Abstract
Type I interferons (IFN-Is) are a group of potent inflammatory and antiviral cytokines. They induce IFN stimulated genes (ISGs), which act as proinflammatory mediators, antiviral effectors, and negative regulators of the IFN-I signaling cascade itself. One such regulator is interferon stimulated gene 15 (ISG15). Humans with complete ISG15 deficiency express persistently elevated levels of ISGs, and consequently, exhibit broad spectrum resistance to viral infection. Here, we demonstrate that IFN-I primed fibroblasts derived from ISG15-deficient individuals are more resistant to infection with single-cycle HIV-1 compared to healthy control fibroblasts. Complementation with both wild-type (WT) ISG15 and ISG15ΔGG (incapable of ISGylation while retaining negative regulation activity) was sufficient to reverse this phenotype, restoring susceptibility to infection to levels comparable to WT cells. Furthermore, CRISPR-edited ISG15ko primary CD4+ T cells were less susceptible to HIV-1 infection compared to cells treated with non-targeting controls. Transcriptome analysis of these CRISPR-edited ISG15ko primary CD4+ T cells recapitulated the ISG signatures of ISG15 deficient patients. Taken together, we document that the increased broad-spectrum viral resistance in ISG15-deficiency also extends to HIV-1 and is driven by a combination of T-cell-specific ISGs, with both known and unknown functions, predicted to target HIV-1 replication at multiple steps.
Author summary
Type I interferons (IFN-Is) are a group of potent inflammatory and antiviral agents. They induce IFN stimulated genes (ISGs), which perform downstream functions to resolve viral infection, mediate the inflammatory response, as well as negatively regulate the IFN-I signaling cascade to prevent hyperinflammation. One such negative regulator is interferon stimulated gene 15 (ISG15). Humans that lack ISG15 have chronic, low levels of antiviral ISGs, and ensuing broad-spectrum resistance to viral infection. We demonstrate that IFN-I priming of ISG15-deficient cells leads to superior resistance to human immunodeficiency virus 1 (HIV-1) infection compared to IFN-I primed healthy control cells. This is true for fibroblast cell lines, as well as primary CD4+ T cells, the main target of HIV-1. Analysis of the gene expression profiles show that ISG15-knockout CD4+ T cells express similar inflammatory markers as ISG15-deficient patients. Overall, we show that the broad-spectrum viral resistance in ISG15-deficiency extends to HIV-1.
Introduction
Type I interferons (IFN-Is) are the first line of defense against viral infections. IFN-I signaling induces the Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway, leading to transcription of about 400 IFN-I-stimulated genes (ISGs) [1]. These ISGs act as antiviral effectors, but some also function as negative regulators of IFN-I signaling in order to prevent overt inflammation. One such negative regulator of IFN-I signaling is interferon stimulated gene 15 (ISG15). ISG15 exists as a free intracellular molecule, free extracellular cytokine, and as a ubiquitin-like conjugate through a process called ISGylation [2–5]. Free intracellular ISG15 aids the stabilization of the ubiquitin-specific peptidase 18 (USP18), which downregulates IFN-I signaling by binding to the IFN-I receptor subunit, IFNAR2, and out-competing JAK1 [6]. Thus, a loss of ISG15 leads to the destabilization of USP18, leading to continued IFN-I signaling, and ensuing IFN-I mediated autoinflammation [7]. Free extracellular ISG15 serves to induce IFN-γ in natural killer (NK) and T cells, in synergy with IL-12. Individuals with complete ISG15 deficiency are thus also more susceptible to Mycobacterial infections [8]. Finally, although ISGylation has been reported to alter the activities and functions of many proteins, the exact role for ISGylation is still debated [9–24].
There are strong differences among species in the role for ISG15 in viral immunity, as unlike human ISG15 which serves to stabilize USP18, the murine orthologue does not perform such activity [25]. Thereby, mice lacking ISG15 were shown to be modestly more susceptible to some but not all viruses [11,26–33], while humans with ISG15 deficiency have overactive antiviral responses and no documented susceptibility to viral infections [6–8,25,34–36]. In turn, these patients suffer from clinical sequalae of chronic inflammation marked by increased levels of ISGs [6–8,34]. To recapitulate the persistent IFN-I signaling observed in ISG15-deficient patients in vivo, we have optimized an in vitro protocol which permits testing of viral susceptibility in a physiologically-relevant cell culture model [6]. Replication of RNA and DNA viruses such as VSV, HSV-1, Influenza A virus, Sendai virus, Nipah virus, and Rift Valley Fever virus was attenuated in ISG15-deficient, IFN-I-primed cells as compared to healthy control IFN-I-primed cells [6]. These data suggest that ISG15-deficiency may actually provide for increased resistance to severe viral infections.
IFN-I signaling and the role of ISG15 in the context of HIV-1 infection is poorly understood. As expected, IFN-I stimulation has been shown to inhibit HIV-1 replication, due to the induction of several ISGs that function as general antivirals [37–52]. Among these, ISGylation of Gag was demonstrated to inhibit HIV-1 budding [11,53–55]. Yet, more recently, a proviral role for USP18 in primary human macrophages was described [56,57]. Given these conflicting data regarding the HIV-1-specific antiviral or proviral effects of ISG15, we sought to investigate the role of ISG15 in regulating susceptibility to HIV-1 infection. Specifically, we investigated these functions under the physiological low-level expression of ISGs observed in ISG15-deficiency.
Results
ISG15 deficient, IFN-primed cells are less susceptible to HIV-1 infection
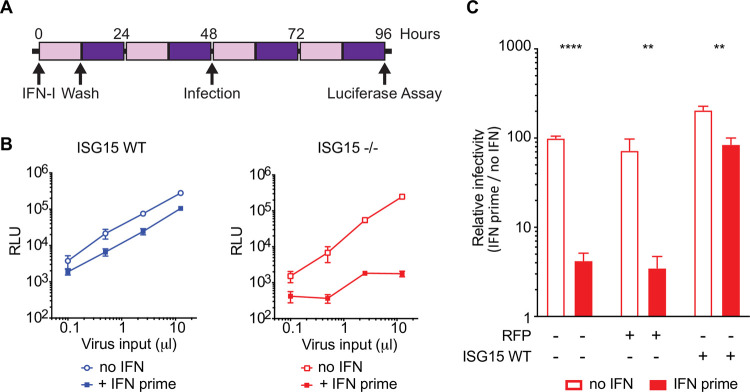
To determine HIV-1 susceptibility of ISG15-deficient, IFN-I-primed cells, we used hTERT-immortalized fibroblasts derived from ISG15-deficient patients to generate a tractable cell culture model system [7]. We used healthy control or ISG15-deficient patient fibroblasts transduced with either RFP-luciferase (negative control) or wildtype (WT) ISG15 [7]. Cells were prime-rested with IFN-I by treating them with 1000 IU/mL IFNα2b for 12 hours, upon which cells were washed and allowed to rest for 36 hours (Fig 1A). At this timepoint, cells with WT ISG15 are no longer signaling an IFN-I response, whereas cells lacking ISG15 remain in an elevated inflammatory state, which recapitulates chronic IFN-I signaling that persists in patients with ISG15 deficiency [6]. After this prime-rest, cells were infected with increasing doses of a single-cycle HIV-1 NL4-3 Δenv expressing Firefly Luciferase in the position of Nef and pseudotyped with a VSVg envelope (HIV-1e- Luc/VSVg). At day 2 post infection (96h in Fig 1A), the level of infection was measured by quantifying luciferase expression. Single-cycle HIV-1 infection was reduced by more than 100-fold in IFN-I-primed ISG15-deficient cells compared to unprimed ISG15-deficient cells, while only a modest 2.8-fold difference was observed in WT IFN-primed cells compared to WT unprimed cells (Fig 1B). This result suggests that, indeed, the absence of ISG15 limits HIV-1 infection.
