Abstract
Although the nasopharyngeal swab (NPS) is considered the gold standard for the diagnosis of the SARS-CoV-2 infection, the Nasal Mid-Turbinate swab (NMTS) is often used due to its higher tolerance among patients. We compared the diagnostic performance of the NPS and the NMTS for the Panbio™ COVID-19 antigen-detecting rapid diagnostic test (Ag-RDT). Two hundred and forty-three individuals were swabbed three times by healthcare professionals: a NMTS and a NPS specimen for the Ag-RDT and an oropharyngeal swab for real time RT-PCR. Forty-nine participants were RNA-SARS-CoV-2 positive by real time RT-PCR: 45 and 40 were positive by the Ag-RDT with NPS and NMTS, respectively. The overall sensitivity and specificity were 91.8% (95% CI: 83.2–100.0) and 99.5% (95% CI: 98.2–100.0) for Ag-RDT with NPS, and 81.6% (95% CI: 69.8–93.5) and 100.0% (95% CI: 99.7–100.0) for the Ag-RDT with NMTS. The Cohen’s kappa index was 0.92 (95% CI: 0.85–0.98). Among asymptomatic individuals, the Ag-RDT with both sampling techniques showed a high sensitivity [100.0% (95% CI: 95.5–100.0) with NPS; 90.9% (95% CI: 69.4–100.0) with NMTS], while the performance of the test decreased in samples with Ct≥ 30 and in patients tested after the first 7 days from symptom onset. Although the NMTS yielded a lower sensitivity compared to NPS, it might be considered a reliable alternative, as it presents greater adherence among patients, enabling scaling of antigen testing strategies, particularly in countries with under-resourced health systems.
Introduction
The main in vitro tests used for the diagnosis of SARS-CoV-2 infection in clinical microbiology laboratories are based on the detection of viral RNA in nasopharyngeal swab (NPS) specimens. The most widely used technique is real time RT-PCR [1], a costly method that requires highly trained technical personnel, adequate infrastructure and sophisticated equipments. Therefore, it is not feasible to implement and perform it on a massive scale in most laboratories and health centers, especially in highly exposed developing countries, such as some low and middle-income regions from South America. The availability of timely, cost-effective and easy detection point-of-care (POC) diagnostic tests in testing centers is essential for an early diagnosis, which will allow the optimization of patient management and the implementation of measures to prevent further spread of the virus in the context of the COVID-19 pandemic. Thus, the use of antigen-detecting rapid diagnostic tests (Ag-RDTs) for SARS-CoV-2 has increased within the last months, as these test represent a quick and easy-to-perform alternative for virus detection [2].
The PanbioTM COVID-19 Antigen Rapid Test Device (Abbott) is an in vitro Ag-RDT for the qualitative detection of SARS-CoV-2 antigen, approved through the WHO Emergency Use Listing procedure in 2020 [3]. Although the NPS specimen is considered the gold standard reference specimen type [4, 5], Nasal Mid-Turbinate swab (NMTS) sample is also frequently used, since it represents a less invasive sampling method and, therefore, more comfortable for individuals, offering new opportunities for SARS-CoV-2 testing strategies [6].
In the present diagnostic accuracy study, two professional-collected specimen types (Nasal Mid-Turbinate and Nasopharyngeal swabs) were evaluated and compared using the PanbioTM COVID-19 Ag Rapid Test Device assay (Abbott).
Materials and methods
This manufacturer-independent study took place on the 19th of February, 2021, and it enrolled 243 individuals that attended an outpatient screening center for the detection of SARS-CoV-2 infection within the framework of the COVID-19 pandemic in Córdoba, the second most populated city from Argentina. Patients were invited to voluntarily participate in this study. By that date, in Córdoba, 20.3% of adult critical care beds were occupied.
All participants showed suspected SARS-CoV-2 infection according to the local governmental testing criteria, which included adults with suggestive symptoms of COVID-19 and/or asymptomatic individuals that had a recent exposure to a SARS-CoV-2 confirmed case or were subjected to the test for other reasons (labor requirement, travel, etc.). Each individual was swabbed three times by a healthcare professional with a standardized sampling technique, following the manufacturer’s instructions [7]: 1-a NMTS sample in both nostrils for the PanbioTM COVID-19 Ag Rapid Test Device (Nasal Mid-Turbinate); 2-a NPS sample for the PanbioTM COVID-19 Ag Rapid Test Device (Nasopharyngeal); and 3-an oropharyngeal swab (OS) specimen for real time RT-PCR.
The clinical and epidemiological data collected for each patient was: age, sex, day from symptom onset, potential close contact with a confirmed positive COVID-19 individual within the last 14 days, symptoms [(cough/sore throat, myalgia, fever, anosmia/dysgenesis, gastrointestinal symptoms (diarrhea), and headache] (S3 Table).
Both Ag-RDTs were performed by trained personnel immediately after sample collection -following the manufacturer’s instructions [8]- and read out at 15–20 minutes.
RNA was isolated from OS samples using the MegaBio plus Virus RNA Purification Kit II on the GenePure Pro Nucleic Acid Purification System NPA-32P (Bioer). Extracted RNA was analyzed using the DisCoVery SARS-CoV-2 Nucleic Acid Detection RT-PCR Kit (Cy5/ROX) (Multiplex Real Time RT-PCR—Ap Biotech), targeting the open reading frame (ORF1-ab) gene and the nucleocapsid protein (N) gene according to the manufacturer’s instructions [9]. The cut-off cycle threshold (Ct) value was 38 for both genes, and if the Ct values of both genes were ≤38 the specimen was defined as positive.
