Abstract
Truncated fragments of the phenoxazinone synthase gene, phsA, were prepared by the PCR. The resulting fragments were cloned into conjugative plasmid pKC1132 and transferred to Streptomyces antibioticus by conjugation from Escherichia coli. Two of the resulting constructs were integrated into the S. antibioticus chromosome by homologous recombination, and each of the resulting strains, designated 3720/pJSE173 and 3720/pJSE174, contained a disrupted phsA gene. Strain 3720/pJSE173 grew poorly, and Southern blotting suggested that genetic changes other than the disruption of the phsA gene might have occurred during the construction of that strain. Strain 3720/pJSE174 sporulated well and grew normally on the medium used to prepare inocula for antibiotic production. Strain 3720/pJSE174 also grew as well as the wild-type strain on antibiotic production medium containing either 1 or 5.7 mM phosphate. Strain 3720/pJSE174 was shown to be devoid of phenoxazinone synthase (PHS) activity, and PHS protein was undetectable in this strain by Western blotting. Despite the absence of detectable PHS activity, strain 3720/pJSE174 produced slightly more actinomycin than did the wild-type parent strain in medium containing 1 or 5.7 mM phosphate. The observation that strain 3720/pJSE174, lacking detectable PHS protein or enzyme activity, retained the ability to produce actinomycin supports the conclusion that PHS is not required for actinomycin biosynthesis in S. antibioticus.
The actinomycins are chromopeptide antibiotics produced by a number of Streptomyces strains and by some strains of Micromonospora (14, 17). In actinomycin D, the pentapeptide chains contain several methylated amino acids and one d-amino acid, but it was shown some years ago that the l-forms of the relevant amino acids serve as precursors for the synthesis of the forms found in the antibiotic (19). These pioneering studies by Katz and Weissbach also demonstrated that the chromophore of actinomycin D, a phenoxazinone ring, is derived from the catabolism of tryptophan (19). Thus, 3-hydroxykynurenine is converted to 3-hydroxyanthranilic acid, and the latter intermediate is methylated to form 4-methyl-3-hydroxyanthranilic acid, the precursor of the phenoxazinone ring (28).
The enzymes required for the methylation of 3-hydroxyanthranilic acid, for the activation of the chromophore precursor, and for the activation of the amino acids in the pentapeptide chains and their incorporation into those chains have been isolated and characterized (7, 22, 23). Schauwecker and coworkers have recently cloned the gene cluster for actinomycin production from Streptomyces chrysomallus (27). Thus, many of the biochemical and molecular genetic details regarding the biosynthesis of actinomycin have been elucidated. One unanswered question regards the synthesis of the actinomycin chromophore. Some years ago, Katz and Weissbach (18) identified an enzyme in Streptomyces antibioticus that catalyzes the oxidative condensation of 2-aminophenol derivatives to produce phenoxazinones (Fig. 1). This enzyme, phenoxazinone synthase (PHS), was subsequently purified to homogeneity (5), and its gene has been cloned and sequenced (11, 13). PHS production in S. antibioticus is subject to glucose repression, as is overall actinomycin production (9, 12), and this observation, along with the catalytic activity of PHS, suggested strongly that PHS was the enzyme responsible for the production of the actinomycin chromophore. However, PHS has been isolated only from S. antibioticus. Repeated attempts to identify the enzyme in other actinomycin producers have been unsuccessful (16).
FIG. 1.
The reaction catalyzed by phenoxazinone synthase. The reaction involves the oxidative condensation of 2-aminophenols and their derivatives, the reactants shown in the figure, to produce the phenoxazinone ring.
It was therefore important to determine definitively whether PHS is required for the production of actinomycin in the one actinomycin-producing organism in which it has been identified, S. antibioticus. To this end, the phsA gene, encoding the PHS subunit, has been disrupted, and the properties of the resulting disruptant strain have been analyzed. This strain lacks detectable PHS activity and PHS protein. Despite the absence of detectable PHS, this strain produces actinomycin as effectively as the wild type. The results lead to the conclusion that PHS is not required for actinomycin production in S. antibioticus.
MATERIALS AND METHODS
Growth of organisms.