Fig 1. ISG15 deficient, IFN-primed cells are less susceptible to HIV-1 infection.
A: hTERT-immortalized fibroblasts were primed with type I IFN (IFNα2b, 1000 units/mL) for 12 hours, washed and rested for 36 hours followed by infection with HIV-1-VSV-Luc. Cells were lysed 48 hours after infection, upon which HIV-1-luciferase expression was measured. B: Serially diluted HIV-1 (0.1, 0.5, 2.5 and 12.5 μL) was used to infect WT and ISG15-/- cells with and without IFN priming. Infections were done using cell lines from 2 unrelated WT controls and 2 unrelated ISG15-/- patients. Representative experiments are shown. RLU: relative light units. C: The parental ISG15-/- cell lines were complemented with RFP or ISG15 and infected as described above. Complementation with ISG15 abolishes the restriction. All infections were conducted in triplicates. Error bars represent SD. Significance was determined using t-tests (Prism 7 software; **** denotes P<0.0001; ** denotes P<0.01).
To confirm that the effect observed was specifically dependent on ISG15 deficiency, we complemented the ISG15-deficient or healthy control cells with WT ISG15 or RFP as a negative control and infected these cells following the same prime-rest procedure outlined in Fig 1A. Consistent with Fig 1B (right), ISG15-deficient, IFN-I-primed cells are less susceptible when compared to ISG15-deficient unprimed cells (Fig 1C). Complementation of ISG15-deficient cell lines with WT ISG15 yields IFN-I-primed cells that are more susceptible to HIV-1 infection compared to RFP-complemented ISG15-deficient cells. Taken together, these data suggest that ISG15-deficient cells restrict HIV-1 in an ISG15-dependent manner.
Complementation with WT or ΔGG ISG15 is sufficient to restore HIV-1 susceptibility
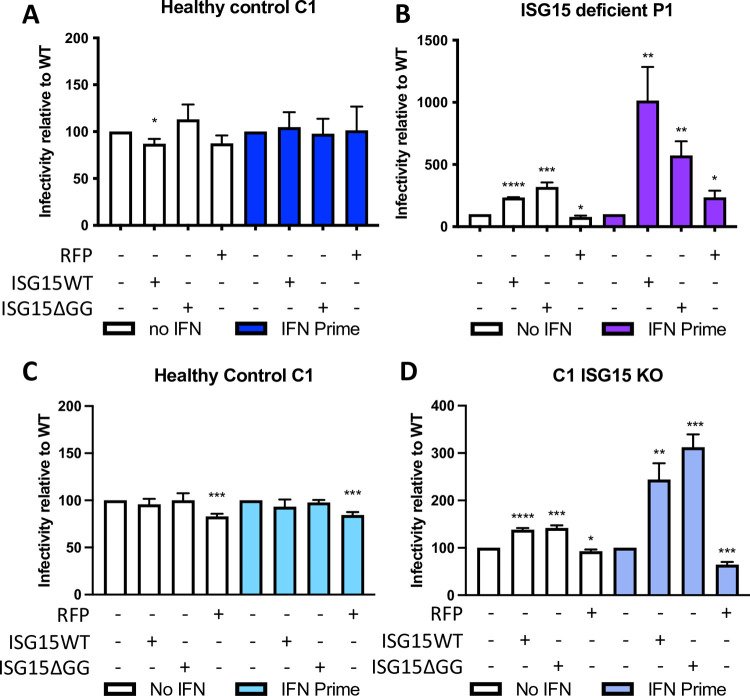
To test whether ISGylation or the negative regulatory function of free ISG15 is necessary for the observed proviral phenotype, we used healthy control and ISG15-deficient patient-derived fibroblasts, transduced with either empty vector containing RFP-luciferase (negative control), WT ISG15, or ISG15ΔGG [7]. The ISG15ΔGG mutation was previously shown to maintain the negative regulatory capacity due to its retained ability to bind and stabilize USP18, while failing to modify targets by ISGylation [7]. Fibroblasts were prime-rested as previously described in Fig 1A and infected with single cycle HIV-1e- GFP/VSVg (in this case, luciferase was replaced by GFP to allow for enumeration of infected cells by flow cytometry) as described above. GFP-positive cells were quantified by flow cytometry analysis, and infectivity was calculated as percent of GFP-positive cells relative to C1 healthy control cells (Fig 2A and 2B). As anticipated, ISG15 complementation of control cells, which endogenously express ISG15, did not change susceptibility to infection with or without IFN-I-priming (Fig 2A). Complementation with WT ISG15 in the ISG15-deficient patient cells significantly increased the percent of GFP-positive cells in IFN-I-primed cells (Fig 2B). Similarly, complementation with ISG15ΔGG in the ISG15-deficient patient cells also significantly increased infection in IFN-primed cells (Fig 2A and 2B), while complementation with RFP did not change the number of HIV-1 infected cells. To confirm these results in an isogenic system, we also compared an ISG15 CRISPR knock out (KO) cell line (C1 ISG15KO) generated on a WT background to their parental line (healthy control C1), by infecting each with single-cycle HIV-1e- GFP/VSVg and quantifying infectivity by flow cytometry analysis on day 3 post infection (Fig 2C and 2D). Again, complementation with WT ISG15 or ISG15ΔGG in C1 ISG15KO cells, but not control C1 cells, increased infection in IFN-I-primed cells. Together these findings indicate that the proviral role of ISG15 in HIV-1 infection is an ISGylation independent process.
Fig 2. ISG15 complementation restores HIV-1 susceptibility.
A, B,: hTERT-immortalized fibroblasts from healthy controls (A) and ISG15-deficient patients (B) were complemented with WT ISG15, ISGylation defective ISG15, or Luciferase RFP as a negative control. Cells stimulated with IFN-I overnight and rested for 36 hours were infected with HIV-1-VSV-GFP reporter virus. Flow cytometry was performed to quantify GFP-positive HIV-1-infected cells. Infectivity normalized to the parental cell-line is shown for mock primed (no IFN) and IFN primed cells. Error bars denote SD. (N = 4) **** denotes P<0.0001; *** denotes P<0.001; ** denotes P<0.01; * denotes P < 0.05 as determined by student’s two-tailed t-test. C, D: hTERTimmortalized fibroblasts from healthy controls (C) or ISG15 knockout healthy controls (D) were complemented with WT ISG15, ISGylation defective ISG15 (ISG15ΔGG), or Luciferase RFP as a negative control. Cells stimulated with IFN-I overnight and rested for 36 hours were infected with HIV-1-VSV-GFP reporter virus. Flow cytometry was performed to quantify GFP-positive HIV-1-infected cells. Infectivity normalized to the parental cell-line is shown for mock primed (no IFN-I) and IFN-I primed cells. Error bars denote SD. (N = 3) **** denotes P<0.0001; *** denotes P<0.001; ** denotes P<0.01; * denotes P<0.05 as determined by a student’s two-tailed T test.