The diagnostic performance of the antigen was assessed using sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). Ag-RDT sensitivity and specificity with 95% confidence intervals (95% CI) were determined relative to real time RT-PCR, as the reference standard technique. Sensitivity was evaluated for the whole study population and according to the presence of symptoms, the Ct values for N and Orf-1 genes and the days from symptom onset. Agreement between techniques was evaluated using Cohen’s kappa score; and the positive percent agreement (PPA), and negative percent agreement (NPA) between NMTS and NPS samples on the Ag-RDT (including one false-positive), were also calculated. All analyzes were performed using Epidat 3.1 [10].
Descriptive statistics were employed to summarize data. A Two-Proportion Z-Test was used to compare test sensitivities obtained from symptomatic and asymptomatic individuals, and the McNemar’s test for paired data was used for the other categories (Ct values; days from symptom onset; etc). These analyzes were performed using STATA version 14.0 (Stata Corp., College Station, TX, USA). Statistical significance was defined as p<0.05. The sample size was calculated based on a type I error of 5%, a power of 80% and an expected sensitivity in accordance with the performance data reported by the manufacturer [8]. The study was conducted according to the guidelines of the Declaration of Helsinki (1964, amended most recently in 2008) of the World Medical Association, and in accordance with specific local ethics regulations, established by the Ministry of Health of Córdoba province, Argentina.
Oral informed consent was obtained from all subjects involved in the study and from parents or guardian of minor participants (age<18). A database with their answers was registered in the informatic system of the local government. The Government of the Province of Córdoba wives the written informed consent, based on the need for rapid surveillance, which allows rapid and effective decision-making in public health.
Results
During the research, 243 participants were included. Our study population had a median age of 32 years (3–79 years) and 46.5% (113/243) were male. In total, 51.0% (124/243) were symptomatic on the day of testing with a median duration of symptoms of 3 days (1–11 days). From the total of asymptomatic participants (119/243; 49.0%), 72.3% (86/119) had had a close contact with another patient with an RT-PCR- confirmed SARS-CoV-2 infection. Clinical and diagnostic characteristics of individuals are shown in Table 1.
Table 1. Characteristics of the study population.
| CHARACTERISTICS | RESULTS |
|---|---|
| Number of individuals (n) | 243 |
| SARS-CoV-2 Positive cases by real time RT-PCR | 49/243 (20.2%) |
| Age (years) | |
| Median (Range) | 32 (3–79) |
| Sex | |
| Male | 113 (46.5%) |
| Risk Factors | |
| Close contact | 177 (72.8%) |
| Diagnosis | |
| Asymptomatic | 119 (49.0%) |
| Symptomatic | 124 (51.0%) |
| Cough/Sore Throat | 15 (12.1%) |
| Headache | 11 (8.9%) |
| Myalgia | 10 (8.1%) |
| Fever | 3 (2.4%) |
| Anosmia/Dysgeusia | 2 (1.6%) |
| Diarrhea | 1 (0.8%) |
| Unspecified | 33 (26.6%) |
| Combination of symptoms | 49 (39.5%) |
| Test from symptom onset (days) | |
| Median (range) | 3 (1–11) |
| Real time RT-PCR Results | |
| Ct values N gene | |
| Mean Ct ± Standar deviation (range) | 25.6 ± 3.9 |
| Ct values Orf-1ab gene | |
| Mean Ct ± Standard deviation (range) | 28.3 ± 3.9 |
| NMTS Ag-RDT Results | |
| Positive | 40/49 (81.6%) |
| Negative | 9/49 (18.4%) |
| NPS Ag-RDT Results | |
| Positive | 45/49 (91.8%) |
| Negative | 4/49 (8.2%) |
Forty-nine out of 243 participants were RNA-SARS-CoV-2 positive by real time RT-PCR (20.2%), of whom 38 were symptomatic (77.6%). Out of 243 individuals, 45 (18.5%) and 40 (16.4%) were positive by the Ag-RDT with NPS and NMTS, respectively (Table 2). Four specimens (1.6%) had a false-negative result with both, the NPS and NMTS, and all of them were symptomatic, with a mean Ct value of 32.6 (30.8–35.1) for the N gene and of 35.1 (33.5–37.4) for the ORF-1ab gene. One sample (0.41%) was false-positive only by the Ag-RDT with NPS. Five specimens (2.1%) were positive by the Ag-RDT with NPS but negative with NMTS, all of them had Ct values >28, and a mean Ct value of 30.7 (28.3–32.6) for the N gene and 33.6 (30.5–36.2) for the ORF-1ab gene, and 1 individual was asymptomatic. Detailed results are available in Table 2. Table 3 shows features of samples with discordant results.
