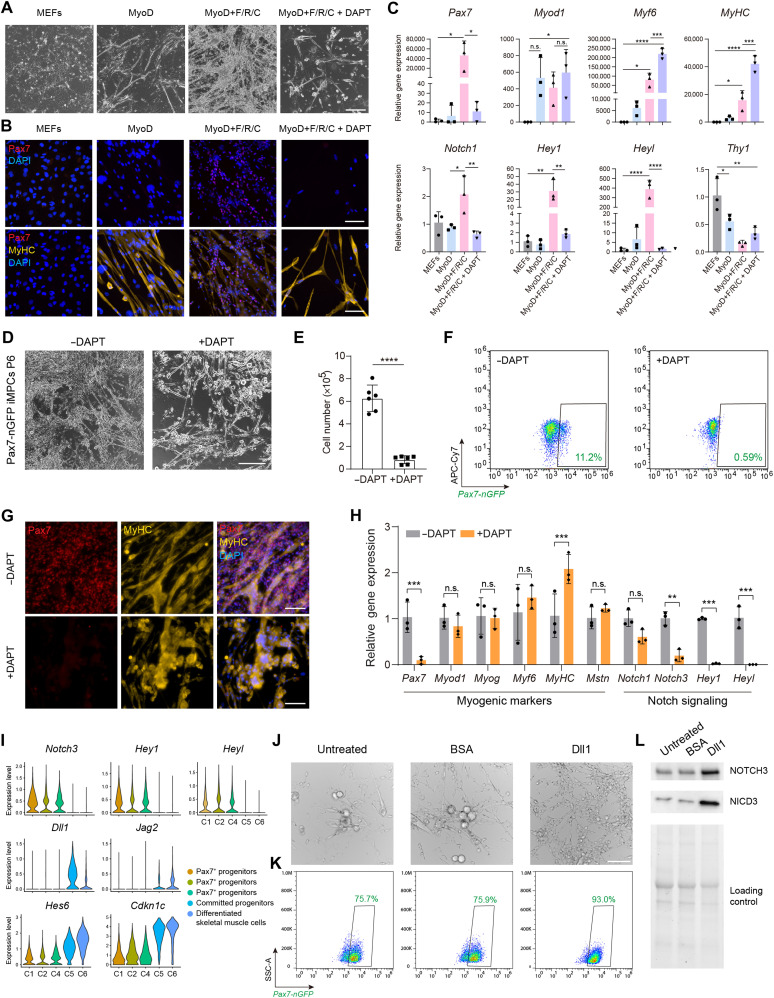
Our transcriptional analysis thus far underpinned the involvement of the Notch pathway during iMPC formation. Hence, we hypothesized that the Notch pathway is critical for iMPC formation as previously reported for the proliferation of endogenous satellite cells in vivo (39–41). We detected up-regulation of Notch1, Notch3, Hey1, and Heyl only under the MyoD+F/R/C condition during the reprogramming course (fig. S10A). To test whether the Notch pathway is critical for iMPC formation, we treated MEFs undergoing MyoD+F/R/C conversion with DAPT, an inhibitor of Notch target γ-secretase (42). We observed complete absence of iMPC formation and cell proliferation at day 10 of MyoD+F/R/C + DAPT treatment in comparison to MyoD+F/R/C-treated cells, albeit multinucleated myotubes formed under both conditions (Fig. 7A).
Fig. 7. Notch signaling is indispensable for the formation and maintenance of iMPCs.
(A) Bright-field images of MEFs subjected to the indicated conditions at day 10. Scale bar, 400 μm. (B) Immunofluorescence images of MEFs subjected to the indicated conditions at day 10. Scale bars, 100 μm. (C) qRT-PCR analysis for the indicated genes and conditions. Relative gene expression is shown as means ± SD. N = 3 cell lines per group. Statistical significance was determined by one-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). (D) Representative bright-field images of iMPCs (P6) either treated or nontreated with DAPT for 5 days. Scale bar, 400 μm. (E) Quantification of cell number based on manual cell counting. Statistical significance was determined by a two-tailed unpaired t test (****P < 0.0001). N = 2 repeats for three cell lines per group. (F) Flow cytometry analysis of Pax7-nGFP iMPCs either treated or nontreated with DAPT for 5 days. (G) Immunofluorescence images of iMPCs treated with DAPT for 5 days. Scale bars, 100 μm. (H) qRT-PCR analysis for DAPT-treated iMPCs at day 5. The data are shown as means ± SD. N = 3 cell lines per group. Statistical significance was determined using one-way ANOVA (**P < 0.01 and ***P < 0.001). (I) Violin plots based on scRNA-seq of iMPCs. (J) Bright-field images of Pax7-nGFP+ iMPCs cultured using the indicated conditions for 6 days. Scale bar, 100 μm. (K) Flow cytometry analysis for the indicated conditions at day 6. (L) Western blot for the indicated proteins.
To confirm the lack of iMPC formation following DAPT treatment, we assessed the expression of Pax7 and MyHC in MEFs subjected to MyoD+F/R/C + DAPT treatment for 10 days. Unlike the MyoD+F/R/C condition at this time point, we did not detect Pax7+ cells under the MyoD+F/R/C + DAPT condition, albeit MyHC+ myotubes did form (Fig. 7B). In accordance with the RNA-seq data, qRT-PCR revealed that the expression of Pax7, Notch1, Hey1, and Heyl was higher under the MyoD+F/R/C condition than under the MyoD condition; however, DAPT treatment abrogated the expression of these genes to levels reminiscent of MEFs or MEFs subjected to MyoD alone (Fig. 7C). DAPT treatment did not reduce the expression of other myogenic markers such as Myf5, Myod1, Mstn, and Myog as well as the fibroblast-specific marker Thy1, suggesting that Notch inhibition preferentially affects the Pax7+ stem cell population (Fig. 7C and fig. S10B).
We next assessed the effect of Notch inhibition on the self-renewal of stable iMPC clones. To this end, we cultured iMPCs in the presence of DAPT for five consecutive days and observed a marked decrease in cell density and extensive depletion of Pax7-nGFP+ cells (Fig. 7, D to F). We also confirmed the absence of Pax7+ cells via immunofluorescence and noted that MyHC+ myofibers were detected under both conditions (Fig. 7G). To investigate further the effect of DAPT treatment, we conducted qRT-PCR for myogenic and Notch-related genes in DAPT-treated and nontreated iMPCs. This analysis revealed a marked reduction of Pax7 expression in concert with down-regulation of Hey1, Heyl, Notch1, and Notch3 in DAPT-treated iMPCs, whereas Myod1, Myog, and Mstn were expressed at about a similar level (Fig. 7H).
Building upon these findings, we next set out to decipher the Notch receptor-ligand interactions between various cell populations that comprise a stable iMPC clone via the scRNA-seq dataset using the CellChat software (43). By way of this analysis, we recorded strong Notch signaling pathway network interactions between the Pax7+ cell populations and the committed myogenic progenitors or differentiated cells in an iMPC clone (fig. S10C). In accordance with this cell-cell communication analysis, we documented that Pax7+ cells highly expressed the Notch receptor genes Notch3 and Notch1 in addition to the canonical Notch targets Heyl and Hey1 (Fig. 7I and fig. S10D). Most notably, the progenitor and differentiated cell populations of iMPCs expressed the canonical Notch ligands Dll1, Jag2, and Dlk1 in addition to the Notch inhibitors Cdkn1c and Hes6 (Fig. 7I and fig. S10D).