ISG15-deficient primary CD4+ T cells were less susceptible to HIV-1 infection
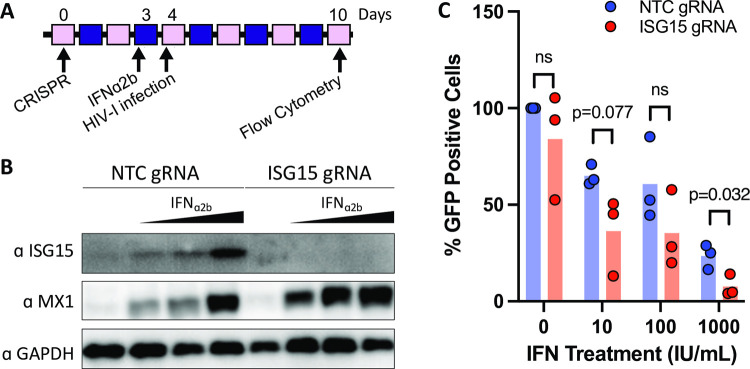
To more closely approximate natural HIV-1 infection, we next investigated the role for ISG15 with and without an IFN-I prime in primary human CD4+ T cells, the physiological target of HIV-1 infection. We generated ISG15 knockout CD4+ T cells using CRISPR-Cas9 nucleofection (outlined in Fig 3A). CD4+ T cells were edited with either non-target control guide 1 (NTCg1) or ISG15 guide 1 (ISG15g1) CRISPR Cas9 ribonucleoproteins (RNPs). Efficient ISG15 knockout in primary CD4+ T cells (with and without 24 hours of IFN-I treatment) was confirmed by Western blot (Fig 3B). Four days after CRISPR genome editing (and 24 hours after IFN-I treatment), we observed a low level of basal ISG15 expression in NTCg1-targeted cells, which increased upon IFN-I treatment (Fig 3B, left). In contrast, ISG15g1-targeted cells did not express detectable levels of ISG15 protein with or without IFN-I treatment (Fig 3B, right).
Fig 3. ISG15-deficient primary CD4+ T cells are less susceptible to HIV-1 infection.
A: Primary CD4+ T cells were targeted with either non-targeting control (NTC gRNA) or ISG15 (ISG15 gRNA) CRISPR RNPs. On day 3, cells were treated with 0, 10, 100, or 1000 IU/mL IFNα2b for 24 hours. On day 4 (24 hours after IFN treatment) cells were infected with replication-competent HIV-1-GFP. Flow cytometry was performed on Day 10 (6 days post HIV-1 infection) to quantify GFP-positive HIV-1-infected cells. B: ISG15 knockout efficiency in primary human CD4+ T cells was assessed by Western blotting. Representative Western blot of CRISPR-targeted CD4+ T cells lysed 24 hours post IFNα2b treatment. C: Infection of IFNα2b-primed, CRISPR-targeted CD4+ T cells using HIV-1-GFP was performed in duplicate. Duplicates were averaged and percent infectivity was calculated relative to NTC-targeted non-IFN-I treated. Three donors are shown. GFP-positive cells were quantified 6 days post infection. Multiple, unpaired T tests were performed and P value is denoted in the figure. NS denotes T test result was non-significant.
In order to determine the role of ISG15 in HIV-1 infection in primary CD4+ T cells, we infected the ISG15KO CD4+ T cells with HIV-1. Briefly, CRISPR-targeted cells that were stimulated with anti-CD3/CD28 were rested for 3 days, after which they were stimulated with 0, 10, 100, or 1000 IU/mL of IFNɑ2b (Fig 3A). Samples of cells were lysed for Western blot analysis to validate ISG15 knockout (Fig 3B). After 24 hours of IFN-I stimulation (day 4, Fig 3A), cells were infected with replication-competent HIV-1 NL4-3 Nef-IRES-GFP (HIV-1-GFP) for 8 hours. Infection was measured by quantification of GFP-positive cells by flow cytometry at day 6 post infection (day 10 after CRISPR, Fig 3A). Individual donor GFP-positive percentage results are shown in S1 Fig. To combine results of three donors, duplicates were averaged and percent infectivity was calculated relative to NTC-targeted cells without IFN-I treatment. Our results demonstrate that ISG15 KO cells are less susceptible to HIV infection when pretreated with 1000 IU/mL of IFNɑ2b (Fig 3C). This is true for the 10 and 100 IU/mL doses as well, but the donor variation limits the statistical significance.
Interestingly, MX1, an antiviral host protein, expression is increased in IFN-I stimulated, ISG15 gRNA targeted cells compared to NTC gRNA targeted controls (Fig 3B), suggesting ISG15-deficient cells upon IFN-I stimulation may be in a heightened antiviral state compared to control IFN-I stimulated cells. Taken together, the loss of ISG15 expression restricted HIV-1 infection, not only in hTERT-immortalized fibroblasts, but also in primary human CD4+ T cells, the main target cell population of HIV-1.
Loss of ISG15 in primary CD4+ T cells lead to an increase in select interferon stimulated genes
To investigate the mechanisms of how ISG15 deficiency shapes the transcriptomic landscape to enhance HIV-1 immunity of T cells, we performed RNA sequencing on CRISPR-edited primary CD4+ T cells. Non-electroporated, NTCg1- and ISG15g1-targeted CD4+ T cells were incubated with IFNβ (1,000 IU/mL) for 24 hours and lysed prior to RNA extraction and RNA sequencing (RNAseq). Unstimulated non-electroporated, NTCg1- and ISG15g1-targeted CD4+ T cells were used as controls.
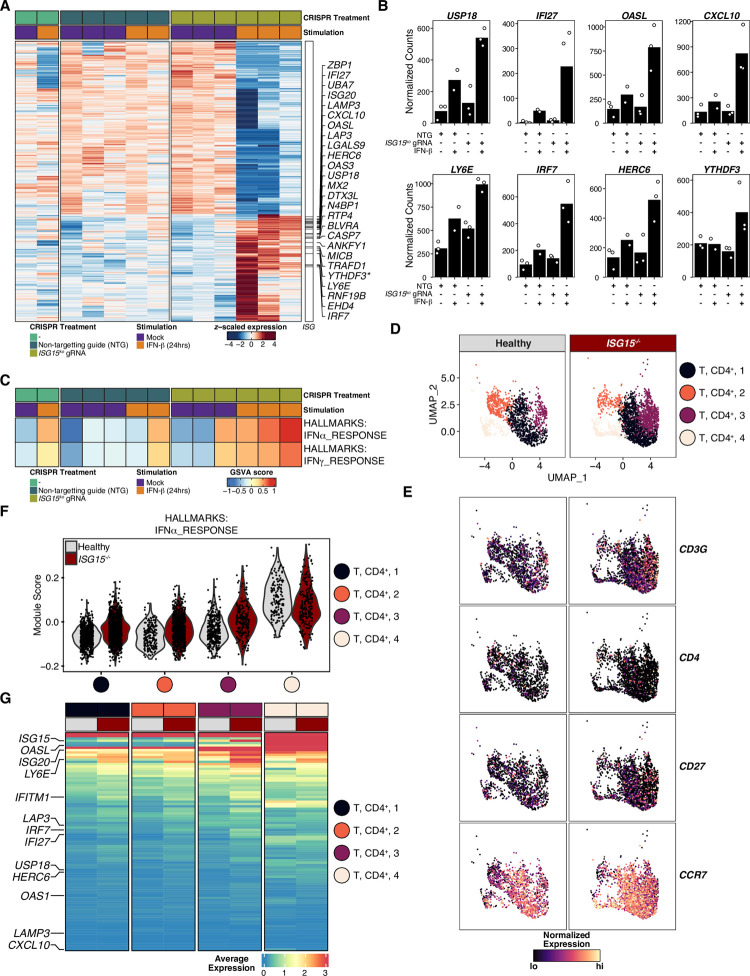
The transcriptome analysis revealed that the knock-down of ISG15 in primary CD4+ T cells resulted in upregulation of a specific ISG signature upon IFNβ stimulation (Fig 4A and 4B). Among these, IFI27, ISG20, HERC6, MX2, N4BP1, MICB, YTHDF3, and IRF7 have previously been described to negatively affect HIV-1 replication, [42–44,58–63]. In addition, our study also indicated a putative role for novel mediators of HIV-1 immunity, namely ZBP1, LAMP3, CXCL10, LGALS9, and ANKYF1, all of which have been associated with HIV-1/AIDS disease progression [64–71]. LY6E was shown to differentially regulate HIV-1 infection depending on cell surface CD4 expression levels [71,72]. Although not a canonical ISG, we document YTHDF3 expression as increased in ISG15-deficient CD4+ T cells stimulated with IFN-I (Fig 4A). We and others previously reported that YTHDF3 negatively affects HIV-1 replication by limiting HIV-1 reverse transcription [62,73]. Further, unsupervised gene set variation analysis showed expected expression signatures in IFNβ stimulated primary CD4+ T cells (Fig 4C). Together, these results suggest that there is likely a set of ISGs in ISG15-deficient CD4+ T cells that is sufficient to reduce the susceptibility of primary CD4+ T cells to HIV-1 infection.