Table 2. Agreement between the Ag-RDT with both sampling techniques and PCR for all samples, and divided into symptomatic and asymptomatic.
| Overall | Symptomatic | Asymptomatic | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PCR | PCR | PCR | |||||||||
| + | - | + | - | + | - | ||||||
| NPS Ag-RDT | + | 45 | 1 | NPS Ag-RDT | + | 34 | 1 | NPS Ag-RDT | + | 11 | 0 |
| - | 4 | 193 | - | 4 | 85 | - | 0 | 108 | |||
| NMTS Ag-RDT | + | 40 | 0 | NMTS Ag-RDT | + | 30 | 0 | NMTS Ag-RDT | + | 10 | 0 |
| - | 9 | 194 | - | 8 | 86 | - | 1 | 108 | |||
Table 3. Cases with discordant results between Ag-RDTs and real time RT-PCR.
| Sample ID | Symptoms | Test after 7 days from symptom onset | Ct value real time RT-PCR | Ag-RDT NPS | Ag-RDT NMTS | Interpretation | |
|---|---|---|---|---|---|---|---|
| N | ORF-1ab | ||||||
| 1 | Yes | No | 30.8 | 33.5 | Negative | Negative | False negative for both Ag detection. |
| 2 | Yes | No | 35.1 | 37.4 | Negative | Negative | False negative for both Ag detection. |
| 3 | No | - | 32.6 | 36.2 | Positive | Negative | False negative for Ag-NMTS detection. |
| 4 | Yes | No | 28.3 | 30.5 | Positive | Negative | False negative for Ag-NMTS detection. |
| 5 | Yes | Yes | 31 | 34 | Positive | Negative | False negative for Ag-NMTS detection. |
| 6 | Yes | Yes | 29.7 | 32.6 | Positive | Negative | False negative for Ag-NMTS detection. |
| 7 | Yes | No | 32.8 | 35.3 | Negative | Negative | False negative for both Ag detection. |
| 8 | Yes | Yes | 32.2 | 34.8 | Positive | Negative | False negative for Ag-NMTS detection. |
| 9 | Yes | Yes | 31.8 | 34.2 | Negative | Negative | False negative for both Ag detection. |
| 10 | Yes | No | - | - | Positive | Negative | False positive for Ag-NPS detection. |
Sensitivity and specificity were calculated for the whole study population and according to the presence or absence of symptoms, days from symptom onset, and Ct values for the N and ORf-1ab genes. The overall sensitivity was 91.8% (95% CI: 83.2–100.0) for the Ag-RDT with NPS and 81.6% (95% CI: 69.8–93.5) with NMTS (no statistically significant difference was observed between these percentages, p = 0.063). For the above-mentioned SARS-CoV-2 prevalence (20.2%), the overall positive predictive values for the Ag-RDT with NPS and NMTS were 97.8% (95% CI: 92.5–100.0) and 100.0% (95% CI: 98.8–100.0), respectively. The overall negative predictive values were 98.0% (95% CI: 95.8–100.0) and 95.6% (95% CI: 92.5–98.6), for the Ag-RDT with NPS and NMTS, respectively.
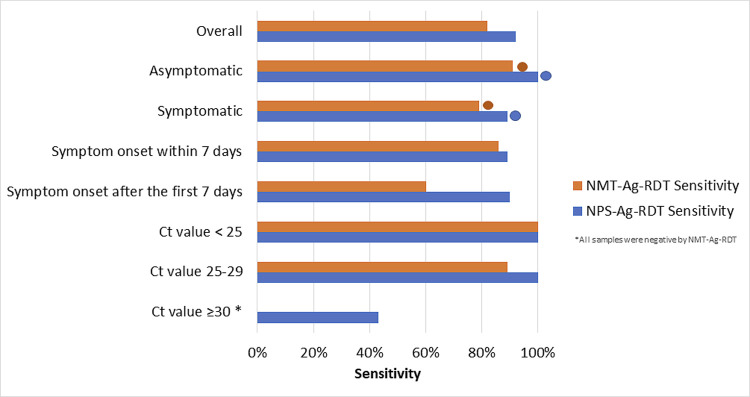
Among the categories analyzed, sensitivity percentages varied between 42.9% and 100.0% (Fig 1). The global specificity was 99.5% (95% CI: 98.2–100.0) for NPS sampling and 100.0% (95% CI: 99.7–100.0) for NMTS sampling, and were higher than 98.5% for all the categories analyzed (S1 Table). The global Cohen’s kappa index between both, NPS and NMTS, was 0.92 (95% CI: 0.85–0.98). We additionally computed the pair of agreement statistical measures: the PPA and NPA between NMTS and NPS samples on Ag-RDT, whose values were 86.96 (95% CI: 76.1–97.8) for the PPA (including one false-positive with NPS) and 100.0% (95% CI: 99.8–100.0) for the NPA.
Fig 1. Sensitivity comparison according to the type of swabbing and the population group.
*All samples in this group were negative by NMTS-Ag-RDT. Cts correspond to the N gene. Circles represent significative differences between sensitivities among symptomatic and asymptomatic individuals with both sampling techniques.
When participants were classified in symptomatic and asymptomatic, it was observed that in symptomatic patients, Ag-RDT with NPS and NMTS sampling yielded a sensitivity of 89.5% (95% CI: 78.4–100.0) and 79.0% (95% CI: 64.7–93.2), respectively (p = 0.125) (Fig 1). Specificity was 98.8% (95% CI: 96.0–100.0) for NPS and 100.0% (95% CI: 92.4–100.0) for NMTS sampling. On the other hand, sensitivity of the Panbio test among asymptomatic individuals was 100.0% (95% CI: 95.5–100.0) for NPS and 90.9% (95% CI: 69.4–100.0) for NMTS sampling (S1 Table) (p = 1.000). The Cohen’s kappa index between the Ag-RDT with both sampling techniques was 0.90 (95% CI: 0.81–0.98) for symptomatic individuals and 0.95 (95% CI: 0.85–1.00) for the asymptomatic ones. Differences in test sensitivities between symptomatic and asymptomatic individuals were statistically significant with both sampling techniques (p value<0.001 with NPS and NMTS) (Fig 1).