Given the substantial expression of Notch3 and Dll1, we opted to interrogate further whether the predicted interaction may have an effect on the self-renewal and differentiation capacities of Pax7+ iMPCs. To this end, we FACS-purified Pax7-nGFP+ cells from iMPC clones and propagated the cells on Dll1-coated plates in the presence of F/R/C. Noticeably, Pax7-nGFP+ cells propagated via this method proliferated robustly with little to no myofiber formation, consisting of 93% Pax7-nGFP+ cells, whereas cells cultured on untreated or bovine serum albumin (BSA)–coated plates readily formed myofiber networks while down-regulating the Pax7 reporter expression in a subset of cells (Fig. 7, J and K). In support of this observation, the expression level of NOTCH3 and its cleaved intracellular domain NICD3 proteins was highly elevated in Pax7-nGFP+ iMPCs cultured on Dll1 compared to controls (Fig. 7L). These observations jointly imply a potential cross-talk via Notch3 and Dll1 between the stem and differentiated cell populations of iMPCs.
Together, we conclude that activation of the Notch pathway is a unique feature of Pax7+ cell formation during MyoD+F/R/C reprogramming. DAPT-mediated Notch pathway inhibition during reprogramming and in iMPC clones precludes the formation of Pax7+ cells and derails the self-renewal of stable iMPC clones. However, myocytes and myotubes can still form following DAPT treatment, suggesting that Notch inhibition solely blunts the formation of myogenic stem cells during reprogramming and in established iMPC clones. Furthermore, heterogeneous iMPCs contain both Pax7+ stem cells that express canonical Notch receptors and targets, as well as differentiated progenies that express Notch pathway–associated ligands and inhibitors, thus recapitulating in vitro their expression during in vivo muscle regeneration. Capitalizing on this observation enabled us to propagate more homogeneously Pax7+ iMPCs using the Notch ligand Dll1.
DISCUSSION
Direct lineage reprogramming of somatic cells into multipotent stem or progenitor cells affords an attractive approach to generate desired cell types for basic research or therapeutic applications. This approach entails several advantages in comparison to transdifferentiation, which typically involves direct conversion of one differentiated cell type into another. Namely, directly reprogrammed progenitors may exhibit self-renewal and multipotency, rendering them more attractive for cell-based therapies. However, most studies to date have reported on protocols to directly transdifferentiate cells, and only a handful of studies documented direct conversion of somatic cells into multipotent progenitors (9). Furthermore, numerous studies have characterized the molecular transitions and mechanisms governing transdifferentiation, yet very few interrogated how the molecular landscape metamorphoses during direct conversion into multipotent progenitor cells (9).
In this study, we set out to address this objective using the skeletal muscle lineage as a model system. To this end, we dissected the molecular changes that accompany fibroblast conversion into myogenic stem and progenitor cells by way of sustained MyoD overexpression in concert with administration of the three small molecules Forskolin, RepSox, and CHIR99210 (F/R/C) (20). Using multiomics approaches, we contrasted this lineage conversion to that of canonical MyoD-mediated transdifferentiation and delineated an array of genes, proteins, and signaling pathways that are unique to each cell fate conversion. We demonstrate that reprogramming to iMPCs occurs via a gradual, stepwise reprogramming process, whereas transdifferentiation into myotubes is typically fast and direct. In addition, we report that the two cell conversions share phenotypical characteristics, including rapid up-regulation of several skeletal muscle differentiation genes, albeit they are also distinct, as only F/R/C administration manifests a robust up-regulation of satellite cell genes and key skeletal muscle differentiation markers in conjunction with opening of the chromatin in promoters of key myogenic genes. We then further compared the transcriptome and proteome of FACS-purified Pax7-nGFP+ iMPCs to that of Pax7-nGFP+ primary myoblasts and documented their congruent and divergent molecular traits. Last, we identified Notch as a molecular pathway that is absolutely essential for the formation of iMPCs in addition to governing their self-renewal.
Several observations emanate from this study. One notable finding pertains to the molecular comparison of Pax7-nGFP+ iMPCs to that of Pax7-nGFP+ myoblasts. These two cell types share molecular attributes at the mRNA and protein levels; however, Pax7-nGFP+ iMPCs also uniquely express a cohort of genes that are indicative of satellite cell activation and proliferation in vivo, in addition to increased expression of cell cycle regulators and unique signaling pathways. We postulate several reasons that may account for these disparities. First, the Pax7-nGFP+ iMPCs are cultivated in the vicinity of neighboring committed progenitors, connective tissue–like cells, and a highly contractile myofiber network, in contrast to primary myoblasts that are typically cultured as dispersed mononucleated cells that are passaged before fusion into myotubes. As such, the heterogeneity of iMPCs may recapitulate, to an extent, the microenvironment activated satellite cells encounter during skeletal muscle regeneration in vivo. Moreover, recent works have reported a cross-talk between resident muscle cells, myofibers, and satellite cells during homeostasis and regeneration (44–47). Similarly, signaling molecules secreted from the multinucleated myofibers of iMPCs may affect the gene expression of Pax7-nGFP+ iMPCs, potentially rendering them more akin to activated satellite cells. An additional explanation for the discrepancies between Pax7+ iMPCs and Pax7+ myoblasts involves the different culture conditions used to cultivate these cell types. Whereas primary myoblasts were cultured on Matrigel-coated plates in medium containing basic fibroblast growth factor (bFGF) and high serum (48), the iMPCs were cultured directly on plastic dishes using cell medium containing serum replacement, serum, bFGF, and, most notably, F/R/C. These cell media supplements may affect molecular attributes and elicit repression or activation of multiple myogenic genes and signaling pathways. In support of this hypothesis, exposure of dissociated skeletal muscle tissue fragments to iMPC media containing F/R/C treatment facilitated the formation of heterogeneous cultures consisting of myogenic progenitor cells and a contractile myofiber network that resembled fibroblast-derived iMPCs (20).
Direct reprogramming via MyoD+F/R/C triggers extensive molecular transformations. One notable pathway that was uniquely up-regulated following F/R/C treatment is Notch, which was similarly reported to be unique to iMPC reprogramming in a recent study (27). This pathway has been extensively investigated in the context of satellite cell quiescence and activation and shown to be critical for their stemness and self-renewal (34, 49–51). In this study, we demonstrate that this pathway is redundant for myotube formation via MyoD overexpression; however, it is critical for the generation of Pax7+ iMPCs and their maintenance. Moreover, we documented a high expression of Notch pathway–associated genes such as Notch3, Heyl, and Hey1 in the Pax7+ subset of iMPCs, whereas Notch ligands such as Dll1, Jag2, and Dlk1 were expressed in downstream differentiated myoblasts and myocytes. This observation is reminiscent of the Notch receptor/ligand interaction in vivo, suggesting a potential role for the differentiated cell population of iMPCs in triggering elevated Notch-related gene expression in Pax7+ iMPCs. Building upon this finding enabled us to homogeneously cultivate Pax7+ iMPCs on the Notch ligand Dll1 as previously shown for satellite cells (52, 53). It will be of interest to further investigate whether Pax7+ iMPCs cultured on Dll1 in the presence of F/R/C maintain activated satellite cell gene expression in vitro and may further augment their engraftment potential in vivo.
How each small molecule exerts its effect during reprogramming or supports the proliferation of established iMPC clones is yet to be fully explored. Previous works have established the effect of Forskolin on enhancing myoblast proliferation and engraftment capacities in mice, and TGF-β inhibition was recently shown to promote myoblast fusion (54–56). When administered together, these molecules have also been reported to enhance satellite cell quiescence in a three-dimensional skeletal muscle tissue bioconstruct (57). It is of interest to further explore their individual effects during reprogramming and whether F/R/C administration may augment the propensity of satellite cells to repair skeletal muscles in vivo as recently shown for F/R supplementation (58).