Fig 4. RNA-seq analysis of CRISPR-Cas9 ISG15 knockout in primary CD4+ T cells reveals parallels to human ISG15 deficiency.
A: Differential gene expression heatmap between IFNβ stimulated (1000 IU/mL) CRISPR-Cas9 ISG15 knockout primary CD4+ T cells and non-targeting/non-edited controls shows elevated levels of interferon-stimulated genes (ISGs) in ISG15ko primary CD4+ T cells. Represented genes are the union of differentially expressed genes identified during IFNβ stimulation of CRISPR-Cas9 ISG15ko and IFNβ stimulation of the non-targeting controls, respectively. Differentially expressed genes (DEGs) were further filtered against a p-value of p<0.01. Flagged genes represent differentially expressed interferon-stimulated genes. YTHDF3 is not an ISG and is denoted by a star in the heatmap. B: Normalized count bar plots derived from RNA-seq data of select differentially expressed genes during IFN-b stimulation reveal elevated levels of ISGs in ISG15ko primary CD4+ T cells relative to non-targeted guide control cells. Bars represent mean normalized count values. C: Gene set variation analysis of differentially expressed genes identified during IFN-b stimulation of CRISPR-Ca9 ISG15ko primary CD4+ T cells confirms an IFN-I response signature in ISG15ko primary CD4+ T cells. D: UMAP representation of unsupervised clustering results of single-cell RNA of 3,780 ISG15-/- patient (n = 2357 cells) and age-matched healthy control (n = 1423 cells) peripheral blood mononuclear cells. Points are colored by CD4+ T cell subset supervised annotation. E: Gene expression UMAPs of classical CD4+ T cell marker gene RNA transcripts in ISG15-/- patient and age matched healthy control PBMCs. Point color intensity is scaled between the 1st percentile (lo) and the 99th percentile (hi) of natural log normalized gene expression count values. F: Gene set score distributions for IFNα signaling in CD4+ T cell subsets. The HALLMARKS:IFNa_RESPONSE gene set was extracted from MSigDB Hallmark collection and used as input in Seurat’s AddModuleScore function. G: Average gene expression heatmap of the HALLMARKS: IFNa_RESPONSE gene set in ISG15-/- patient and healthy control CD4+ T cell subsets. Heatmap color intensity represented log normalized average expression. Average expression value for each patient-cluster condition calculated from Seurat’s AverageExpression function.
To assess whether the elevated ISG expression signatures detected were comparable to patterns identified in ISG15-deficient patients, we re-analyzed a single cell RNA-sequencing dataset [34,74] of PBMCs isolated from an ISG15-deficient patient and a healthy control (Fig 4D). A closer investigation into the CD4+ T-cell compartment (defined based on the expression of canonical markers), our supervised analysis identifies four clusters present in both ISG15-deficient patient and healthy controls (Fig 4D and 4E). Gene set score analysis of the IFN-alpha response signature at single cell resolution reveals elevated levels of ISGs in ISG15-deficient patient CD4+ T cell subsets, with significant differences in clusters 1 and 3 (Fig 4F). Evaluation of average expression values of IFN-I related genes in the CD4+ T cell compartment suggests that our CRISPR edited ISG15KO primary CD4+ T cell model are transcriptionally similar to patient derived CD4+ T cells (Fig 4G). Indeed, the transcriptional program of CD4+ T cells from an ISG15-deficient patient noted here is likely to result in increased resistance to HIV-1 replication.
Discussion
The function of ISG15 in HIV-1 immunity remains an area of active investigation [6,8,36]. We observed here that ISG15-deficient patient fibroblasts were more resistant to HIV-1 infection than WT cells (Figs 1 and 2). This susceptibility was reversed when cells were complemented with WT or ΔGG ISG15 (Figs 1 and 2), suggesting that ISGylation was dispensable for this process. We found ISG15 deficiency limits HIV-1 infection not only in fibroblasts, but also in human primary CD4+ T cells (Fig 3).
Our results point to ISG15 as a predominantly positive regulator of HIV-1 infection (“a proviral ISG”) in human cells, unlike previous studies suggesting an antiviral role for ISGylation. It was previously reported that ISG15 expression is elevated in PBMCs of patients infected with HIV-1 compared to healthy controls, and that ISG15 expression levels correlated with higher plasma viral loads [75]. While this elevation is likely a result of the elevated systemic inflammation in HIV-1 positive individuals, these elevated levels of ISG15 may be advantageous for HIV-1 replication as it enhances the inhibition of IFN-I signaling.
Since IFN-I pre-treatment limited HIV-1 infection in WT primary human CD4+ T cells (Fig 3C and 3D), and to a greater extent in ISG15KO cells, upregulation of a combination of differentially expressed genes is likely responsible for the HIV-1 resistance demonstrated here. Of the upregulated genes, a large fraction are ISGs (Fig 4A, right). Among these ISGs, many of them have previously been reported to be antiviral against HIV-1 or antiviral in general and likely contribute to limiting HIV-1 replication in the absence of ISG15 (Fig 4A–4C).
The results of this study align with previous results demonstrating that ISG15 deficiency restricts different viral infections [6–8,25,34–36]. Interestingly, ISG15 has previously been shown to act as an antiviral HIV-1 restriction factor. First, ISGylation was shown to restrict HIV-1 by limiting accumulation of misfolded p53 [56]. This manuscript demonstrates that the protease activity of USP18 stabilizes misfolded mutated p53, which requires ISG15 for its degradation. Second, ISGylation of Gag by the E3 ligase HERC5 was shown to restrict HIV-1 particle production in U2OS cells [54]. Finally, overexpression of ISG15 was shown to restrict HIV release in 293T cells [53]. While all these well controlled studies indicate that ISG15 can be antiviral, these studies did not examine the role for ISG15 in the context of its negative regulatory capacity of IFN-I. In the naturally occurring ISG15 deficiency, the primary role for ISG15 is that of a negative regulator of IFN-I, which minimizes ISG15’s function as an antiviral molecule.