Regarding Ct values for both genes, a comparison analyzes was made organizing the total of real time RT-PCR positive samples (n = 49) in sub-groups: Ct<25 (23/49); Ct between 25–30 (19/49); Ct≥30 (7/49) for the N gene. Test sensitivities for each category is detailed in S1 Table and in Fig 1. Approximately 96.0% of all viable specimens with Cts<30 for the N gene and the 100.0% of the samples with Cts<30 for the ORF-1ab gene were detected by the Ag-RDT with both specimen types. Alongside an increase in Ct values for the N gene, it was observed a reduction in the number of real time RT-PCR positive samples detected by the Ag-RDT with both sampling techniques: in specimens with Cts≥ 30, 3 out of 7 samples were positive by the Ag-RDT with the NPS and none of the 7 samples were detected by the Ag-RDT with the NMTS. Similar results were observed for the ORF-1ab gene: in specimens with Cts≥ 30, 11 out of 15 were positive by the Ag-RDT with the NPS and 6 out of 15 by the Ag-RDT with the NMTS.
When samples were stratified according to the days from symptom onset, both NPS and NMTS-Ag-RDT, showed a modest improvement in the sensitivity percentages among cases tested within 7 days from symptom onset (89.7 Vs. 90.0% for NPS Ag-RDT; and 85.7 Vs. 60.0% for NMST Ag-RDT, although these differences were not statistically significant) (Fig 1) (S1 Table).
Additionally, a double stratification of the samples was made, according to their Ct values and the presence of symptoms. Sensitivities and specificities were higher than 98.8% for the NPS-Ag-RDT, and higher than 80.0% for NMTS-Ag-RDT in samples with Cts<30, independently of the clinical status (S2 Table). A more remarkable drop in the identification of real time samples by Ag-RDT with NPS was observed in symptomatic individuals with Cts≥30 (33.3%), and no RT-PCR positive sample could be detected by the Ag-RDT with NMTS in symtomatic or asymptomatic population with Cts≥30. In order to compare our findings with previous similar studies, Table 4 shows the results obtained from the literature review regarding researches that assessed the diagnostic performance of different Ag-RDTs using nasal swab (Nasal Mid Turbinate and Anterior Nasal).
Table 4. Comparative studies that assessed the diagnostic performance of different instrument-free Ag-RDTs using nasal swab.
Sensitivity and specificity results for nasal specimen in comparison to the reference technique were included in the table.
| Reference, first author | Ag-RDT | Study location | Sample size | Study population | Sample types | SARS-CoV-2 prevalence (%) | Reference standard technique | Sensitivity (95% IC) | Specificity (95% IC) |
|---|---|---|---|---|---|---|---|---|---|
| Alqahtani et al., 2022 | Panbio, Abbot | Bahrain | 4183 | Mildly symptomatic | NMTS and NPS | 17.5 | RT-PCR in NPS | 82.1 (79.2–84.8) | 99.1 (98.8–99.4) |
| Begum et al., 2021 | InTec | Bangladesh | 214 | Symptomatic participants with known COVID-19 status | NS and NPS | 52.3 | RT-PCR in NS | 80.0 (70.8–87.3) | 97.4 (92.5–99.5) |
| STANDARD Q, SD Biosensor | 78.0 (68.6–85.7) | 94.7 (88.9–98.0) | |||||||
| Cassuto et al., 2021. | Autotest COVID-VIRO | France | 234 | Volunteers with mild to moderate symptoms lasting less than 7 days | Self-collected ANS /NPS | 13.7 | RT-PCR in NPS | 96.9 (83.8–99.9) | 100.0 (98.2–100.0) |
| Chiu et al., 2021 | INDICAID,PHASE Scientific International Ltd. | San Francisco, Oakland and San Fernando, United States | 349 | Symptomatic Populations within 5 days from symptom onset | ANS | 22.8 | RT-PCR | 85.3 (75.6–91.6) | 94.9 (91.6–96.9) |
| Hong Kong | 22994 | Asymptomatic individuals | 0.16 | 84.2 (69.6–92.6) | 99.9 (99.9–100.0) | ||||
| Homza et al., 2021 | Test 1* | Karvina, Czech Republic | 488 | Mildly symptomatic and asymptomatic with a confirmed SARS-CoV-2 close contact | NPS/ANS (one nostril) | 26.4 | RT-PCR in NPS | 46.5 (37.7–55.5) | 99.4 (98.0–99.9) |
| Test 2* | 406 | 42.4 | 54.1 (46.3–61.7) | 97.4 (94.5–9.1) | |||||
| Kronberg et al., 2021 | STANDARD Q, SD Biosensor | Copenhagen, Denmark | 7074 | Screening in general population with non-characteristic COVID-19 symptoms | ANS/OPS | 0.9 | RT-PCR in OPS | 48.5 | 100.0 |
| James et al., 2021 | BinaxNOW, Abbot | Arkansas | 2339 | Symptomatic and asymptomatic clinical and non-clinical employees of an acute care Hospital | NS | 6.5 | RT-PCR in NS | 56.6 (48.7–4.5) | 99.9 (99.7–00) |
| Klein et al., 2021 | Panbio, Abbot | Heidelberg, Germany | 290 | Adults with symptoms or a recent contact with a confirmed SARS-CoV-2 case | Self-collected NMTS/NPS | 15.5 | RT-PCR in NPS | 84.4 (71.2–2.3) | 99.2 (97.1–9.8) |
| Lindner et al., 2021 (1) | STANDARD Q, SD Biosensor | Berlin, Germany | 146 | Symptomatic adults | Self-collected NMTS/ Professional-collected NPS | 27.4 | RT-PCR in OPS/NPS | 82.5 (68.1–1.3) | 100.0 (96.5–00) |