To date, the conversion of somatic cells into iMPCs has mainly been reported for wild-type fibroblasts. As iMPCs recapitulate a unique myogenic differentiation program in vitro, it is of interest to attempt their establishment from fibroblasts of murine muscular dystrophy models. Namely, generation of iMPCs from a mouse model of Duchenne muscular dystrophy can provide a population of proliferative progenitors and contractile myofibers that lack dystrophin expression, thus affording an innovative approach to model disease pathology in vitro, a platform for drug screens, or a source for autologous cell therapy following genetic correction. Last, several recent studies reported on direct conversion of mouse somatic cells into iMPCs using a variety of transcription factors and/or small molecules (18–20). Reports on the conversion of human fibroblasts directly into myogenic stem and progenitor cells with satellite cell attributes and robust engraftment capacities are still lacking, and further work is highly warranted to establish such cell lines. We envision that, with success, human iMPCs could serve as a complementary toolbox in the skeletal muscle field for basic research and disease modeling and as a potential source for cell-based therapies.
METHODS
Animals
Mice carrying a Pax7-nGFP reporter (Tg:Pax7-nGFP/C57BL6;DBA2) (24) or Pax7-CreERT2; Rosa26-LSL-ntdTomato (26) system were used in this study. All mice used in this study were housed with three to four littermates and maintained under specific pathogen–free–like conditions. Mice were fed standard food and water and handled in accordance with the Swiss Federal Law on Animal Protection. All animal procedures were approved by the Zurich Cantonal Animal Welfare Committee (license number ZH108/18).
Plasmid construction
The plasmids used in this study were generated by VectorBuilder. LV-EF1α-rtTA3/PGK-Neomycin and LV-tetO-MyoD/PGK-Puromycin denote the respective plasmids: pLV[Exp]-Neo-EF1A>Tet3G (VectorBuilder, VB170530-1031pbc) and pLV[Tet]-Puro-TRE3G>mMyod1[NM_010866. 2] (VectorBuilder, VB181022-1110vfj).
Virus production and storage
About 60 to 70% confluent human embryonic kidney (HEK) 293T cells were prepared in a 15-cm culture plate. For plasmid transfection, Δ8.9 (16.5 μg), vesicular stomatitis virus glycoprotein envelope (11 μg), and 22 μg of the target plasmid (either pLV[Exp]-Neo-EF1A>Tet3G or pLV[Tet]-Puro-TRE3G>mMyod1) were mixed in 150 mM NaCl at a final volume of 1 ml, followed by 10 min of incubation with 1 ml of polyethylenimine (PEI; 2 mg/ml) (Polysciences, POL23966-1). As the next step, HEK-293T cells were incubated with a DNA/PEI mixture solution. After 1 day, the DNA/PEI mixture was replaced with fresh medium. At days 2 and 3, medium containing virus was collected and filtered through a 0.45-μm syringe filter (Corning, 431220). For virus precipitation, filtered medium was incubated with 5× PEG-it solution (System Biosciences, LV825A-1) overnight and centrifuged at 1500g for 30 min at 4°C. The virus pellet was then resuspended in phosphate-buffered saline (PBS; Thermo Fisher Scientific, 10010015) containing 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (Thermo Fisher Scientific, 15630056) in 1/10 to 1/100 of the original volume and stored at −80°C until use.
Generation of Thy1+ Rep-MEFs
MEFs were isolated from Pax7-nGFP mice and cultured in “MEF medium” containing high-glucose Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher Scientific, 41966029) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, 10270106), 1% GlutaMAX (Thermo Fisher Scientific, 35050061), 1% nonessential amino acids (Thermo Fisher Scientific, 11140050), 1% penicillin-streptomycin (Pen-Strep; Thermo Fisher Scientific, 15140122), and 0.1% 2-mercaptoethanol (Thermo Fisher Scientific, 21985023). To generate dox-inducible Rep-MEFs, cells were passaged and, once confluent, transduced with LV-EF1α-rtTA3/PGK-Neomycin plus LV-tetO-MyoD/PGK-Puromycin at 1:1 ratio and supplemented with polybrene (6 μg/ml; Sigma-Aldrich, TR-1003-G). Transduced MEFs were expanded and selected by sequential antibiotic treatment with MEF medium containing either G418 solution (1 mg/ml; Sigma-Aldrich, 4727878001) or puromycin (1 μg/ml; Thermo Fisher Scientific, A1113803) for a total of 4 days. To establish a homogeneous population of fibroblasts from Rep-MEF cultures, we FACS-purified these cultures with an antibody recognizing the fibroblast-specific surface marker CD90.2 (Thy 1.2) (Thermo Fisher Scientific, 48-0902-80). Cells were FACS-purified or analyzed using an SH800S FACS-Sorter (Sony Biotechnology Inc.).
Reprogramming of Thy1+ Rep-MEFs
Approximately 2.5 × 105 to 3.0 × 105 Thy1+ MEFs were seeded onto six-well plates and treated by the following conditions 1 day after seeding. For the MyoD condition, Thy1+ Rep-MEFs were cultured with dox (2 μg/ml; Sigma-Aldrich, D9891) in “iMPC medium” containing KnockOut DMEM (Thermo Fisher Scientific, 10829018) supplemented with 10% KnockOut Serum Replacement (Thermo Fisher Scientific, 10828028), 10% FBS, 1% GlutaMAX, 1% nonessential amino acids, 1% Pen-Strep, 0.1% 2-mercaptoethanol, and bFGF (10 ng/ml; R&D Systems, 233-FB). For the MyoD+F/R/C condition, Thy1+ Rep-MEFs were cultured in iMPC medium containing dox (2 μg/ml) and administered with three small molecules: 5 μM Forskolin (F) (R&D Systems, 1099/50), 5 μM RepSox (R) (R&D Systems, 3742/50), and 3 μM CHIR99021 (C) (R&D Systems, 4423/50).
Establishment and maintenance of stable iMPC clones
To establish iMPC clones, Thy1+ Rep-MEFs were cultured under the MyoD+F/R/C condition for 10 days, followed by another 2 to 3 days of culture in iMPC medium that contained only Forskolin, RepSox, and CHIR99210 (without dox). For maintenance of stable clones, P0 iMPC clones were trypsinized and further expanded in iMPC medium containing the three small molecules.
Satellite cell isolation
Whole-body skeletal muscles were harvested from Pax7-nGFP mice and minced thoroughly with surgical scissors. PBS was added to minced muscles, followed by centrifugation at 350g for 2 min. Cell pellets were incubated with 0.2% collagenase type 2 (Thermo Fisher Scientific, 17101015) in high-glucose DMEM for 90 min in a 37°C shaking water bath. After collagenase digestion, cell pellets were washed once with “wash buffer” consisting of Ham’s F-10 Nutrient mix (Thermo Fisher Scientific, 22390025) supplemented with 10% horse serum (Thermo Fisher Scientific, 16050122). This was followed by Dispase digestion with F10 containing 0.4% Dispase II (Thermo Fisher Scientific, 17105041) and 0.2% collagenase type 2 for 30 min in a 37°C shaking water bath. After 30 min of incubation, the remaining cells were filtered and washed several times. The final cell pellet was resuspended in PBS containing 2% FBS, and satellite cells carrying Pax7-nGFP reporter were purified using an SH800S FACS-Sorter.
Myoblast culture
Myoblasts were seeded onto Matrigel-coated plates and cultured in “myoblast medium.” To prepare Matrigel-coated plates, 4% of Matrigel (Corning, 356237) diluted in low-glucose DMEM (Thermo Fisher Scientific, 31885023) was applied to the plate that was placed on ice. After 7 min of incubation on ice, Matrigel was removed from the plate, which was further incubated for 1 hour at 37°C. For myoblast medium, high-glucose DMEM and F-10 were mixed at a 1:1 ratio and supplemented with 20% FBS, 10% horse serum, 1% Pen-Strep, and bFGF (10 ng/ml) as previously described (59). Only P2-P4 myoblasts were used for the reported analyses.