Furthermore, and in line with our work, two independent studies show that USP18 knockdown restricts HIV replication [52,57]. Given that ISG15 acts to stabilize USP18, which negatively regulates JAK-STAT signaling, this makes ISG15 deficiency a de facto partial USP18 deficiency [7], which results in restriction of HIV-1 replication.
Overall, our findings confirm the principal role for ISG15 as a negative regulator of IFN-I signaling and points to novel ISGs in primary human CD4+ T cells whose effector function in the context of HIV-1 replication remains to be explored.
Materials and methods
Ethics statement
Peripheral blood lymphocytes were purchased from New York Blood Center from anonymous donors. The investigators had no direct interactions with blood donors or influence on the selection of PBMCs. This work is regarded as non-human subject research.
Cell culture
hTERT immortalized fibroblasts were stably complemented with ISG15, ISG15ΔGG or luciferase by lentiviral transduction [6,7]. hTERT immortalized fibroblasts, and the TZM-bl reporter cell-line (cat#8129, NIH AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health [76–80]) were maintained in Dulbecco’s modified Eagle medium (DMEM, Corning) in the presence of 10% fetal bovine serum (FBS; GemCell), 100 IU penicillin, and 100 μg/mL streptomycin (D-10) at 37°C and 5% CO2.
Primary human CD4+ T cells were purified from peripheral blood lymphocytes obtained from anonymous healthy blood donors (New York Blood Center). Ficoll (Ficoll Hystopaque; Sigma) density centrifugation was performed as per the manufacturer’s instructions. CD4+ T cells were negatively selected using magnetic beads (CD4+ T-cell Isolation Kit I; Miltenyi Biotec) and were maintained in RPMI 1640 (Corning) supplemented with 10% FBS (Gibco), 100 IU penicillin, 100 μg/mL streptomycin, 1x Glutamax, 10 mM HEPES, and 20 U/mL recombinant human IL-2 (NIH AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health) (R-10-IL2) at 37°C and 5% CO2.
Genome editing
Primary human CD4+ T cell Cas9 Ribonucleoprotein (RNP)-mediated gene editing experiments were carried out as previously described [62,81,82]. Primary CD4+ T cells (2.5 × 106 cells/mL) were stimulated for 3 days at a volume of 500 μL per well with plate-bound anti-CD3 (Clone UCHT1; Tonbo) and suspended anti-CD28 (5 μg/mL; Clone CD28.2; Tonbo). Guide RNA (gRNA) was designed using the Benchling tool to target ISG15 (ISG15g1: 5’-CCGCCAGCATCTTCACCGTC-3’). The non-targeting control guide (NTCg1) used was previously published [82]. Generation of Cas9 RNPs and nucleofection of primary CD4+ T cells was performed exactly as previously described [62].
Western blots
Primary CD4+ T cells were lysed in RIPA buffer (Thermo Fisher Scientific) supplemented with 1x protease/phosphatase inhibitor cocktail (Cell Signaling Technology). Lysates were incubated with DTT and NuPAGE LDS sample buffer (Invitrogen). Lysates were run on 12% Criterion TGX gels (BioRad) and transferred to Amersham Hybond polyvinylidene difluoride (PVDF) blotting membranes (Cytiva). Western blots were blocked in 5% BSA in TBS 0.1% Tween and washed in TBS 0.2% Tween. Western blots were incubated with secondary antibody in 5% milk in TBS 0.1% Tween. Western blot chemiluminescence was detected with SuperSignalTM West Femto Substrate (Thermo Scientific). Imaging of the Western blot bands was performed using AlphaView software (ProteinSimple). The following antibodies were used for immunoblots: α-ISG15 (clone F9, 1:500, Santa Cruz Biotech, Cat# sc-166755), α-MX1 (1:1000, Abcam, ab95926), α-GAPDH (clone 6C5, 1:10,000, Sigma Aldrich, MAB374).
Plasmids
The HIV-1 NL4-3 Δenv Luc construct was kindly provided by N. R. Landau [83]. HIV-1 NL4-3 Δenv eGFP (reagent #11100, [84]) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health. The VSVg envelope was expressed from plasmid phCMV G [85]. The HIV-1 NL4-3 Nef-IRES-GFP (HIV-1-GFP) construct was kindly provided by B. K. Chen [86].
Production of viral stocks
Single-cycle viruses HIV-1 NL4-3 Δenv-eGFP and HIV-1 NL4-3 Δenv-Luc were generated and pseudotyped with VSVg by transfection of HEK293T cells with Polyethylenimine (PEI) (Polysciences). Three days after transfection, culture supernatants were harvested, clarified at 500 x G, filtered (0.45 μm), aliquoted, and stored at -80°C.
Replication competent (HIV-1-GFP) virus was generated by transfection of HEK293T cells with PEI (Polysciences). Two days after transfection, culture supernatants were harvested, filtered (0.45 μm), aliquoted, and stored at -80°C.
TZM-bl cells containing the β-galactosidase reporter gene driven by the HIV-1 long terminal repeat, were used to determine viral titers (TCID50/mL) of viral stocks produced as previously described [87].
IFN-I priming
Fibroblasts were prime-rested as described previously [6], by treating with IFNα2b (Merck Intron A 0085-4350-01) (1000 IU/mL) in D-10 media for 12 hours, upon which cells were washed three times with PBS, fresh D-10 media was added, and the cells were allowed to rest for 36 hours before infection.
CRISPR-targeted CD4+ T cells were stimulated with IFNɑ2b (10, 100, 1000 IU/mL) in R-10-IL2 media for 24 hours.
HIV-1 infection experiments
Prime-rested fibroblasts were infected with increasing doses of HIV-1-VSV-Luc. The level of infection was determined 48 hours post infection by quantifying Firefly Luciferase using the Luciferase Assay System kit (Promega).
Prime-rested fibroblasts were infected in triplicate with HIV-1-VSV-GFP overnight in the presence of polybrene (5 μg/mL). D-10 media was changed the next morning. The level of infection was determined on day 4 post infection by quantifying GFP-positive fibroblasts by flow cytometry (Luminex Guava easyCyte). Flow cytometry data was analyzed with Luminex Guava InCyte software.
IFN-primed CD4+ T cells were infected with HIV-1-GFP for 8 hours in the presence of polybrene (2 μg/mL). After 8 hours, viral supernatant was removed and R-10-IL2 culture media was replenished. Media was changed on days 3 and 5 post infection. The level of infection was determined on day 6 post infection by quantifying GFP-positive CD4+ T cells by flow cytometry (Luminex Guava easyCyte). Flow cytometry data was analyzed with Luminex Guava InCyte software.
RNAseq
Primary CD4+ T cells were nucleofected with NTCg1 or ISG15 CRISPR RNPs as described above for 3 days and then either stimulated or not with 1000 U/mL of IFNβ1b (PBL Assay Science, cat. 11420–1) for 24h. Cells were lysed in TRIzol Reagent (Invitrogen) and RNA was extracted per the manufacturer’s instructions. Total RNA was treated with RNase-Free DNase (QIAGEN 79254) and prepped for RNAseq with the TruSeq RNA Sample Prep Kit v2 (Illumina). 75 bp unpaired reads were generated on a NextSeq 550 (Illumina). Raw bcl image processing, fastq generation, reference genome alignment and feature counting were conducted using the RNA Express module in the BaseSpace suite (Illumina). Raw gene-sample counts matrices were extracted and read into the R statistical environment and further analyzed using the DESeq2 package [88]. Differentially gene expression testing was conducted contrasting CRISPR ISG15ko primary CD4+ T cells during IFN stimulation. Similar DGE testing was done contrasting the non-targeting guide controls. Resulting gene lists were filtered on a p-value of p<0.01 and expression values visualized using the ComplexHeatmap and ggplot2 packages.