| Lindner et al. 2021 (2) | STANDARD Q, SD Biosensor | Berlin, Germany | 289 | Symptomatic adults | Self-collected NMTS/Professional-collected NPS and OPS | 13.5 | RT-PCR in OPS/NPS | 74.4 (58.9–85.4) | 99.2 (97.1–99.8) |
| Lindner et al. 2021 (3) | STANDARD Q, SD Biosensor | Berlin, Germany | 179 | Symptomatic adults | Professional-collected NMTS | 22.9 | RT-PCR in OPS/NPS | 80.5 (66.0–89.8) | 98.6 (94.9–99.6) |
| Masiá et al., 2021 | Panbio, Abbot | Spain | 659 | patients, either with COVID-19 signs/symptoms or asymptomatic contacts | NS (one nostril) /NPS | 21.5 | RT-PCR in NPS | 44.7 (36.1–3.6) | 100.0 (9.1–00.0) |
| Okoye et al., 2021 | BinaxNOW, Abbot | Utah, United States | 2645 | Asymptomatic students | Self-collected NMTS | 1.7 | RT-PCR in self-collected NMTS | 53.3 (39.1–67.1) | 100.0 (99.9–100.0) |
| Pilarowski et al., 2020 | BinaxNOW, Abbot | California, United States | 878 | Screening among symptomatic and asymptomatic individuals | ANS | 3.0 | RT-PCR in ANS | 57.7 (6.9–76.6) | 100.0 (9.6–100%) |
| Pollock et al., 2020 | BinaxNOW, Abbot | Massachusetts, United States | 2308 | Symptomatic and asymptomatic individuals | ANS | 12.7 | RT-PCR in ANS | *77.4 (72.2–82.1) | 99.4 (99.0–99.7) |
| Pollock et al., 2020 | Access Bio CareStart | Massachusetts, United States | 1498 | Symptomatic and asymptomatic individuals | ANS | 15.6 | RT-PCR in ANS | 57.7 (51.1–64.1) | 98.3 (7.5–99.0) |
| Stohr et. Al, 2021 | BD-Veritor System | Tilburg, Noord-Brabant, the Netherlands. | 1595 | Adults with symptoms or a recent contact with a confirmed SARS-CoV-2 case | Self-collected NMTS/ professional NPS and OPS | 11.8 | RT-PCR in OPS and NPS | 49.1 (41.7–56.5) | 99.9 (99.7–100.0) |
| STANDARD Q, SD Biosensor | 1606 | 61.5 (54.6–68.3) | 99.7 (99.4–99.9) | ||||||
| Takeuchi et al., 2021 | QuickNavi | Tsukuba, Japan. | 862 | Symptomatic and asymptomatic individuals | ANS/NPS | 5.9 | RT-PCR in NPS | 72.5 (58.3–84.1) | 100.0 (9.3–100) |
| Van der MoerenID et al., 2021 | BD Veritor System | West-Brabant, the Netherlands. | 352 | Symptomatic adults | OPS-NS | 4.8 | RT-PCR in OPS and NS | 94.1 (71.1–100) | 100 (98.9–100) |
NS: Nasal Swab; NMTS: Nasal Mid Turbinate swab; ANS: Anterior Nasal Swab; OPS: Oropharyngeal Swab
* Names not mentioned in the study
Discussion
The overall sensitivities and specificities of the PanbioTM Ag-RDT with both sampling techniques (nasopharyngeal swab and Nasal Mid-Turbinate swab) obtained during this study meet the WHO criteria for acceptable performance for SARS-CoV-2 Ag-RDTs (minimum requirements of 80% sensitivity and 97% specificity) [11].
Two previous similar studies up to date reported comparable diagnostic assessment for nasal specimen in Panbio Ag-RDT, whose results are consistent with our findings (Table 4). Klein et al. [6] performed a head-to-head comparison of the Panbio Ag-RDT with self-collected NMTS vs. professional-collected NPS samples. The overall sensitivities obtained were 84.4% with NMTS and 88.9% with NPS. In addition, the performance in our study is corroborated by Alqahtani et al. [12]. They evaluated the analytical accuracy of the Panbio Ag-RDT in nasal swab specimens in a group of patients with mild symptoms, with reference to the real time RT-PCR in NPS samplings, and the calculated sensitivity and specificity were 82.1% and 99.1%, respectively. Moreover, other studies targeting different SARS-CoV-2 Ag-RDTs, such as the Standard Q (SD Biosensor), showed similar results in the use of nasal swab sampling regarding efficacy and sensitivity [13–15] (Table 4).
On the other hand, another report by Másia et al. [16] informed that the Panbio test performance was not satisfactory for the nasal swab sampling and its sensitivity was much lower with this type of sample than with the nasopharyngeal one (44.7% vs. 60.5%, respectively). Discrepancies in diagnostic performance of this research compared to our results could be due to multiple reasons, including different clinical and epidemiological scenarios in which both studies were performed; variations in sampling techniques and handling procedures, which depends on health care personal in charge of sample collection in each study, such as different depths of insertion for nasal mid-turbinate swab and swabbing one or both nostrils [17]), inadequate quality of the sample collected, etc. Furthermore, because the readout of this type of assays is by visual inspection, results may be subjective, especially when bands are faint or partial. Additionally, viral kinetics in patients and the moment of the infection that they were swabbed can also be factors that contribute to discrepancies in sensitivities obtained in both studies.