EdU assay
5-ethynyl-2′-deoxyuridine (EdU) assay was performed using the Click-iT EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Thermo Fisher Scientific, C10424) according to the manufacturer’s guideline. Shortly, cells were incubated in medium containing 10 μM EdU for 1.5 hours and washed with 1% BSA in PBS. Cells were harvested and fixed for 15 min in 100 μl of Click-iT fixative. Cells were then permeabilized in 100 μl of Click-iT saponin-based permeabilization solution and incubated for 30 min in 500 μl of Click-iT reaction cocktail containing fluorescent dye. Cells were then washed twice with permeabilization solution, and EdU+ cells were analyzed by FACS-Sorter.
Quantitative real-time PCR
Total RNA was extracted using an RNeasy mini kit (Qiagen, 74104). The RNA was subjected to deoxyribonuclease treatment (Qiagen, 79254), and its concentration was measured with a Tecan plate reader. Complementary DNAs (cDNAs) were synthesized using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, 4368814) according to the manufacturer’s protocol. qRT-PCR was performed in a 10-μl reaction containing 10 ng of cDNA, 5 μl of Applied Biosystems PowerUp SYBR Green Master Mix (Thermo Fisher Scientific, A25741), and 0.4 μl of each 10 μM forward and reverse primer for the target genes. Pgk was used as a housekeeping gene. The sequence of the primers for each target gene is described in table S1. For Pax7, Myf5, and Myog presented in Fig. 1C and fig. S1B, probe-based qPCR was conducted according to the manufacturer’s protocol using PrimeTime qPCR assays purchased from Integrated DNA Technologies (Coralville, USA); mGapdh (Mm.PT.39a.1), mPax7 (Mm.PT.58.12398641), mMyf5 (Mm.PT.58.5271235), and mMyog (Mm.PT.58.30712483.gs).
Immunofluorescence staining and imaging
Cells were fixed using 4% paraformaldehyde (Alfa Aesar, 43368) for 10 min and incubated in blocking PBS solution containing 2% BSA (AppliChem, 9048-46-8) and 0.2% Triton X-100 (Sigma-Aldrich, 9002-93-1) for 30 min to permeabilize cell membrane and block unspecific antigen binding. As the next step, the cells were incubated with primary antibodies in blocking solution for 2 hours, followed by 30 min of incubation with secondary antibodies and 4′,6-diamidino-2-phenylindole (DAPI) for nuclear staining (1:1000; Thermo Fisher Scientific, 62248). Stained cells were imaged using a Nikon microscope (ECLIPSE Ti2). The following primary antibodies have been used in this study: anti-human/mouse/rat/chicken Pax7 (5 μg/ml; R&D Systems, MAB1675), anti-human/mouse Myod1 (5.8A) (1:100; Thermo Fisher Scientific, MA512902), anti-mouse MyHC (1:1000; R&D Systems, MAB4470), and anti–Ki-67 (SP6) (1:250; Thermo Fisher Scientific, MA514520). The following secondary antibodies were used in this study at 1:500 dilution: anti-mouse immunoglobulin G1 (IgG1; goat, Alexa Fluor 647) (Thermo Fisher Scientific, A21240), anti-mouse IgG2B (goat, Alexa Fluor 546) (Thermo Fisher Scientific, A21240), and anti-rabbit IgG (H + L) (donkey, Alexa Fluor 546) (Thermo Fisher Scientific, A10040).
Immunofluorescence image quantification
The quantification of immunofluorescence images was performed by counting positive and negative cells. The percentage of the positive cells for each protein was calculated by normalizing the number of positive cells by that of DAPI-positive cells.
Analysis of Pdgfrb and Thy1 expression in iMPCs
Cells were stained for 15 min in 300 μl of FACS buffer supplemented with 2 μl of allophycocyanin (APC)–conjugated CD140b [platelet-derived growth factor receptor B (PDGFRB)] antibody (Thermo Fisher Scientific, 17-1402-82) or 1.5 μl of eFluor450-conjugated CD90.2 (Thy 1.2) antibody (Thermo Fisher Scientific, 48-0902-80). Cells were then washed with PBS and used for FACS analysis. DAPI was only applied for PDGFRB-stained cells.
Inhibition of Notch via DAPT treatment during reprogramming
For assessment of DAPT treatment during MEF reprogramming, 2.5 × 105 to 3.0 × 105 Rep-MEFs were exposed to either the MyoD or MyoD+F/R/C condition. For the MyoD+F/R/C + DAPT condition, DAPT (Sigma-Aldrich, D5942) was additionally added to the MyoD+F/R/C condition at a final concentration of 15 μM. All analyses associated with MEF reprogramming in the presence of DAPT were performed at day 10 of treatment. For iMPC cultures, identical numbers of iMPCs were seeded onto a 24-well plate and cultured in iMPC medium either without DAPT (−DAPT) or with DAPT (+DAPT) at the same concentration for five consecutive days.
Culture of Pax7-nGFP+ cells on Dll1-coated plates
For preparation of Dll1-coated plates, Dll1 protein (1 μg/ml; R&D Systems, 5026-DL-050) dissolved in 0.1% BSA/PBS was applied to the cell culture plates, followed by incubation overnight at 4°C. The next day, Dll1 solution was aspirated, and Pax-nGFP+ cells were directly sorted from iMPCs onto Dll1-coated plates at a density of 5000 cells/cm2 and cultured for 6 days before analysis.
Western blot assay
Cells were washed twice with cold PBS and lysed on ice using 80 μl of radioimmunoprecipitation assay buffer containing 1× Halt protease inhibitor cocktail (Thermo Fisher Scientific, 87785). Cell lysate was collected and centrifuged at 10,000g for 12 min at 4°C. Supernatant (75 μl) containing total proteins was collected in a new tube and diluted into 1:5 for protein concentration measurement using a DC protein assay kit (Bio-Rad, 5000111). According to the measurement result, samples were normalized in the same concentration and mixed with 4× Laemmli buffer (Bio-Rad, 1610747) supplemented by 2-mercaptoethanol, followed by boiling for 5 min at 95°C. The boiled samples and a protein ladder (Thermo Fisher Scientific, 26620) were loaded into precasted gels (Bio-Rad, 4568094) and electrophoresed at 100 V for the first 10 min and at 130 V for 50 to 60 min. Then, the stain-free gel was imaged for loading control and used for protein blotting onto polyvinylidene difluoride (PVDF) membrane using Trans-Blot Turbo Mini PVDF Transfer Packs (Bio-Rad, 1704156). For NOTCH3 and NICD3 protein detection, protein-transferred membrane was first incubated for 1 hour in 5% dry milk (Carl Roth, T145.1) blocking solution based on TBS-T buffer and then incubated in TBS-T (Tris-buffered saline, 0.1% Tween 20) containing 5% BSA and primary antibody against NOTCH3 (1:1000; Abcam, ab23426) overnight at 4°C. The next day, membrane was washed three times with TBS-T for 10 min and incubated in TBS-T containing 5% dry milk and horseradish peroxidase–linked secondary antibody against rabbit IgG (1:5000; Cell Signaling, 7074S) for 1 hour at room temperature, followed by washing three times with TBS-T for 10 min. For color development, Western Lightning Plus-ECL reagents (PerkinElmer, NEL103E001EA) were applied to the membrane for 1 min. Chemiluminescent images were taken for 10 to 400 s of exposure and combined later with a colorimetric image of membrane to define the size of the band.