Single-cell RNAseq (scRNAseq)
Previously published single-cell RNA-sequencing data of PBMCs from a patient with human ISG15 deficiency (N = 1) and a healthy control (N = 1) from Martin-Fernandez et al. (34) was accessed and analyzed. Raw gene-cell matrices were read into the R (v4.0.4) statistical environment and analyzed using Seurat (v4.0.1) for quality control, integration, clustering, and differential gene expression [89]. For quality control, data were filtered to exclude genes detected in less than three cells (per subject), to exclude cells with < 200 expressed genes (empty droplets) and to exclude cells with > 7.5% UMIs assigned to mitochondrial genes (dying cells). Filtered data were independently normalized using Seurat’s SCTransform function (developer’s default parameters) [90]. To account for subject-specific effects, both data sets were integrated using Seurat’s FindIntegrationAnchors and IntegrateData functions (developer’s default parameters). Dimensional reduction of the integrated data set was performed by principal component analysis (PCA) and first 25 principal components were used for unsupervised graph-based clustering (resolution: 1.2) and visualized by Uniform manifold approximation and projection (UMAP; parameters: n.dims: 25, n.neighbors: 30, metric: cosine). Supervised analysis was then conducted to subset CD4+ T cell clusters using canon immune markers (CD3, CD4, CD27, CCR7). IFNa gene signature scores (input genes were included from MSigDB HALLMARKS: IFNa RESPONSE) were then computed at single-cell resolution using Seurat’s AddModuleScore function.
Supporting information
On day 3, cells were treated with 0, 10, 100, or 1000 IU/mL IFNα2b for 24 hours. On day 4 (24 hours after IFN treatment) cells were infected with replication-competent HIV-1-GFP. Infection of IFNα2b-primed, CRISPR-targeted CD4+ T cells using HIV-1-GFP was performed in duplicate. Flow cytometry was performed on day 6 post infection to quantify GFP positive cells. The percent of GFP positive cells is shown for three donors.
(TIF)
Acknowledgments
We thank E. L. McGregor for FACS expertise. We are also grateful to the Simon, Bogunovic, and Manganaro labs for helpful discussions. We thank our colleagues for generous sharing of reagents directly with our group or through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This research was supported by the United States National Institute of Health (NIH, https://urldefense.proofpoint.com/v2/url?u=https-3A__www.nih.gov_&d=DwIGaQ&c=shNJtf5dKgNcPZ6Yh64b-A&r=Q4Bma9ov07gd-WRbWyoW48pXkah26wiYlToTi2zd09A&m=hVG4zdwGWMT3lhv9BtzQmw6dYKwah-wZ4Dkk9xiajS8&s=_HqwPuCrppLNDNdnohI6jElR3A5Zy19E5HU8vm5suoY&e=) grants and fellowships: AI134366-01 (DB and VS), AI120998 (VS), AI150355 (LCFM), R01AI148963 (DB), R01AI127372 (DB), R01AI151029 (DB), 5T32AI007647-20 (DJ), 5T32AI078892-13 (DJ), 5T32AI007647-22 (RSP), T32HD075735 (CG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5:375–86. doi: 10.1038/nri1604 [DOI] [PubMed] [Google Scholar]
- 2.Reich N, Evans B, Levy D, Fahey D, Knight E, Jr., Darnell JE, Jr. 1987. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc Natl Acad Sci U S A 84:6394–8. doi: 10.1073/pnas.84.18.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loeb KR, Haas AL. 1992. The IFN inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J Biol Chem 267:7806–7813. [PubMed] [Google Scholar]
- 4.Haas AL, Ahrens P, Bright PM, Ankel H. 1987. Interferon induces a 15-ilodalton protein exhibiting marked homology to ubiquitin. J Biol Chem 262:11315–23. [PubMed] [Google Scholar]
- 5.Knight E, Cordova B. 1991. Ifn-Induced 15-Kda Protein Is Released from Human-Lymphocytes and Monocytes. Journal of Immunology 146:2280–2284. [PubMed] [Google Scholar]
- 6.Speer SD, Li Z, Buta S, Payelle-Brogard B, Qian L, Vigant F et al. , 2016. ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun 7:11496. doi: 10.1038/ncomms11496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, et al. , 2015. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature 517:89–93. doi: 10.1038/nature13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogunovic D, Byun M, Durfee LA, Abhyankar A, Sanal O, Mansouri D, et al. , 2012. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science 337:1684–8. doi: 10.1126/science.1224026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 31:119–30. doi: 10.1089/jir.2010.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okumura F, Zou W, Zhang DE. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev 21:255–60. doi: 10.1101/gad.1521607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pincetic A, Kuang Z, Seo EJ, Leis J. 2010. The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol 84:4725–36. doi: 10.1128/JVI.02478-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng Q, Sekula D, Guo Y, Liu X, Black CC, Galimberti F, et al. , 2008. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol Cancer Ther 7:3780–8. doi: 10.1158/1535-7163.MCT-08-0753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou W, Wang J, Zhang D. 2007. Negative Regulation of ISG15 E3 ligase EFP through its autoISGylation. Biochem Biophys Res Commun 354:321–327. doi: 10.1016/j.bbrc.2006.12.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, et al. , 2009. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep 10:374–80. doi: 10.1038/embor.2009.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durfee LA, Lyon N, Seo K, Huibregtse JM. 2010. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol Cell 38:722–32. doi: 10.1016/j.molcel.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, et al. , 2010. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol 30:2424–36. doi: 10.1128/MCB.01466-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang Y, Zhong G, Zhu L, Liu X, Shan Y, Feng H, et al. , 2010. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J Immunol 184:5777–90. doi: 10.4049/jimmunol.0903588 [DOI] [PubMed] [Google Scholar]
- 18.Shah SJ, Blumen S, Pitha-Rowe I, Kitareewan S, Freemantle SJ, Feng Q, et al. , 2008. UBE1L represses PML/RAR{alpha} by targeting the PML domain for ISG15ylation. Mol Cancer Ther 7:905–14. doi: 10.1158/1535-7163.MCT-07-0515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou W, Papov V, Malakhova O, Kim KI, Dao C, Li J, et al. , 2005. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem Biophys Res Commun 336:61–8. doi: 10.1016/j.bbrc.2005.08.038 [DOI] [PubMed] [Google Scholar]
- 20.Takeuchi T, Iwahara S, Saeki Y, Sasajima H, Yokosawa H. 2005. Link between the ubiquitin conjugation system and the ISG15 conjugation system: ISG15 conjugation to the UbcH6 ubiquitin E2 enzyme. J Biochem 138:711–9. doi: 10.1093/jb/mvi172 [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi T, Yokosawa H. 2005. ISG15 modification of Ubc13 suppresses its ubiquitin-conjugating activity. Biochem Biophys Res Commun 336:9–13. doi: 10.1016/j.bbrc.2005.08.034 [DOI] [PubMed] [Google Scholar]
- 22.Im E, Yoo L, Hyun M, Shin WH, Chung KC. 2016. Covalent ISG15 conjugation positively regulates the ubiquitin E3 ligase activity of parkin. Open Biol 6. doi: 10.1098/rsob.160193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park Jung M, Yang Seung W, Yu Kyung R, Ka Seung H, Lee Seong W, Seol Jae H, et al. , 2014. Modification of PCNA by ISG15 Plays a Crucial Role in Termination of Error-Prone Translesion DNA Synthesis. Molecular Cell 54:626–638. doi: 10.1016/j.molcel.2014.03.031 [DOI] [PubMed] [Google Scholar]