SARS-CoV-2 positive samples by real time RT-PCR that were not detected by the antigen test, presented higher Ct values than those that were positive by the antigen tests with both types of specimen collection, indicating that viral load would be crucial to obtaining concordant results. Besides, when samples were divided into sub-groups according to the Ct values, all samples with RT-PCR Ct-values <25 were detectable by the Panbio Ag-RDT with both sampling techniques. Furthermore, it yielded a greater number of discordant results among patients with Ct≥ 30, consistent with findings of previous studies that assessed the Abbott Ag-RDT in different specimen types, but also with reports of commercial Ag-RDTs from other suppliers [6, 7, 12, 15, 16, 18–28]. These results could also be observed when a double stratification of the samples was performed (according to their Ct values and the presence of symptoms) (S2 Table). There is a substantial drop in sensitivity among the group of symptomatic and asymptomatic individuals with Ct≥30. However, it is important to remark that a grvowing body of evidence indicates that subjects with low viral load testing SARS-CoV-2 positive by RT-PCR and negative by Ag-RDTs have a limited capacity for effective transmission [20, 29].
Information with reference to subjects with asymptomatic SARS-CoV-2 infections is relevant because these individuals have played an important role in the pandemic, comprising a significant portion of the infected population and having a substantial impact on the spreading of this virus (32). Contrary to other similar researches that assessed the Panbio Ag-RDT diagnostic performance in asymptomatic individuals [16, 19, 22, 28, 30–32], our results yielded significantly higher sensitivity among this population with NPS and NMTS sampling techniques. One of the possible reasons that could explain this is that in our study the proportion of participants with Ct values ≥30 was higher in symptomatic than in asymptomatic participants. Besides, all samples of the latter group, except one (with a Ct value of 32.6, the only one with a false positive NMTS Ag-RDT result), are within the range of moderate Ct values (25–29; with a mean Ct of 27.5 for the N gene) and were detected by the rapid antigen test with both sampling techniques. Increasing trend of Panbio test sensitivity in asymptomatic participants with lower Ct values was also observed in previous reports [28, 32, 33]. Alemany et al., (2021) revealed that in their study the Panbio Ag-test sensitivity increased with Cts< 25 and < 30 (100.0% and 98.6%, respectively). Torres et al., [32] also found that the Panbio sensitivity in asymptomatic individuals was directly related to the magnitude of SARS-CoV-2 RNA load in NPS specimens. Another previous recent study by Winkel et al. [33] assessed the performance of the Panbio Ag Test in NPS samples from asymptomatic individuals, and sensitivity ranged from 80.0% to 86.7%, similar to our results. Comparable findings have also been documented in previous studies of a wide variety of different SARS-CoV-2 Ag-RDTs [16, 23, 28, 30, 34–37]. The higher sensitivity of the Panbio Ag-RDT in samples with low Ct values, irrespective of the presence of symptoms, indicates that the test is particularly suitable for identifying individuals who are contagious, and can have a high diagnostic yield for transmission relevant infections with limited false positives. Thus, it can be an appropriate epidemiological surveillance tool for universal screening [16, 23, 34, 35].
Viral load kinetics have been confirmed to be largely similar in asymptomatic and symptomatic patients, and high viral loads are expected to be found in asymptomatic subjects during early infection after exposure with close contacts [18, 33, 36, 38, 39]. Information regarding the establishment of the optimal timeframe for upper respiratory tract collection after exposure to the index case seems crucial to accurately determine the sensitivity of the test [19, 33, 34]. Nevertheless, as a limitation of our study, these data was unavailable. Furthermore, the clinical course of asymptomatic cases could not be followed, and information regarding possible posterior development of symptoms later from diagnosis could not be obtained. Hence, we can not rule out that asymptomatic individuals had actually been tested in the presymptomatic window.
Rapid antigen tests can reliably detect symptomatic SARS-CoV-2 infections in the early phase of disease, within the 7 days from symptom onset, thereby identifying the most contagious persons [31]. In this sense, our results showed a good performance for the Ag test with both sampling techniques in this population group (sensitivities≥ 85.7%). However, the sensitivity was considerably lower (although not statistically significant) when the Ag-RDT with NMTS was performed after the first 7 days from symptom onset, so the diagnostic interpretation in this group of patients should be cautious.
The use of NMTS yielded a lower sensitivity in comparison to NPS, although not statistically significant. However, other variables would counterbalance these differences in the diagnostic performance, and the use of NMTS could prove to be a viable alternative to the reference sampling method: while nasopharyngeal swab is considered more invasive and uncomfortable [27, 40], nasal sampling is of higher tolerance for patients [6], increasing adherence and acceptability of a greater number of individuals, which may improve and maximize the capacity to managing and tracing COVID-19 cases and controlling new outbreaks. Moreover, this technique can also be used for the self-sampling method, which allows more widespread and frequent testing.
Moreover, low and middle-income countries, especially during complex epidemio-logical scenarios, might face socioeconomic insufficiencies and lack of public investment in extensive laboratory structures or highly skilled healthcare professionals to implement molecular techniques. In these regions, with scarce laboratory instrumentations or with real time RT-PCR assays insufficiently available, Ag-RDTs are a highly considerable alternative for cost-reduction and large scales test strategies [22, 24, 30, 41]. In terms of operability and efficiency, these tests are easy-to-administer, allowing performance in decentralized settings and with independence from laboratory facilities. Additionally, these devices are also simple to perform and to read, cost-effective and expeditious, incrementing the pace of testing, identifying and treating people for COVID-19 at the POC.