Bulk RNA-seq
Total RNA was extracted using an RNeasy mini kit according to the manufacturer’s protocol. The RNA quality was determined by measuring the 28S/18S ratio with a Fragment Analyzer (Agilent, Santa Clara, CA, USA). Samples with a 28S/18S ratio of more than 1.9 were used for library preparation. Libraries were constructed according to the TruSeq Stranded mRNA protocol (Illumina Inc., California, USA). Briefly, 100 to 1000 ng of total RNA was polyadenylate-enriched and reverse-transcribed into double-stranded cDNA. Then, the cDNA was fragmented, end-repaired, and adenylated before ligation of TruSeq adapters containing unique dual indices for multiplexing. Fragments containing TruSeq adapters on both ends were selectively enriched with PCR. The quality and quantity of the enriched libraries were validated using a Fragment Analyzer, which resulted in an average fragment size of approximately 360 base pairs (bp). The libraries were normalized to 10 nM in tris-Cl buffer (10 mM, pH 8.5) with 0.1% Tween 20 and sequenced with 100-bp single-end reads on NovaSeq 6000 (Illumina Inc., California, USA) according to standard protocol. Around 20 million reads were obtained for each sample.
Bulk RNA-seq data analysis
Raw sequencing reads were preprocessed using the SUSHI framework, which was developed by the Functional Genomics Center Zurich (FGCZ) (60). The reads were trimmed (adapter sequences, low-quality end, and low-quality reads with phred score < 20) first using Trimmomatic v0.36 (61). Pseudoalignment of the trimmed reads was performed against the reference mouse genome assembly GRCm38.p6, and gene expression level (GENCODE release 23) was quantified using Kallisto v0.44 (62). Differential gene expression analysis was conducted between different time points as well as different conditions based on negative binomial distribution using the R package edgeR v3.28 (63). Genes showing variable expression with adjusted (Benjamini-Hochberg method) P < 0.05 and twofold change were considered to be differentially expressed. Gene expression time series data were subjected to soft clustering (64) using the R package Mfuzz v2.50 (65) to reveal the underlying hidden expression patterns under either the MyoD or MyoD+F/R/C condition. For each fuzzy cluster, genes were annotated by gene ontology (GO) terms using the STRING v11 database. Bulk RNA-seq data from Yagi et al. (27) were downloaded from Gene Expression Omnibus (GSE169489) and processed in the same way as mentioned before using the SUSHI framework (60).
Protein extraction and digestion
Cultured cells were collected using a cell scraper, snap-frozen in dry ice for 30 min, and stored at −80°C. For protein extraction and digestion, cell pellets were lysed by using a commercial iST Kit (PreOmics, Germany) according to the updated version of the manufacturer’s protocol. Briefly, 100 to 200 μg of the cell pellets was solubilized in “lyse” buffer, boiled at 95°C for 10 min, and processed with high-intensity focused ultrasound for 30 s under the ultrasonic amplitude to 85%. Then, cell lysates were transferred to the cartridge and digested by adding 50 μl of the “digest” solution. After an hour of incubation at 37°C, 100 μl of “stop” solution was added to halt protein digestion. The cartridge was centrifuged at 3800g, and the throughput was discarded. The peptides remaining in the iST-filter of the cartridge were washed, eluted, dried, and resolubilized in 20 μl of “LC-Load” buffer for MS analysis.
LC-MS analysis
MS analysis was performed on an Orbitrap Fusion Lumos (Thermo Fisher Scientific) equipped with a Digital PicoView source (New Objective) and coupled to an M-Class UPLC (Waters). Channel A was filled with 0.1% formic acid in water, while channel B was filled with 0.1% formic acid and 99.9% acetonitrile. For each sample, 1 to 2 μl of peptides was loaded on a commercial MZ Symmetry C18 Trap Column (100 Å, 5 μm, 180 μm × 20 mm, Waters) followed by nanoEase MZ C18 HSS T3 Column (100 Å, 1.8 μm, 75 μm × 250 mm, Waters). After 3 min of initial hold at 5% B, a gradient from 5 to 22% B in 83 min and 22 to 32% B in an additional 10 min was applied. The column was cleaned after the run by increasing to 95% B and holding 95% B for 10 min before reestablishing loading condition. Samples were acquired in a randomized order. The mass spectrometer was operated in data-dependent acquisition (DDA) mode acquiring a full-scan MS spectrum [300 to 1500 mass/charge ratio (m/z)] at a resolution of 120,000 at 200 m/z after accumulation to a target value of 500,000. Data-dependent tandem MS (MS/MS) was recorded in the linear ion trap using quadrupole isolation with a window of 0.8 Da and higher-energy C-trap dissociation (HCD) fragmentation with 35% fragmentation energy. The ion trap was operated in rapid scan mode with a target value of 10,000 and a maximum injection time of 50 ms. Only precursors with intensity above 5000 were selected for MS/MS, and the maximum cycle time was set to 3 s. In charge state screening, singly assigned and unassigned charge states and charge states higher than seven were rejected. Precursor masses previously selected for MS/MS measurement were excluded from further selection for 20 s, and the exclusion window was set at 10 parts per million. The samples were acquired using internal lock mass calibration on m/z 371.1012 and 445.1200. The data obtained from the MS proteomics were handled using the local laboratory information management system (66).
Protein identification and label-free protein quantification
The acquired raw MS data were processed by MaxQuant (version 1.6.2.3), followed by protein identification using the integrated Andromeda search engine (67). Spectra were searched against a Swissprot Mus musculus reference proteome (taxonomy 10090, version from 9 July 2019), concatenated to its reversed decoyed fasta database and common protein contaminants. Carbamidomethylation of cysteine was treated as a fixed modification, while methionine oxidation and N-terminal protein acetylation were treated as a variable. Trypsin/P was set for enzyme specificity, allowing a minimal peptide length of seven amino acids and a maximum of two missed cleavages. MaxQuant Orbitrap default search settings were used. Peptides with false discovery rate (FDR) < 0.01 and proteins with FDR < 0.05 were processed for further steps. Label-free quantification (LFQ) was carried out by applying a 2-min window for match between runs. LFQ intensity results from MaxQuant were used for hierarchical clustering across all samples implemented in Perseus software (68).
Two-group comparison analysis in proteomics
Each sample file was kept separate in the experimental design to acquire individual quantitative values in the MaxQuant experimental design template. Fold changes of proteins were calculated on the basis of intensity values reported in the proteinGroups.txt file. Filtration for proteins with two or more peptides allowing a maximum of four missing values, normalization with a modified robust z-score transformation, and the t test with pooled variance to compute P values were implemented using the functions in R package SRMService (69). If all measurements of a protein are missing in one of the conditions, a pseudo fold change was calculated, replacing the missing group average by the mean of 10% smallest protein intensities in that condition. Proteins showing variable expression with adjusted P < 0.05 and twofold change were considered to be differentially expressed between the conditions.
Bulk RNA-seq/proteomic correlation analysis
An integrated dataset was created with DEGs overlapping with DEPs based on their common Ensembl identifiers. Pearson correlation coefficient was computed between DEGs and proteins using their log2 fold change values.
Enrichment analysis for functional annotation
For bulk RNA-seq data, GSEA was performed based on genes ranked in order of log2FC and the functional WikiPathways (www.wikipathways.org) database via WEB-based GEne SeT AnaLysis Toolkit (WebGestalt) (70). Only significant (FDR < 0.05) pathway categories with 10 to 500 genes were considered for enrichment analyses. For proteomics and integrated transcriptomic-proteomic datasets, enrichment analysis was performed based on significant DEGs/DEPs (|log2FC| > 1, adjusted P < 0.05) in Metacore (https://clarivate.com/products/metacore/; Clarivate Analytics, London, UK). Only significant (FDR < 0.05) prebuilt process networks were presented. Up-regulated and down-regulated process networks are shown using −log10(FDR) and log10(FDR), respectively. For bulk RNA-seq data comparison between Pax7-nGFP+ iMPCs and Pax7-nGFP+ myoblasts, ORA was performed using the R package goseq v1.42.