The strains and plasmids used or constructed in this study are described in Table 1. S. antibioticus IMRU 3720 was grown on NZ-amine and galactose-glutamic acid (GGA) media as described previously (5, 9). Escherichia coli strains XL-1 Blue (Stratagene, La Jolla, Calif.) and ET12567/pUB307 (8) were grown in L broth containing antibiotics as necessary. Conjugation mixtures were plated on SFM agar (8).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Properties | Reference or source |
|---|---|---|
| Strains | ||
| S. antibioticus IMRU 3720 | Actinomycin-producing wild-type strain | 9 |
| E. coli XL-1 Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIqZΔM15 Tn10) (Tetr) | Stratagene |
| E. coli ET12567/ pUB307 | dam-13::Tn9 dcm-6 hsdM | 8 |
| Plasmids | ||
| pUB307 | Conjugative plasmid bearing a Kanr gene | 8 |
| pKC1132 | Conjugative plasmid bearing AprrreppUC | 2 |
| pJSE173 | pKC1132 bearing 1,688-bp fragment of phsA | This study |
| pJSE174 | pKC1132 bearing 1,325-bp fragment of phsA | This study |
| pJSE175 | pKC1132 bearing 1,032-bp fragment of phsA | This study |
| pJSE176 | pKC1132 bearing 735-bp fragment of phsA | This study |
Construction of strains containing a disrupted phsA gene.
Primers were prepared from the sequence of the phsA gene (11), and PCRs were performed to produce fragments truncated at their 5′ and 3′ ends as indicated in Fig. 2. The PCR primers were designed to contain EcoRI sites to facilitate cloning into EcoRI-digested pKC1132, a 3.5-kb conjugative plasmid bearing an apramycin resistance gene (2) (Table 1). The relevant ligation mixtures were used to transform E. coli XL-1 Blue. Transformants containing the desired recombinant plasmids were identified, and the plasmids were isolated and used to transform E. coli ET12567/pUB307 (8). ET12567/pUB307 is a conjugative strain of E. coli in which the transfer functions and a kanamycin resistance (Kanr) gene are borne by plasmid pUB307 (8). Appropriate transformants were then used for conjugation with S. antibioticus spores as described previously (10, 15). Plates containing putative exconjugants were overlaid with apramycin. As pKC1132 cannot replicate in Streptomyces, apramycin-resistant exconjugants could be produced only by homologous recombination between the inserts of the plasmid derivatives used for conjugation and the phsA gene in the S. antibioticus chromosome. S. antibioticus IMRU 3720 produces the pigment melanin, and this property was used to identify putative exconjugants on SFM agar. Melanin-producing colonies appeared approximately 7 days following plating. These colonies were streaked on GGA agar, and the resulting cultures were allowed to sporulate. Spores were then isolated from the plates and streaked on GGA agar containing apramycin (50 μg/ml). These plates were incubated until spores were formed, and the spores were isolated and used for the studies described herein. Exconjugants were obtained from only two of the conjugations. The strain containing the integrated plasmid bearing the 1,688-bp fragment of phsA was designated 3720/pJSE173, and the strain containing the integrated 1,325-bp fragment was designated 3720/pJSE174.
FIG. 2.
Partial restriction map of the phsA gene and positions of the primers used to generate PCR fragments for mutational cloning. The sizes of the PCR primers are not shown to scale. The numbers to the right of the primers, at the 3′ end of the gene, represent the sizes of the fragments generated by each primer pair. pJSE173 and other plasmid designations represent the plasmids obtained by cloning the PCR fragments into pKC1132.
Preparation of mycelial extracts and assays for PHS.
S. antibioticus mycelium was harvested from GGA cultures 12, 24, 48, 72, and 96 h postinoculation. Mycelium from 10-ml portions of relevant cultures was disrupted by sonication in 1 ml of a buffer composed of 50 mM Tris-HCl (pH 7.5), 5% glycerol, and Complete protease inhibitor cocktail at the concentration specified by the supplier (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Mycelial debris was removed from sonic extracts by centrifugation for 5 min at 12,000 × g and 4°C. The PHS assay was performed as described previously (5), except that the total reaction volume was 1.0 ml, the final concentration of sodium acetate used was 50 mM, and the substrate was 2-aminophenol. An extinction coefficient of 23,200 was used to calculate concentrations of 2-aminophenoxazinone (1). The protein concentration was determined with dye-binding assay reagent (Bio-Rad, Hercules, Calif.) and bovine serum albumin as the standard. We followed the protocol specified by Bio-Rad.
Miscellaneous procedures.