Conclusions
To conclude, our findings corroborate and confirm the increasing evidence base supporting the use of alternative sampling method, such as Nasal Mid-Turbinate swab, in the context of mass screening strategy during the SARS-CoV-2 pandemic, especially in developing countries or regions with under-resourced health systems. The nasopharyngeal sampling method is often uncomfortable and invasive for patients. Our findings reveal that Ag-RDTs with NMTS specimens might be recognized as a reasonable, valuable and reliable diagnostic tool for widespread utilization, particularly among individuals in the first stage of disease with high viral loads. However, these tests should be interpreted cautiously depending on the clinical and epidemiological context, and they might be used in addition to real time RT-PCR as part of the testing strategies for COVID- 19.
Supporting information
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
We acknowledge the work of the team of healthcare professionals for their support on the testing site.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Corman V, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guglielmi G. The explosion of new coronavirus tests that could help to end the pandemic. Nature. 2020;7817(583):506–9. doi: 10.1038/d41586-020-02140-8 [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. Emergency use listing procedure. World Heal Organ. 2020;(January):1–62. [Google Scholar]
- 4.Wang H, Liu Q, Hu J, Zhou M, Yu MQ, Li KY, et al. Nasopharyngeal Swabs Are More Sensitive Than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load. Front Med. 2020;7(June):1–8. doi: 10.3389/fmed.2020.00334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: A systematic review and meta-analysis. J Clin Microbiol. 2021;59(5):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klein JAF, Krüger LJ, Tobian F, Gaeddert M, Lainati F, Schnitzler P, et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Med Microbiol Immunol [Internet]. 2021;210(4):181–6. Available from: doi: 10.1007/s00430-021-00710-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinnes J, Deeks JJ, Adriano A, Berhane S, Davenport C, Dittrich S, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;2020(8). doi: 10.1002/14651858.CD013705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbot. GLOBAL POINT OF CARE [Internet]. 2021. p. https://www.globalpointofcare.abbott/es/product-de. Available from: https://www.globalpointofcare.abbott/es/product-details/panbio-covid-19-ag-antigen-test.html.
- 9.Ap-Biotech. DisCoVery SARS-CoV-2RT-PCR Detection Kit Rox [Internet]. 2019. p. https://apbiotech.com.ar/discovery-sars-cov-2rt-pc. Available from: https://apbiotech.com.ar/discovery-sars-cov-2rt-pcr-detection-kit-rox-apb-cov19r2.html.
- 10.Xunta de Galicia (Consellería de Sanidade—Servizo Galego de Saúde). Epidat 3.1. 2006. p. https://www.sergas.es/Saude-publica/Epidat-3-1-des.
- 11.World Health Organization (WHO). Antigendetection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays [Internet]. Interim Guidance. 2020. Available from: https://www.who.int/publications/i/item/antigen-detection-in-thediagnosis-of-sars-cov-2infection-using-rapid-immunoassays. [Google Scholar]
- 12.Alqahtani M, Abdulrahman A, Mustafa F, Alawadhi AI, Alalawi B, Mallah SI. Evaluation of Rapid Antigen Tests Using Nasal Samples to Diagnose SARS-CoV-2 in Symptomatic Patients. Front Public Heal. 2022;9(January):1–8. doi: 10.3389/fpubh.2021.728969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur Respir J. 2021;57(4):3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nikolai O, Rohardt C, Tobian F, Junge A, Corman VM, Jones TC, et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: does localisation or professional collection matter? Infect Dis (Auckl) [Internet]. 2021;53(12):947–52. Available from: doi: 10.1080/23744235.2021.1969426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindner AK, Nikolai O, Rohardt C, Burock S, Hülso C, Bölke A, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with professional-collected nasal versus nasopharyngeal swab. Eur Respir J. 2021;57(5):3–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masiá M, Fernández-González M, Sánchez M, Carvajal M, García JA, Gonzalo-Jiménez N, et al. Nasopharyngeal Panbio COVID-19 Antigen Performed at Point-of-Care Has a High Sensitivity in Symptomatic and Asymptomatic Patients with Higher Risk for Transmission and Older Age. Open Forum Infect Dis. 2021;8(3):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callesen RE, Kiel CM, Hovgaard LH, Jakobsen KK, Papesch M, von Buchwald C, et al. Optimal insertion depth for nasal mid-turbinate and nasopharyngeal swabs. Diagnostics. 2021. Jul 1;11(7). doi: 10.3390/diagnostics11071257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brümmer LE, Katzenschlager S, Gaeddert M, Erdmann C, Schmitz S, Bota M, et al. Accuracy of novel antigen rapid diagnostics for SARS-CoV-2: A living systematic review and meta-analysis. Vol. 18, PLOS Medicine. 2021. e1003735 p. doi: 10.1371/journal.pmed.1003735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bulilete O, Lorente P, Leiva A, Carandell E, Oliver A, Rojo E, et al. Panbio TM rapid antigen test for SARS-CoV-2 has acceptable accuracy in symptomatic patients in primary health care. J Infect. 2021;82:391–8. doi: 10.1016/j.jinf.2021.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albert E, Torres I, Bueno F, Huntley D, Molla E, Fern?andez-Fuentes MÁ, et al. Field evaluation of a rapid antigen test (PanbioTM COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin Microbiol Infect. 2020;27:472.e7-472e.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gremmels H, Winkel BMF, Schuurman R, Rosingh A, Rigter NAM, Rodriguez O, et al. Real-life validation of the PanbioTM COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine [Internet]. 