- 24.Yoo L, Yoon AR, Yun CO, Chung KC. 2018. Covalent ISG15 conjugation to CHIP promotes its ubiquitin E3 ligase activity and inhibits lung cancer cell growth in response to type I interferon. Cell Death Dis 9:97. doi: 10.1038/s41419-017-0138-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu X, Taft J, Bogunovic D. 2020. Developing Broad-Spectrum Antivirals Using Porcine and Rhesus Macaque Models. J Infect Dis 221:890–894. doi: 10.1093/infdis/jiz549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morales DJ, Lenschow DJ. 2013. The antiviral activities of ISG15. J Mol Biol 425:4995–5008. doi: 10.1016/j.jmb.2013.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, et al. , 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104:1371–6. doi: 10.1073/pnas.0607038104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiang TY, Zhao C, Krug RM. 2009. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol 83:5971–7. doi: 10.1128/JVI.01667-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, Garcia-Sastre A, et al. , 2009. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol 83:1147–51. doi: 10.1128/JVI.00105-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannakopoulos NV, Arutyunova E, Lai C, Lenschow DJ, Haas AL, Virgin HW. 2009. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J Virol 83:1602–10. doi: 10.1128/JVI.01590-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Burke CW, Ryman KD, Klimstra WB. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol 81:11246–55. doi: 10.1128/JVI.01282-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai J, Pan W, Wang P. 2011. ISG15 facilitates cellular antiviral response to dengue and west nile virus infection in vitro. Virol J 8:468. doi: 10.1186/1743-422X-8-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog 4:e1000096. doi: 10.1371/journal.ppat.1000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin-Fernandez M, Bravo Garcia-Morato M, Gruber C, Murias Loza S, Malik MNH, Alsohime F, et al. , 2020. Systemic Type I IFN Inflammation in Human ISG15 Deficiency Leads to Necrotizing Skin Lesions. Cell Rep 31:107633. doi: 10.1016/j.celrep.2020.107633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taft J, Bogunovic D. 2018. The Goldilocks Zone of Type I IFNs: Lessons from Human Genetics. J Immunol 201:3479–3485. doi: 10.4049/jimmunol.1800764 [DOI] [PubMed] [Google Scholar]
- 36.Hermann M, Bogunovic D. 2017. ISG15: In Sickness and in Health. Trends Immunol 38:79–93. doi: 10.1016/j.it.2016.11.001 [DOI] [PubMed] [Google Scholar]
- 37.Gendelman HE, Baca LM, Turpin J, Kalter DC, Hansen B, Orenstein JM, et al. , 1990. Regulation of HIV replication in infected monocytes by IFN-alpha. Mechanisms for viral restriction. J Immunol 145:2669–76. [PubMed] [Google Scholar]
- 38.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A 88:3195–9. doi: 10.1073/pnas.88.8.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunzi MS, Pitha PM. 1996. Role of interferon-stimulated gene ISG-15 in the interferon-omega-mediated inhibition of human immunodeficiency virus replication. J Interferon Cytokine Res 16:919–27. doi: 10.1089/jir.1996.16.919 [DOI] [PubMed] [Google Scholar]
- 40.Chakrabarti LA, Simon V. 2010. Immune mechanisms of HIV control. Curr Opin Immunol 22:488–96. doi: 10.1016/j.coi.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El-Diwany R, Soliman M, Sugawara S, Breitwieser F, Skaist A, Coggiano C, et al. , 2018. CMPK2 and BCL-G are associated with type 1 interferon-induced HIV restriction in humans. Sci Adv 4:eaat0843. doi: 10.1126/sciadv.aat0843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Espert L, Degols G, Lin YL, Vincent T, Benkirane M, Mechti N. 2005. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol 86:2221–2229. doi: 10.1099/vir.0.81074-0 [DOI] [PubMed] [Google Scholar]
- 43.Goujon C, Moncorge O, Bauby H, Doyle T, Ward CC, Schaller T, et al. , 2013. Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502:559–62. doi: 10.1038/nature12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. , 2013. MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502:563–6. doi: 10.1038/nature12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nasr N, Alshehri AA, Wright TK, Shahid M, Heiner BM, Harman AN, et al. , 2017. Mechanism of Interferon-Stimulated Gene Induction in HIV-1-Infected Macrophages. J Virol 91. doi: 10.1128/JVI.00744-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasr N, Maddocks S, Turville SG, Harman AN, Woolger N, Helbig KJ, et al. , 2012. HIV-1 infection of human macrophages directly induces viperin which inhibits viral production. Blood 120:778–88. doi: 10.1182/blood-2012-01-407395 [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, C et al. , 2013. The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14:398–410. doi: 10.1016/j.chom.2013.08.015 [DOI] [PubMed] [Google Scholar]
- 48.Michaelis B, Levy JA. 1989. Hiv Replication Can Be Blocked by Recombinant Human Interferon Beta. Aids 3:27–31. [PubMed] [Google Scholar]
- 49.Smith MS, Thresher RJ, Pagano JS. 1991. Inhibition of human immunodeficiency virus type 1 morphogenesis in T cells by alpha interferon. Antimicrob Agents Chemother 35:62–7. doi: 10.1128/AAC.35.1.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. 1989. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med 169:1137–51. doi: 10.1084/jem.169.3.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bednarik DP, Mosca JD, Raj NB, Pitha PM. 1989. Inhibition of human immunodeficiency virus (HIV) replication by HIV-trans-activated alpha 2-interferon. Proc Natl Acad Sci U S A 86:4958–62. doi: 10.1073/pnas.86.13.4958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor JP, Cash MN, Santostefano KE, Nakanishi M, Terada N, Wallet MA. 2018. CRISPR/Cas9 knockout of USP18 enhances type I IFN responsiveness and restricts HIV-1 infection in macrophages. J Leukoc Biol doi: 10.1002/JLB.3MIA0917-352R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A 103:1440–5. doi: 10.1073/pnas.0510518103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woods MW, Kelly JN, Hattlmann CJ, Tong JG, Xu LS, Coleman MD, et al. , 2011. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology 8:95. doi: 10.1186/1742-4690-8-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuang Z, Seo EJ, Leis J. 2011. Mechanism of inhibition of retrovirus release from cells by interferon-induced gene ISG15. J Virol 85:7153–61. doi: 10.1128/JVI.02610-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osei Kuffour E, Konig R, Haussinger D, Schulz WA, Munk C. 2019. ISG15 Deficiency Enhances HIV-1 Infection by Accumulating Misfolded p53. mBio 10. doi: 10.1128/mBio.01342-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osei Kuffour E, Schott K, Jaguva Vasudevan AA, Holler J, Schulz WA, Lang PA, et al. , 2018. USP18 (UBP43) Abrogates p21-Mediated Inhibition of HIV-1. J Virol 92. doi: 10.1128/JVI.00592-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hubbard JJ, Greenwell-Wild T, Barrett L, Yang J, Lempicki RA, Wahl SM, et al. , 2012. Host gene expression changes correlating with anti-HIV-1 effects in human subjects after treatment with peginterferon Alfa-2a. J Infect Dis 205:1443–7. doi: 10.1093/infdis/jis211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paparisto E, Woods MW, Coleman MD, Moghadasi SA, Kochar DS, Tom SK, et al. , 2018. Evolution-Guided Structural and Functional Analyses of the HERC Family Reveal an Ancient Marine Origin and Determinants of Antiviral Activity. J Virol 92. doi: 10.1128/JVI.00528-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamasoba D, Sato K, Ichinose T, Imamura T, Koepke L, Joas S, R et al. , 2019. N4BP1 restricts HIV-1 and its inactivation by MALT1 promotes viral reactivation. Nat Microbiol 4:1532–1544. doi: 10.1038/s41564-019-0460-3 [DOI] [PubMed] [Google Scholar]