Southern blotting was performed as described previously (13) with the cloned phsA gene as a probe. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed essentially as described by Laemmli (24). PAGE samples were boiled for 15 min in sample buffer prior to electrophoresis. Following electrophoresis, proteins were electroblotted to Hybond P membranes (Amersham Pharmacia Biotech, Piscataway, N.J.). Western blotting was performed with a Bio-Rad Opti-4CN kit and a 1:1,000 dilution of anti-PHS antibody. Actinomycin concentrations were estimated by extracting culture media with ethyl acetate. The absorbance of the extracts was then determined at 452 nm, and actinomycin concentrations were calculated with an extinction coefficient of 24,800 (18). To label actinomycin, 5-ml portions of 72-h cultures were incubated for 60 min with 5 μCi of [methyl-14C]methionine. At the end of the incubation, the reaction mixtures were extracted with ethyl acetate, the extracts were evaporated to dryness, and the resulting residues were dissolved in methanol. Radioactive products were separated by thin-layer chromatography in ethyl acetate-methanol-water (100:5:5 [vol/vol/vol]). Plates were subjected to autoradiography.
RESULTS
Construction and characteristics of strains 3720/pJSE173 and 3720/pJSE174.
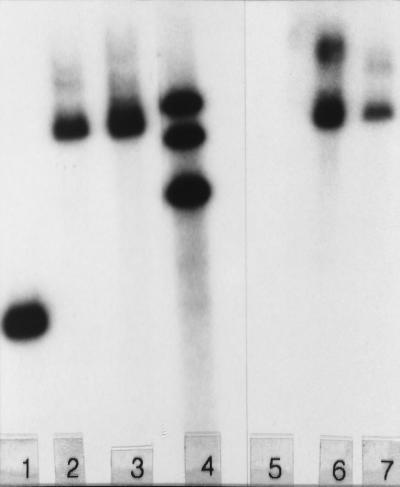
Although gene disruption by gene replacement was used effectively to inactivate the relA gene in S. antibioticus (10), it was not possible in initial experiments to disrupt the phsA gene by this technique. Thus, attempts were made to disrupt the phsA gene by using the technique of mutational cloning (4). As indicated in Materials and Methods, four PCR products, each representing forms of phsA truncated at the 5′ and 3′ ends (Fig. 2), were cloned into conjugative plasmid pKC1132 (2). E. coli strains containing the four plasmids were conjugated with S. antibioticus, and two of the plasmids, pJSE173 and pJSE174, were successfully transferred via a single crossover to the S. antibioticus chromosome by homologous recombination with the resident phsA gene. No exconjugants were obtained when the other two plasmids were used, presumably because the cloned fragments were too small to support recombination in S. antibioticus. The presence of the relevant plasmids in the chromosomes of the disruptant strains was verified by Southern blotting (Fig. 3). In the experiments depicted, chromosomal DNAs were digested with SphI. As pKC1132 contains no SphI site, digestion with this enzyme should produce only one band containing the wild-type or disrupted phsA gene. Lanes 2 and 3 of Fig. 3 show that the 2.3-kb phsA band (lane 1) is replaced by a band of the expected size (ca. 7 kb) in strains 3720/pJSE173 and 3720/pJSE174, respectively. Lanes 6 and 7 of Fig. 3 show that this band hybridizes to cloning vector pKC1132, while chromosomal DNA from the wild-type S. antibioticus strain (lane 5) does not. Thus, the phsA gene was disrupted in strains 3720/pJSE173 and 3720/pJSE174.
FIG. 3.
Southern blot of digests of chromosomal DNAs from S. antibioticus strains IMRU 3720, 3720/pJSE173, and 3720/pJSE174. Approximately 2 μg of DNA was digested with SphI. The filter containing the digests depicted in lanes 1 to 3 was probed with the radioactively labeled phsA gene, while the digests in lanes 5 to 7 were probed with pKC1132. Lanes 1 and 5, IMRU 3720 DNA; lanes 2 and 6, DNA from 3720/pJSE173; lanes 3 and 7, DNA from 3720/pJSE174. Lane 4 contains a set of markers (9.8, 6.3, and 4.4 kb).