2021;31:100677. Available from: doi: 10.1016/j.eclinm.2020.100677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenollar F, Bouam A, Ballouche M, Fuster L, Prudent E, Colson P, et al. Evaluation of the panbio COVID-19 rapid antigen detection test device for the screening of patients with COVID-19. J Clin Microbiol. 2021;59(2):2020–2. doi: 10.1128/JCM.02589-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schildgen V, Demuth S, Lüsebrink J, Schildgen O. Limits and opportunities of sars-cov-2 antigen rapid tests: An experienced-based perspective. Pathogens. 2021;10(1):1–7. doi: 10.3390/pathogens10010038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krüger LJ, Gaeddert M, Tobian F, Lainati F, Gottschalk C, Klein JAF, et al. The Abbott PanBio WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2—Evaluation of the accuracy and ease-of-use. PLoS One. 2021;16(5):1–11. doi: 10.1371/journal.pone.0247918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krüttgen A, Cornelissen CG, Dreher M, Hornef MW, Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J Virol Methods. 2021;288(114024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger A, Nsoga MTN, Perez-Rodriguez FJ, Aad YA, Sattonnet-Roche P, Gayet-Ageron A, et al. Diagnostic accuracy of two commercial SARSCoV- 2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS One. 2021;16(3):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lambert-niclot S, Cuffel A, Pape L, Vauloup-fellous C, Morand-joubert L. Evaluation of a Rapid Diagnostic Assay for Detection of SARS-CoV-2 Antigen in Nasopharyngeal Swabs. J Clin Microbiol. 2020;58:e00977–20. doi: 10.1128/JCM.00977-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alemany A, Baró B, Ouchi D, Rodó P, Ubals M, Corbacho-Monné M, et al. Analytical and clinical performance of the panbio COVID-19 antigen-detecting rapid diagnostic test. J Infect. 2021;82(5):188–90. doi: 10.1016/j.jinf.2020.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.CDC. Discontinuation of Transmission-Based Precautions and Disposition of Patients with COVID-19 in Healthcare Settings [Internet]. Interim Guidance. 2020. Available from: https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html#print. [Google Scholar]
- 30.Baro B, Rodo P, Ouchi D, Bordoy AE, Amaro Saya EN, Salsench S V, et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: a head-to-head benchmark comparison. J Infect. 2021;82:269–75. doi: 10.1016/j.jinf.2021.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linares M, Pérez-Tanoira R, Carrero A, Romanyk J, Pérez-García F, Gómez-Herruz P, et al. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J Clin Virol. 2020;133(104659). doi: 10.1016/j.jcv.2020.104659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torres I, Poujois S, Albert E, Colomina J, Navarro D. Evaluation of a rapid antigen test (PanbioTM COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin Microbiol Infect. 2020;636(January):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winkel B, Schram E, Gremmels H, Debast S, Schuurman R, Wensing A, et al. Screening for SARS-CoV-2 infection in asymptomatic individuals using the Panbio COVID-19 antigen rapid test (Abbott) compared with RT-PCR: a prospective cohort study. BMJ Open. 2021;11(10):e048206. doi: 10.1136/bmjopen-2020-048206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuit E, Veldhuijzen IK, Venekamp RP, Van Den Bijllaardt W, Pas SD, Lodder EB, et al. Diagnostic accuracy of rapid antigen tests in asymptomatic and presymptomatic close contacts of individuals with confirmed SARS-CoV-2 infection: Cross sectional study. BMJ. 2021;374. doi: 10.1136/bmj.n1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiyasu Y, Takeuchi Y, Akashi Y, Kato D, Kuwahara M, Muramatsu S, et al. Prospective analytical performance evaluation of the QuickNaviTM-COVID19 Ag for asymptomatic individuals. J Infect Chemother. 2021;27(January):1489–92. doi: 10.1016/j.jiac.2021.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wachinger J, Olaru ID, Horner S, Schnitzler P, Klaus Heeg, Denkinger CM. The potential of SARS-CoV-2 antigen-detection tests in the screening of asymptomatic persons. Clin Microbiol Infect. 2021;(January). doi: 10.1016/j.cmi.2021.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Favresse J, Gillot C, Oliveira M, Cadrobbi J, Elsen M, Eucher C, et al. Head-to-head comparison of rapid and automated antigen detection tests for the diagnosis of SARS-CoV-2 infection. J Clin Med. 2021;10(2):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee S, Kim T, Lee E, Lee C, Kim H, Rhee H, et al. Clinical Course and Molecular Viral Shedding among Asymptomatic and Symptomatic Patients with SARS-CoV-2 Infection in a Community Treatment Center in the Republic of Korea. JAMA Intern Med. 2020;180(11):1447–52. doi: 10.1001/jamainternmed.2020.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones TC, Biele G, Mühlemann B, Veith T, Schneider J, Beheim-Schwarzbach J, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science (80-). 2021;373(6551). doi: 10.1126/science.abi5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagura-ikeda M, Imai K, Tabata S, Miyoshi K, Murahara N, Mizuno T, et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription–Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J Clin Microbiol. 2020;58(9):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pollock NR, Lee F, Ginocchio CC, Yao JD, Humphries RM. Considerations for Assessment and Deployment of Rapid Antigen Tests for Diagnosis of Coronavirus Disease 2019. Open Forum Infect Dis. 2021;8(6):1–4. doi: 10.1093/ofid/ofab110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.