ATAC-seq
Libraries for ATAC-seq were constructed using the Omni-ATAC protocol as previously reported (71). Briefly, 50,000 to 60,000 cells were lysed on ice by cold lysis buffer containing 0.1% NP-40 (Sigma-Aldrich, 98379), 0.1% Tween 20 (Sigma-Aldrich, P3416), 0.01% Digitonin (Promega, G9441), 10 mM tris-HCl (pH 7.5), 10 mM NaCl, and 3 mM MgCl2 in nuclease-free water. Nuclei were collected by centrifugation and subjected to transposition reaction in 1× Tagment DNA buffer containing 5% Tn5 transposase (Illumina, 20034197), 0.1% Tween 20, and 0.01% Digitonin at 37°C for 30 min on a thermomixer, followed by DNA purification using a MinElute PCR purification kit (Qiagen, 28004). To identify samples, index PCR amplification was performed by mixing 10 μl of transposed DNA with 10 μl of nuclease-free water, 25 μl of NEBNext High-Fidelity 2X PCR Master Mix, and 2.5 μl of each 25 μM Ad.1 and Ad.2, as previously published (72). Total PCR cycle was determined according to the amplification graph after the first 12 cycles. To remove primer dimers and larger fragments, double-sided bead purification was carried out using AMPure XP (Beckman Coulter, A63881). Library quality was validated using TapeStation (Agilent Technologies) and sequenced on HiSeq-2500 (Illumina Inc., California, USA) with paired-end read of 70 bp. All libraries had more than 50 million reads.
ATAC-seq data analysis
ATAC-seq was performed on a HiSeq-2500 instrument at FGCZ. After initial quality control (adapter and low-quality base trimming) using fastp v0.20 (73), raw sequencing reads were mapped against the reference mouse genome assembly GRCm38.p6 using Bowtie2 v2.4.1 (74). PCR duplicates were removed using the MarkDuplicates tool from Picard (https://broadinstitute.github.io/picard/). Peak calling was performed using MACS2 (75) with -nomodel -f BAMPE -gsize mm -keep-dup all -extsize 200 options. Called peaks were annotated using the R package ChIPseeker. For each sample, fragment count matrix was generated using the R package chromVAR based on the promoter region, defined as −1 to 1 kb around the transcription start site. The R package edgeR v3.28 (63) was used to perform differential accessible region analysis between different conditions using the fragment count matrices. Genes showing variable expression with P < 0.01 and onefold change were considered to have an open chromatin in the defined promoter region.
Bulk RNA/ATAC-seq correlation analysis
An integrated dataset was created with DEGs overlapping with the genes with differential accessible regions in the promoter region based on their common Ensembl identifiers. Pearson correlation coefficient was calculated for the overlapping genes based on their log2 fold change values.
Single-cell RNA-seq
An early-passage iMPC clone and Rep-MEFs were trypsinized and filtered using a 40-μm cell strainer (VWR, 734-0002) to filter out debris or fragments of myofibers. Filtered cells were washed with PBS, and the number of cells was counted manually using a hemocytometer with trypan blue (Sigma-Aldrich, T8154) staining. Next, the cell pellet was resuspended in PBS at a concentration of 1000 cells/μl and immediately used for 10x single-cell library construction. The 10x library was built using a single-cell 3′ reagent kit v3 (10x Genomics, Pleasanton, CA) according to the manufacturer’s protocol. Briefly, cells were loaded in chromium chip B targeting ~10,000 recovered cells. Generated Gel Beads-in-emulsion (GEMs) were cleaned, and cDNA was amplified by PCR, followed by cDNA fragmentation, end repair, A-tailing, adapter ligation, index PCR, and double-sized selection. The library was sequenced on a full SP flowcell of NovaSeq 6000 (Illumina Inc., California, USA), which allows obtaining 560 million reads for around 10,000 cells of iMPCs and 215 million for more than 6000 cells of MEFs.
scRNA-seq data analysis
CellRanger v4.0.0 pipeline was used for demultiplexing the samples, mapping raw reads to the mouse reference genome (build GRCm38.p6), and generating feature-barcode count matrices (76). The count matrices were further analyzed using Seurat v3.2.3 pipeline (28, 77). Droplets with low feature counts (<250) were discarded from both samples for quality control. Furthermore, iMPC droplets with feature counts >7200 and mitochondrial gene counts >5% and MEF droplets with feature counts >6000, UMI counts >40,000, and mitochondrial gene counts >6% were filtered out. The filtered data were globally scaled via log normalization. The scaled data were dimensionally reduced via PCA using 2000 highly variable genes. Cells were clustered based on the first 20 principal components (PCs) using the Louvain algorithm with a resolution of 0.5. Uniform manifold approximation and projection (UMAP) method was applied using the same PCs to visualize the clustered cells in low-dimensional place. Cluster markers were identified using the Wilcoxon rank sum test (adjusted P < 0.05). The cell-cell communication in the iMPC sample was analyzed using the R package CellChat v1.1.3 (43). Nonreprogrammed fibroblast clusters were not considered for the cell-cell communication network analysis.
Bulk RNA-seq/scRNA-seq correlation analysis
To generate pseudo bulk data from scRNA-seq at the sample level, raw counts were aggregated across clusters of similar cell types (fibroblasts and progenitors). Pearson correlation coefficient was computed between the overlapping genes expressed in bulk and pseudo bulk RNA-seq data based on their log2-normalized expression values.
scRNA-seq trajectory analysis
Unsupervised single-cell trajectory analysis was performed using Monocle3 (78). The cells were ordered in pseudotime along the learned trajectory with cluster 1 being the root. Because of the presence of a strong feature outside the focus of interest that might influence unsupervised analysis, we also performed semisupervised single-cell trajectory analysis using Monocle2 (29). Six genes were defined as markers, namely, Ccnb1 for cycling progenitors, Myog for committed progenitors, Myog and Tnnt2 for myocytes, Pdgfrb and Fbln2 for connective tissue/fibroblast cells, and Pax7 for Pax7+ progenitors. A set of differential genes was selected, which covaries with these markers. Cells were ordered using the top 1000 DEGs based on their adjusted P. Each cell was assigned a pseudotime value to capture its progress during the biological process. Branch expression analysis modeling was performed to find the branch-dependent genes that could identify the mechanism underlying the cell fate decisions. Branch-dependent genes categorized by each cluster were annotated with GO term using the online tool DAVID v.6.8.
Statistical analysis
We used three cell lines per group that were isolated from different mice for bulk RNA-seq and other experiments including iMPC generation and Notch inhibition, whereas four cell lines for LC-MS were used. We used the software Prism to perform statistical analysis for gene expression results such as qRT-PCR data as indicated in the figure legends. Statistical analysis for multiomics datasets was carried out as described in each respective section.
Acknowledgments
We wish to thank K. De Bock and K. Hochedlinger as well as members of the Regenerative and Movement Biology laboratory for constructive comments and feedback. We are grateful to S. Tajbakhsh for providing the Pax7-nGFP mouse strain. We acknowledge the use of the Functional Genomics Center Zurich (FGCZ) facility and infrastructure and are grateful to L. Opitz and L. Kunz for assistance with data analysis of RNA-seq and LC-MS.
Funding: This study was supported by start-up funds from ETH Zurich and an Eccellenza Grant (grant no. PCEGP3_187009) from the Swiss National Science Foundation to O.B.-N. Other support for the Bar-Nur laboratory include grants from The Good Food Institute Foundation (#16936), The Novartis Foundation for Medical-Biological Research (#20B147), The Helmut Horten Foundation (#18598), and the National Centre of Competence in Research (NCCR) Robotics of the Swiss National Science Foundation (30-2467-1). Work in the von Meyenn group is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 803491).