As shown in Fig. 3, lane 6, pKC1132 hybridized strongly to a second band in the digest of DNA from strain 3720/pJSE173. This observation, coupled with the fact that the strain grew very poorly on actinomycin production medium (GGA) (data not shown), suggested that changes in addition to the disruption of the phsA gene occurred during the construction of this strain. Moreover, restoring an intact copy of the phsA gene to this strain using either high- or low-copy-number plasmids (21, 25) had no effect on growth or on the ability of the strain to produce actinomycin. Therefore, subsequent experiments reported below were done with only strain 3720/pJSE174. Strain 3720/pJSE174 sporulated well on GGA agar and SFM agar; spores of this strain appeared normal to the unaided eye. The strain grew vigorously on NZ-amine medium, the medium used to prepare inocula for antibiotic production.
Strain 3720/pJSE174 lacks PHS.
Because pJSE174 contains a truncated form of phsA, integration of this plasmid via homologous recombination into the S. antibioticus chromosome should inactivate the resident phsA gene. Strain 3720/pJSE174 was grown on actinomycin production medium (GGA), and mycelial samples were removed periodically to assay for PHS. Because previous studies suggested the possibility that both PHS activity and actinomycin production are regulated by phosphate (10; unpublished results), analyses were performed on cultures grown in either 1 or 5.7 mM phosphate. As shown in Fig. 4, extracts of the wild-type strain contained significant levels of PHS activity, as has been documented previously (18). The maximum specific activity in cultures grown in either 1 or 5.7 mM phosphate was achieved ca. 48 h postinoculation. It is interesting to note, however, that the maximum specific activity observed was about threefold lower in extracts of cultures grown in 1 mM phosphate (Fig. 4B) than in extracts of cultures grown in 5.7 mM phosphate (Fig. 4A). The data in Fig. 4 further indicate that no detectable PHS activity was present in strain 3720/pJSE174, containing a disrupted form of the phsA gene.
FIG. 4.
PHS activity in S. antibioticus strains IMRU 3720 and 3720/pJSE174. PHS assays were performed as described in Materials and Methods. Results are expressed as nanomoles of 2-aminophenoxazinone formed per minute per milligram of protein under the assay conditions described in the text.
It seemed possible that, despite the absence of detectable levels of PHS activity in 3720/pJSE174, the disruptant strain might still produce PHS protein. To explore this possibility, the extracts used for the experiments shown in Fig. 4 were fractionated by SDS-PAGE and analyzed by Western blotting with antibody to PHS. The results of such an analysis performed with two different concentrations of extract protein (20 and 50 μg) failed to reveal the presence of PHS protein in extracts of mycelia of strain 3720/pJSE174 (data not shown). Under the same conditions, PHS was easily detectable in the wild-type strain.
Growth characteristics of strains 3720/pJSE173 and 3720/pJSE174.
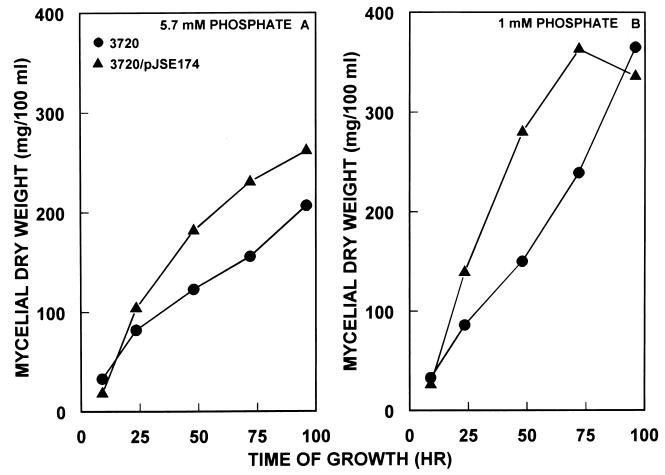
The growth characteristics on antibiotic production medium (GGA) of the wild-type S. antibioticus strain and strain 3720/pJSE174 are shown in Fig. 5. Strain 3720/pJSE174 grew somewhat more vigorously than did the parental strain, S. antibioticus 3720, at both 1 mM and 5.7 mM phosphate.
FIG. 5.
Growth of S. antibioticus IMRU 3720, 3720/pJSE173, and 3720/pJSE174 on GGA medium at different phosphate concentrations. Growth and assay conditions were as described in Materials and Methods. Apramycin was included in NZ-amine medium, used to prepare inocula for growth on GGA medium, but the antibiotic was not included in the latter medium.
Actinomycin production by the parental S. antibioticus strain and by strain 3720/pJSE174.