- 61.Zhou Y, Rong L, Lu J, Pan Q, Liang C. 2008. Insulin-like growth factor II mRNA binding protein 1 associates with Gag protein of human immunodeficiency virus type 1, and its overexpression affects virus assembly. J Virol 82:5683–92. doi: 10.1128/JVI.00189-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jurczyszak D, Zhang W, Terry SN, Kehrer T, Bermudez Gonzalez MC, McGregor E, et al. , 2020. HIV protease cleaves the antiviral m6A reader protein YTHDF3 in the viral particle. PLoS Pathog 16:e1008305. doi: 10.1371/journal.ppat.1008305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sirois M, Robitaille L, Allary R, Shah M, Woelk CH, Estaquier J, et al. , 2011. TRAF6 and IRF7 control HIV replication in macrophages. PLoS One 6:e28125. doi: 10.1371/journal.pone.0028125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayashi T, Nishitsuji H, Takamori A, Hasegawa A, Masuda T, Kannagi M. 2010. DNA-dependent activator of IFN-regulatory factors enhances the transcription of HIV-1 through NF-kappaB. Microbes Infect 12:937–47. doi: 10.1016/j.micinf.2010.06.003 [DOI] [PubMed] [Google Scholar]
- 65.Li G, Dziuba N, Friedrich B, Murray JL, Ferguson MR. 2011. A post-entry role for CD63 in early HIV-1 replication. Virology 412:315–24. doi: 10.1016/j.virol.2011.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu EQ, Pan L, Xie YH, Mu DG, Liu W, Jin FG, et al. , 2015. Tetraspanin CD63 is a regulator of HIV-1 replication. International Journal of Clinical and Experimental Pathology 8:1184–1198. [PMC free article] [PubMed] [Google Scholar]
- 67.Lane BR, King SR, Bock PJ, Strieter RM, Coffey MJ, Markovitz DM. 2003. The C-X-C chemokine IP-10 stimulates HIV-1 replication. Virology 307:122–134. doi: 10.1016/s0042-6822(02)00045-4 [DOI] [PubMed] [Google Scholar]
- 68.Colomb F, Giron LB, Premeaux TA, Mitchell BI, Niki T, Papasavvas E,et al. , 2019. Galectin-9 Mediates HIV Transcription by Inducing TCR-Dependent ERK Signaling. Front Immunol 10:267. doi: 10.3389/fimmu.2019.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abdel-Mohsen M, Chavez L, Tandon R, Chew GM, Deng X, Danesh A, et al. , 2016. Human Galectin-9 Is a Potent Mediator of HIV Transcription and Reactivation. PLoS Pathog 12:e1005677. doi: 10.1371/journal.ppat.1005677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, et al. , 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe 4:495–504. doi: 10.1016/j.chom.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 71.Yu J, Liang C, Liu SL. 2017. Interferon-inducible LY6E Protein Promotes HIV-1 Infection. J Biol Chem 292:4674–4685. doi: 10.1074/jbc.M116.755819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu J, Liang C, Liu SL. 2019. CD4-Dependent Modulation of HIV-1 Entry by LY6 E. J Virol 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. Elife 5. doi: 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pacella I, Spinelli FR, Severa M, Timperi E, Tucci G, Zagaglioni M, et al. , 2020. ISG15 protects human Tregs from interferon alpha-induced contraction in a cell-intrinsic fashion. Clin Transl Immunology 9:e1221. doi: 10.1002/cti2.1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Scagnolari C, Monteleone K, Selvaggi C, Pierangeli A, D’Ettorre G, Mezzaroma I, et al. , 2016. ISG15 expression correlates with HIV-1 viral load and with factors regulating T cell response. Immunobiology 221:282–90. doi: 10.1016/j.imbio.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 76.Takeuchi Y, McClure MO, Pizzato M. 2008. Identification of gammaretroviruses constitutively released from cell lines used for human immunodeficiency virus research. J Virol 82:12585–8. doi: 10.1128/JVI.01726-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Platt EJ, Bilska M, Kozak SL, Kabat D, Montefiori DC. 2009. Evidence that ecotropic murine leukemia virus contamination in TZM-bl cells does not affect the outcome of neutralizing antibody assays with human immunodeficiency virus type 1. J Virol 83:8289–92. doi: 10.1128/JVI.00709-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 Cell Surface Concentrations on Infections by Macrophagetropic Isolates of Human Immunodeficieny Virus Type I. J Virol 72:2855–2864. doi: 10.1128/JVI.72.4.2855-2864.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, et al. , 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, et al. , 2000. Sensitivity of Human Immunodeficiency Virus Type 1 to the Fusion Inhibitor T-20 Is Modulated by Coreceptor Specificity Defined by the V3 Loop of gp120. J Virol 74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hultquist JF, Hiatt J, Schumann K, McGregor MJ, Roth TL, Haas P, et al. , 2017. A CRISPR-Cas9 Genome Engineering Platform in Primary CD4+ T Cells for the Interrogation of HIV Host Factors. Nat Protoc doi: 10.1101/205500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hultquist JF, Schumann K, Woo JM, Manganaro L, McGregor MJ, Doudna J,et al. , 2016. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep 17:1438–1452. doi: 10.1016/j.celrep.2016.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is Required for Efficient Replication of Human Immunodeficiency Virus Type-1 in Mononuclear Phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016 [DOI] [PubMed] [Google Scholar]
- 84.Zhang H, Zhou Y, Alcock C, Kiefer T, Monie D, Siliciano J, et al. , 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol 78:1718–29. doi: 10.1128/jvi.78.4.1718-1729.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yee J-K, Friedmann T, Burns JC. 1994. Chapter 5 Generation of High-Titer Pseudotyped Retroviral Vectors with Very Broad Host Range, p 99–112, Protein Expression in Animal Cells doi: 10.1016/s0091-679x(08)60600-7 [DOI] [PubMed] [Google Scholar]
- 86.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL, et al. , 1999. The Selective Downregulation of Class I Major Histocompatibility Complex Proteins by HIV-1 Protects HIV-Infected Cells from NK Cells. Immunity 10:661–671. doi: 10.1016/s1074-7613(00)80065-5 [DOI] [PubMed] [Google Scholar]
- 87.Ooms M, Majdak S, Seibert CW, Harari A, Simon V. 2010. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J Virol 84:7961–9. doi: 10.1128/JVI.00754-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, L et al. , 2020. Integrated analysis of multimodal single-cell data. bioRxiv doi: 10.1101/2020.10.12.335331:2020.10.12.335331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hafemeister C, Satija R. 2019. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20:296. doi: 10.1186/s13059-019-1874-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
On day 3, cells were treated with 0, 10, 100, or 1000 IU/mL IFNα2b for 24 hours. On day 4 (24 hours after IFN treatment) cells were infected with replication-competent HIV-1-GFP. Infection of IFNα2b-primed, CRISPR-targeted CD4+ T cells using HIV-1-GFP was performed in duplicate. Flow cytometry was performed on day 6 post infection to quantify GFP positive cells. The percent of GFP positive cells is shown for three donors.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.