Author contributions: Experiment design: I.K., A.G., and O.B.-N. Conceptualization: I.K., A.G., and O.B.-N. Investigation: I.K., A.G., and E.P. Methodology: I.K., N.B., and L.H. Supervision: O.B.-N. and F.v.M. Writing—original draft: O.B.-N. and I.K. Writing—review and editing: I.K., A.G., N.B., L.H., E.P., F.v.M., and O.B.N.
Competing interests: O.B.-N. is an inventor on a pending patent related to this work filed by the General Hospital Corporation (WO2017177050A1 filed on 6 April 2017, published 30 May 2019). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Plasmids generated in this study can be obtained from the lead contact or VectorBuilder (ID VB170530-1031pbc and ID VB181022-1110vfj). Raw datasets for the bulk RNA-seq, ATAC-seq, and scRNA-seq generated in this study are available in Gene Expression Omnibus (GEO) repository (GSE169053, GSE186271, and GSE169054, respectively). The MS proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (79) partner repository with the dataset identifier PXD029379.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Table S1
REFERENCES AND NOTES
- 1.Wosczyna M. N., Rando T. A., A muscle stem cell support group: Coordinated cellular responses in muscle regeneration. Dev. Cell 46, 135–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Comai G., Tajbakhsh S., Molecular and cellular regulation of skeletal myogenesis. Curr. Top. Dev. Biol. 110, 1–73 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Yin H., Price F., Rudnicki M. A., Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M. A., Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Dumont N. A., Bentzinger C. F., Sincennes M. C., Rudnicki M. A., Satellite cells and skeletal muscle regeneration. Compr. Physiol. 5, 1027–1059 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Montarras D., Morgan J., Collins C., Relaix F., Zaffran S., Cumano A., Partridge T., Buckingham M., Direct isolation of satellite cells for skeletal muscle regeneration. Science 309, 2064–2067 (2005). [DOI] [PubMed] [Google Scholar]
- 7.S. A. Domenig, A. S. Palmer, O. Bar-Nur, in Organ Tissue Engineering (Springer International Publishing, 2020), chap. 19–1, pp. 1–62. [Google Scholar]
- 8.Judson R. N., Rossi F. M. V., Towards stem cell therapies for skeletal muscle repair. NPJ Regen. Med. 5, 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J., Du Y., Deng H., Direct lineage reprogramming: Strategies, mechanisms, and applications. Cell Stem Cell 16, 119–134 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Davis R. L., Weintraub H., Lassar A. B., Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 11.Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D., Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M., Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L., Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Sekiya S., Suzuki A., Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Lalit P. A., Salick M. R., Nelson D. O., Squirrell J. M., Shafer C. M., Patel N. G., Saeed I., Schmuck E. G., Markandeya Y. S., Wong R., Lea M. R., Eliceiri K. W., Hacker T. A., Crone W. C., Kyba M., Garry D. J., Stewart R., Thomson J. A., Downs K. M., Lyons G. E., Kamp T. J., Lineage reprogramming of fibroblasts into proliferative induced cardiac progenitor cells by defined factors. Cell Stem Cell 18, 354–367 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lujan E., Chanda S., Ahlenius H., Sudhof T. C., Wernig M., Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 109, 2527–2532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ring K. L., Tong L. M., Balestra M. E., Javier R., Andrews-Zwilling Y., Li G., Walker D., Zhang W. R., Kreitzer A. C., Huang Y., Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 11, 100–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito N., Kii I., Shimizu N., Tanaka H., Takeda S., Direct reprogramming of fibroblasts into skeletal muscle progenitor cells by transcription factors enriched in undifferentiated subpopulation of satellite cells. Sci. Rep. 7, 8097 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato T., Higashioka K., Sakurai H., Yamamoto T., Goshima N., Ueno M., Sotozono C., Core transcription factors promote induction of PAX3-positive skeletal muscle stem cells. Stem Cell Reports 13, 352–365 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bar-Nur O., Gerli M. F. M., di Stefano B., Almada A. E., Galvin A., Coffey A., Huebner A. J., Feige P., Verheul C., Cheung P., Payzin-Dogru D., Paisant S., Anselmo A., Sadreyev R. I., Ott H. C., Tajbakhsh S., Rudnicki M. A., Wagers A. J., Hochedlinger K., Direct reprogramming of mouse fibroblasts into functional skeletal muscle progenitors. Stem Cell Reports 10, 1505–1521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cacchiarelli D., Qiu X., Srivatsan S., Manfredi A., Ziller M., Overbey E., Grimaldi A., Grimsby J., Pokharel P., Livak K. J., Li S., Meissner A., Mikkelsen T. S., Rinn J. L., Trapnell C., Aligning single-cell developmental and reprogramming trajectories identifies molecular determinants of myogenic reprogramming outcome. Cell Syst. 7, 258–268.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Dall’Agnese A., Caputo L., Nicoletti C., di Iulio J., Schmitt A., Gatto S., Diao Y., Ye Z., Forcato M., Perera R., Bicciato S., Telenti A., Ren B., Puri P. L., Transcription factor-directed re-wiring of chromatin architecture for somatic cell nuclear reprogramming toward trans-differentiation. Mol. Cell 76, 453–472.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G. J., Parker M. H., MacQuarrie K. L., Davison J., Morgan M. T., Ruzzo W. L., Gentleman R. C., Tapscott S. J., Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev. Cell 18, 662–674 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambasivan R., Gayraud-Morel B., Dumas G., Cimper C., Paisant S., Kelly R. G., Tajbakhsh S., Distinct regulatory cascades govern extraocular and pharyngeal arch muscle progenitor cell fates. Dev. Cell 16, 810–821 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Polo J. M., Anderssen E., Walsh R. M., Schwarz B. A., Nefzger C. M., Lim S. M., Borkent M., Apostolou E., Alaei S., Cloutier J., Bar-Nur O., Cheloufi S., Stadtfeld M., Figueroa M. E., Robinton D., Natesan S., Melnick A., Zhu J., Ramaswamy S., Hochedlinger K., A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151, 1617–1632 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy M. M., Lawson J. A., Mathew S. J., Hutcheson D. A., Kardon G., Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yagi M., Ji F., Charlton J., Cristea S., Messemer K., Horwitz N., Stefano B. D., Tsopoulidis N., Hoetker M. S., Huebner A. J., Bar-Nur O., Almada A. E., Yamamoto M., Patelunas A., Goldhamer D. J., Wagers A. J., Michor F., Meissner A., Sadreyev R. I., Hochedlinger K., Dissecting dual roles of MyoD during lineage conversion to mature myocytes and myogenic stem cells. Genes Dev. 35, –1209, 1228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H. A., Trapnell C., Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L., Kubota M., Nakamura A., Kaji T., Seno S., Uezumi A., Andersen D. C., Jensen C. H., Fukada S. I., Dlk1 regulates quiescence in calcitonin receptor-mutant muscle stem cells. Stem Cells 39, 306–317 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Almada A. E., Horwitz N., Price F. D., Gonzalez A. E., Ko M., Bolukbasi O. V., Messemer K. A., Chen S., Sinha M., Rubin L. L., Wagers A. J., FOS licenses early events in stem cell activation driving skeletal muscle regeneration. Cell Rep. 34, 108656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yue L., Wan R., Luan S., Zeng W., Cheung T. H., Dek modulates global intron retention during muscle stem cells quiescence exit. Dev. Cell 53, 661–676.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Leung C., Murad K. B. A., Tan A. L. T., Yada S., Sagiraju S., Bode P. K., Barker N., Lgr5 marks adult progenitor cells contributing to skeletal muscle regeneration and sarcoma formation. Cell Rep. 33, 108535 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Baghdadi M. B., Castel D., Machado L., Fukada S. I., Birk D. E., Relaix F., Tajbakhsh S., Mourikis P., Reciprocal signalling by Notch-Collagen V-CALCR retains muscle stem cells in their niche. Nature 557, 714–718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Noguchi Y.