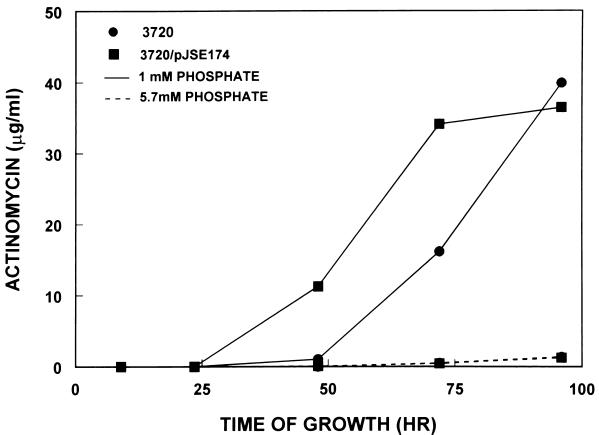
Actinomycin production by the two strains of interest in this study was examined by two procedures. In the first, portions of the culture medium were extracted with ethyl acetate, a procedure shown previously to remove actinomycin preferentially from the growth medium (19). The amount of actinomycin present in the extracts was estimated spectrophotometrically. The results of this analysis are shown in Fig. 6. Levels of actinomycin produced by the parental S. antibioticus strain were low in medium containing 5.7 mM phosphate but significantly higher in medium containing 1 mM phosphate. Strain 3720/pJSE174 produced levels of actinomycin that were higher than or equal to those produced by the parental strain under both sets of conditions (Fig. 6).
FIG. 6.
Actinomycin production by strains IMRU 3720 and 3720/pJSE174. Actinomycin was assayed spectrophotometrically as described in Materials and Methods. Because of the scale used for this figure, the levels of actinomycin production by strains IMRU 3720 and 3720/pJSE174 in medium containing 5.7 mM phosphate appear identical, but strain 3720/pJSE174 produced slightly larger amounts of the antibiotic under these conditions.
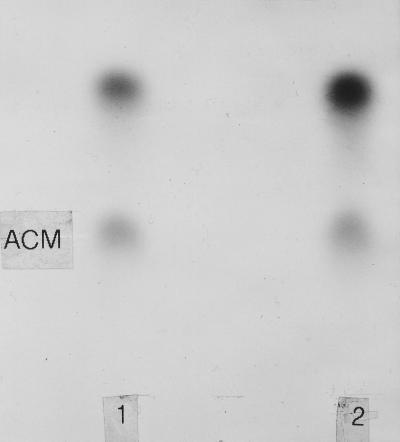
The second method used to assess actinomycin production involved the incubation of mycelia (72 h postinoculation) with a radioactive precursor of actinomycin. Incubation mixtures were extracted with ethyl acetate, and the extracts were concentrated and analyzed by thin-layer chromatography. The chromatogram shown in Fig. 7 confirms that the parental S. antibioticus strain and strain 3720/pJSE174 produced authentic actinomycin.
FIG. 7.
Autoradiogram of a thin-layer chromatogram of neutral ethyl acetate-extracted products following incubation of strains IMRU 3720 (lane 1) and 3720/pJSE174 (lane 2) with [methyl-14C]methionine. ACM, actinomycin.
DISCUSSION
The data presented above confirm the successful disruption of the PHS gene in S. antibioticus. While four plasmid constructs were prepared for mutational cloning, only two of these were successfully integrated into the S. antibioticus chromosome, presumably because the inserts in the other two recombinant constructs were too small to allow homologous recombination to occur. The disruption of the phsA gene and its consequences have implications for the mechanism of actinomycin production, discussed in greater detail below. It can also be concluded from the experiments described here that the phsA gene is not essential to the viability of S. antibioticus.
Strain 3720/pJSE174, produced by using a 1,325-bp truncated fragment of phsA (Fig. 2), sporulated well, grew well on NZ-amine and GGA media, and produced actinomycin in cultures containing either 1 or 5.7 mM phosphate. In general, strain 3720/pJSE174 was similar in its properties to the parental strain, except for the absence of detectable levels of PHS activity and PHS protein (Fig. 4 and data not shown). Because strain 3720/pJSE174 was capable of producing actinomycin in the absence of detectable PHS, it is reasonable to conclude that the phsA gene and the PHS enzyme are not required for actinomycin production in S. antibioticus IMRU 3720. Consistent with this conclusion is the observation, shown in Fig. 4 and 6, that conditions which produced the highest PHS specific activities in S. antibioticus mycelial extracts (growth at 5.7 mM phosphate) produced only low levels of actinomycin. By comparison, growth at 1 mM phosphate produced 3-fold lower PHS specific activities but up to 30-fold higher levels of actinomycin. That PHS is not required for actinomycin production does not mean that the wild-type strain does not use this enzyme if it is available. It is known that PHS can utilize 4-methyl-3-hydroxyanthraniloyl peptides, suspected precursors of actinomycin in vivo, as substrates (20, 26).
At least two significant questions are raised by the results presented in this report. First, if PHS is not required for actinomycin production, what is it function in S. antibioticus? A possible answer to this question was suggested by comparisons of the amino acid sequence of the PHS subunit to those of proteins in the GenBank sequence database. PHS is quite similar to enzymes of the blue copper family (11), as it is a blue copper enzyme itself. Interestingly, PHS is also quite similar to the CotA protein of Bacillus subtilis (3). CotA is a component of B. subtilis spores and may play a role in the synthesis of a spore pigment (6). Because of this similarity between PHS and CotA, we examined S. antibioticus spores to determine whether they contain a PHS-like protein. These analyses showed that sonic extracts of purified spores contain a protein with the electrophoretic and immunological properties of the PHS subunit and significant amounts of a protein with a lower molecular weight. This protein, designated PHS*, is apparently produced by proteolysis of the PHS subunit (unpublished data). Thus, there is some evidence to suggest that PHS may be involved in the process of spore formation in S. antibioticus, and this may represent its major role in that organism. It is noteworthy that both PHS and PHS* are absent from the spores of strain 3720/pJSE174 (data not shown). This observation strongly suggests that PHS and PHS* are both products of the phsA gene. Studies are in progress to further elucidate the role of PHS in S. antibioticus.
The second question raised by the observations described above concerns the mechanism of actinomycin biosynthesis. If PHS is not responsible for the formation of the actinomycin chromophore, how is that moiety synthesized? There are at least two possibilities. There may be another enzyme elaborated by S. antibioticus and by other actinomycin producers that is capable of catalyzing the condensation of actinomycin half molecules to yield the antibiotic. Alternatively, the condensation may occur nonenzymatically, under the influence of pH and divalent cations. With regard to this possibility, it is perhaps noteworthy that the physical map of the actinomycin cluster from S. chrysomallus does not include a gene for the formation of the chromophore (27). Experiments are in progress to distinguish between the possibilities for the synthesis of the chromophore.
ACKNOWLEDGMENTS
The studies reported here were supported by grant 1 RO1 GM51589 from the National Institutes of Health to G.H.J.
REFERENCES
- 1.Barry C E, III, Parmesh G, Nayar P G, Begley T. Phenoxazinone synthase: mechanism for the formation of the phenoxazinone chromophore of actinomycin. Biochemistry. 1989;28:6323–6333. doi: 10.1021/bi00441a026. [DOI] [PubMed] [Google Scholar]
- 2.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 3.Borriss R, Porwollik S, Schroeter R. The 52 degrees-55 degrees segment of the Bacillus subtilis chromosome: a region devoted to purine uptake and metabolism, and containing the genes cotA, gabP and guaA and the pur gene cluster within a 34960 bp nucleotide sequence. Microbiology. 1996;142:3027–3031. doi: 10.1099/13500872-142-11-3027. [DOI] [PubMed] [Google Scholar]
- 4.Chater K F, Bruton C J. Mutational cloning in Streptomyces and the isolation of antibiotic production genes. Gene. 1983;26:67–78. doi: 10.1016/0378-1119(83)90037-9. [DOI] [PubMed] [Google Scholar]
- 5.Choy H A, Jones G H. Phenoxazinone synthase from Streptomyces antibioticus: purification of the large and small enzyme forms. Arch Biochem Biophys. 1981;211:55–65. doi: 10.1016/0003-9861(81)90429-x. [DOI] [PubMed] [Google Scholar]
- 6.Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fawaz F, Jones G H. Actinomycin synthesis in Streptomyces antibioticus. Purification and properties of 3-hydroxyanthranilate-4-methyltransferase. J Biol Chem. 1988;263:4602–4606. [PubMed] [Google Scholar]