-T., Nakayama H., Kaji T., Tsujikawa K., Ikemoto-Uezumi M., Uezumi A., Okada Y., Doi T., Watanabe S., Braun T., Fujio Y., Fukada S.-I., The CalcR-PKA-Yap1 axis is critical for maintaining quiescence in muscle stem cells. Cell Rep. 29, 2154–2163.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Kawabe Y., Wang Y. X., McKinnell I. W., Bedford M. T., Rudnicki M. A., Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell 11, 333–345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knopp P., Figeac N., Fortier M., Moyle L., Zammit P. S., Pitx genes are redeployed in adult myogenesis where they can act to promote myogenic differentiation in muscle satellite cells. Dev. Biol. 377, 293–304 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Schmidt K., Glaser G., Wernig A., Wegner M., Rosorius O., Sox8 is a specific marker for muscle satellite cells and inhibits myogenesis. J. Biol. Chem. 278, 29769–29775 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Bjornson C. R. R., Cheung T. H., Liu L., Tripathi P. V., Steeper K. M., Rando T. A., Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30, 232–242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mourikis P., Sambasivan R., Castel D., Rocheteau P., Bizzarro V., Tajbakhsh S., A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Bröhl D., Vasyutina E., Czajkowski M. T., Griger J., Rassek C., Rahn H.-P., Purfürst B., Wende H., Birchmeier C., Colonization of the satellite cell niche by skeletal muscle progenitor cells depends on Notch signals. Dev. Cell 23, 469–481 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Evin G., Sernee M. F., Masters C. L., Inhibition of γ-secretase as a therapeutic intervention for Alzheimer’s disease. CNS Drugs 20, 351–372 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Jin S., Guerrero-Juarez C. F., Zhang L., Chang I., Ramos R., Kuan C. H., Myung P., Plikus M. V., Nie Q., Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eliazer S., Muncie J. M., Christensen J., Sun X., D’Urso R. S., Weaver V. M., Brack A. S., Wnt4 from the niche controls the mechano-properties and quiescent state of muscle stem cells. Cell Stem Cell 25, 654–665.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sampath S. C., Sampath S. C., Ho A. T. V., Corbel S. Y., Millstone J. D., Lamb J., Walker J., Kinzel B., Schmedt C., Blau H. M., Induction of muscle stem cell quiescence by the secreted niche factor oncostatin M. Nat. Commun. 9, 1531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Latroche C., Weiss-Gayet M., Muller L., Gitiaux C., Leblanc P., Liot S., Ben-Larbi S., Abou-Khalil R., Verger N., Bardot P., Magnan M., Chrétien F., Mounier R., Germain S., Chazaud B., Coupling between myogenesis and angiogenesis during skeletal muscle regeneration is stimulated by restorative macrophages. Stem Cell Reports 9, 2018–2033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lukjanenko L., Karaz S., Stuelsatz P., Gurriaran-Rodriguez U., Michaud J., Dammone G., Sizzano F., Mashinchian O., Ancel S., Migliavacca E., Liot S., Jacot G., Metairon S., Raymond F., Descombes P., Palini A., Chazaud B., Rudnicki M. A., Bentzinger C. F., Feige J. N., Aging disrupts muscle stem cell function by impairing matricellular WISP1 secretion from fibro-adipogenic progenitors. Cell Stem Cell 24, 433–446.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rando T. A., Blau H. M., Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275–1287 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mourikis P., Tajbakhsh S., Distinct contextual roles for Notch signalling in skeletal muscle stem cells. BMC Dev. Biol. 14, 2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Low S., Barnes J. L., Zammit P. S., Beauchamp J. R., Delta-like 4 activates Notch 3 to regulate self-renewal in skeletal muscle stem cells. Stem Cells 36, 458–466 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Verma M., Asakura Y., Murakonda B. S. R., Pengo T., Latroche C., Chazaud B., Loon L. K. M., Asakura A., Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and Notch signaling. Cell Stem Cell 23, 530–543.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakai H., Fukuda S., Nakamura M., Uezumi A., Noguchi Y. T., Sato T., Morita M., Yamada H., Tsuchida K., Tajbakhsh S., Fukada S. I., Notch ligands regulate the muscle stem-like state ex vivo but are not sufficient for retaining regenerative capacity. PLOS ONE 12, e0177516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerli M. F. M., Moyle L. A., Benedetti S., Ferrari G., Ucuncu E., Ragazzi M., Constantinou C., Louca I., Sakai H., Ala P., de Coppi P., Tajbakhsh S., Cossu G., Tedesco F. S., Combined Notch and PDGF signaling enhances migration and expression of stem cell markers while inducing perivascular cell features in muscle satellite cells. Stem Cell Reports 12, 461–473 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu C., Tabebordbar M., Iovino S., Ciarlo C., Liu J., Castiglioni A., Price E., Liu M., Barton E. R., Kahn C. R., Wagers A. J., Zon L. I., A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 155, 909–921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girardi F., Taleb A., Ebrahimi M., Datye A., Gamage D. G., Peccate C., Giordani L., Millay D. P., Gilbert P. M., Cadot B., Le Grand F., TGFβ signaling curbs cell fusion and muscle regeneration. Nat. Commun. 12, 750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melendez J., Sieiro D., Salgado D., Morin V., Dejardin M.-J., Zhou C., Mullen A. C., Marcelle C., TGFβ signalling acts as a molecular brake of myoblast fusion. Nat. Commun. 12, 749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quarta M., Brett J. O., DiMarco R., de Morree A., Boutet S. C., Chacon R., Gibbons M. C., Garcia V. A., Su J., Shrager J. B., Heilshorn S., Rando T. A., An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol. 34, 752–759 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fang J., Sia J., Soto J., Wang P., Li L. A. K., Hsueh Y. Y., Sun R., Faull K. F., Tidball J. G., Li S., Skeletal muscle regeneration via the chemical induction and expansion of myogenic stem cells in situ or in vitro. Nat. Biomed. Eng. 5, 864–879 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bosnakovski D., Xu Z., Li W., Thet S., Cleaver O., Perlingeiro R. C. R., Kyba M., Prospective isolation of skeletal muscle stem cells with a Pax7 reporter. Stem Cells 26, 3194–3204 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hatakeyama M., Opitz L., Russo G., Qi W., Schlapbach R., Rehrauer H., SUSHI: An exquisite recipe for fully documented, reproducible and reusable NGS data analysis. BMC Bioinformatics 17, 228 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Futschik M. E., Carlisle B., Noise-robust soft clustering of gene expression time-course data. J. Bioinform. Comput. Biol. 3, 965–988 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Kumar L., Futschik M. E., Mfuzz: A software package for soft clustering of microarray data. Bioinformation 2, 5–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.C. Türker, F. Akal, D. Joho, C. Panse, S. Barkow-Oesterreicher, H. Rehrauer, R. Schlapbach, B-Fabric: The Swiss Army Knife for life sciences, in Proceedings of the 13th International Conference on Extending Database Technology, Lausanne, Switzerland (2010). [Google Scholar]
- 67.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M. Y., Geiger T., Mann M., Cox J., The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 69.J. G. W. Wolski, C. Panse, SRMService - r-package to report quantitative mass spectrometry data (2018); http://github.com/protViz/SRMService.
- 70.Zhang B., Kirov S., Snoddy J., WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nusbaum C., Myers R. M., Brown M., Li W., Liu X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng G. X. Y., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., McFarland C., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D. M., Hill A. J., Zhang F., Mundlos S., Christiansen L., Steemers F. J., Trapnell C., Shendure J., The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yılmaz Ş., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., Vizcaíno J. A., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Table S1