- 8.Flett F, Mersenias V, Smith C P. High efficiency intergenic conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1998;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 9.Gallo M, Katz E. Regulation of secondary metabolite biosynthesis: catabolite repression of phenoxazinone and actinomycin formation by glucose. J Bacteriol. 1971;109:659–667. doi: 10.1128/jb.109.2.659-667.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoyt S, Jones G H. relA is required for actinomycin production in Streptomyces antibioticus. J Bacteriol. 1999;181:3824–3829. doi: 10.1128/jb.181.12.3824-3829.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh C-J, Jones G H. Nucleotide sequence, transcriptional analysis, and glucose regulation of the phenoxazinone synthase gene (phsA) from Streptomyces antibioticus. J Bacteriol. 1995;177:5740–5747. doi: 10.1128/jb.177.20.5740-5747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones G H. Regulation of phenoxazinone synthase expression in Streptomyces antibioticus. J Bacteriol. 1985;163:1215–1221. doi: 10.1128/jb.163.3.1215-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones G H, Hopwood D A. Molecular cloning and expression of the phenoxazinone synthase gene from Streptomyces antibioticus. J Biol Chem. 1984;259:14151–14157. [PubMed] [Google Scholar]
- 14.Jones G H, Keller U. Biochemistry and genetics of actinomycin production. In: Strohl W R, editor. Biotechnology of antibiotics. New York, N.Y: Marcel Dekker, Inc.; 1997. pp. 335–361. [Google Scholar]
- 15.Jones G H, Paget M S B, Chamberlin L, Buttner M J. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol Microbiol. 1997;23:169–178. doi: 10.1046/j.1365-2958.1997.2001566.x. [DOI] [PubMed] [Google Scholar]
- 16.Katz E. Biogenesis of the actinomycins. In: Waksman S A, editor. Actinomycin: natural formation and activities. New York, N.Y: Wiley Interscience; 1968. pp. 45–68. [Google Scholar]
- 17.Katz E. Actinomycin. In: Gottlieb D, Shaw P D, editors. Antibiotics II. New York, N.Y: Springer-Verlag; 1976. pp. 276–341. [Google Scholar]
- 18.Katz E, Weissbach H. Biosynthesis of the actinomycin chromophore: enzymatic conversion of 4-methyl 3-hydroxyanthranilic acid to actinocin. J Biol Chem. 1962;237:882–886. [PubMed] [Google Scholar]
- 19.Katz E, Weissbach H. Incorporation of C14-labeled amino acids into actinomycin and protein by Streptomyces antibioticus. J Biol Chem. 1963;238:666–675. [PubMed] [Google Scholar]
- 20.Katz E, Lloyd H A, Mauger A B. Enzymatic synthesis of actinomycin and analogues containing N-methylalanine from synthetic pentapeptide lactone precursors. J Antibiot. 1990;43:731–733. doi: 10.7164/antibiotics.43.731. [DOI] [PubMed] [Google Scholar]
- 21.Katz E, Thompson C J, Hopwood D A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983;129:2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- 22.Keller U. Actinomycin synthetases: multifunctional enzymes responsible for the synthesis of the peptide chains of actinomycin. J Biol Chem. 1987;262:5852–5856. [PubMed] [Google Scholar]
- 23.Keller U, Schlumbohm W. Purification and characterization of actinomycin synthetase I, a 4-methyl-3-hydroxyanthranilic acid:AMP ligase from Streptomyces chrysomallus. J Biol Chem. 1992;267:11745–11752. [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lydiate D J, Malpartida F, Hopwood D A. The Streptomyces plasmid SCP2*: its functional analysis and development into useful cloning vectors. Gene. 1985;35:223–235. doi: 10.1016/0378-1119(85)90001-0. [DOI] [PubMed] [Google Scholar]
- 26.Salzman L, Weissbach H, Katz E. Enzymatic synthesis of actinocinyl peptides. Arch Biochem Biophys. 1969;130:536–546. doi: 10.1016/0003-9861(69)90067-8. [DOI] [PubMed] [Google Scholar]
- 27.Schauwecker F, Pfennig F, Schröder W, Keller U. Molecular cloning of the actinomycin synthetase gene cluster from Streptomyces chrysomallus and functional heterologous expression of the gene encoding actinomycin synthetase II. J Bacteriol. 1998;180:2468–2474. doi: 10.1128/jb.180.9.2468-2474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Troost T, Katz E. Phenoxazinone biosynthesis: accumulation of a precursor, 4-methyl-3-hydroxyanthranilic acid, by mutants of Streptomyces parvulus. J Gen Microbiol. 1979;111:121–132. doi: 10.1099/00221287-111-1-121. [DOI] [PubMed] [Google Scholar]