Abstract
The SARS-CoV-2 Omicron variant can escape neutralization by vaccine-elicited and convalescent antibodies. Memory B cells (MBCs) represent another layer of protection against SARS-CoV-2, as they persist after infection and vaccination and improve their affinity. Whether MBCs elicited by mRNA vaccines can recognize the Omicron variant remains unclear. We assessed the affinity and neutralization potency against the Omicron variant of several hundred naturally expressed MBC-derived monoclonal IgG antibodies from vaccinated COVID-19-recovered and -naive individuals. Compared with other variants of concern, Omicron evaded recognition by a larger proportion of MBC-derived antibodies, with only 30% retaining high affinity against the Omicron RBD, and the reduction in neutralization potency was even more pronounced. Nonetheless, neutralizing MBC clones could be found in all the analyzed individuals. Therefore, despite the strong immune escape potential of the Omicron variant, these results suggest that the MBC repertoire generated by mRNA vaccines still provides some protection against the Omicron variant in vaccinated individuals.
Keywords: Omicron, SARS-CoV-2, COVID-19, mRNA vaccine, memory B cells, MBC, variants, VOC, B cells, germinal center, affinity
Graphical abstract
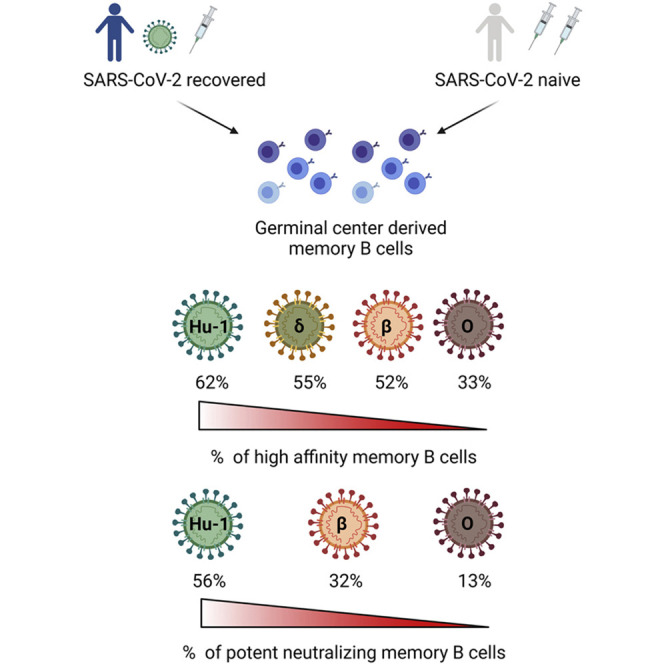
Whether memory B cells (MBCs) elicited by SARS-CoV-2 mRNA vaccines can recognize the Omicron variant remains unclear. Sokal et al. show that compared with other variants of concern, Omicron evaded recognition and neutralization by a larger proportion of MBC-derived antibodies elicited after infection and/or vaccination. Nonetheless, Omicron-neutralizing MBC clones could be found in the repertoire of all the analyzed individuals.
Introduction
The Omicron variant of SARS-CoV-2 has overcome the previously dominant Delta lineage in most countries, suggesting a strong selective advantage. The spike protein of SARS-CoV-2 Omicron harbors 32 mutations as compared with the ancestral strain (Hu-1) originally identified in Wuhan, with particular hotspots of mutations in the angiotensinogen-converting enzyme 2 (ACE2) receptor-binding domain (RBD) (15-amino-acid substitutions) and in the N-terminal domain (NTD) (3 deletions, 1 insertion, and 4 substitutions). Of particular concern, the Omicron variant displays not only key mutations previously associated with immune escape (K417N, E484A, and T478K in the RBD) or enhanced infectivity (N501Y, P681H) but also numerous mutations rarely detected in previous variants. SARS-CoV-2 Omicron may have thus emerged after extensive selection based on beneficial combinatorial effects, as was predicted in silico for the Q498R mutation, for example (Zahradník et al., 2021). The overall mutational profile of the Omicron variant thus suggests both increased immune escape and increased infectivity.
Despite sizable immune evasion by some of the previous variants of concern (VOCs), SARS-CoV-2 mRNA vaccines have so far maintained strong protection in recently vaccinated individuals due to an initially broad and strong serum immunoglobulin G (IgG) response. This response is nonetheless waning with time. We and others have been able to show that SARS-CoV-2-specific memory B cells (MBCs) represent a potent layer of additional immune protection (Dugan et al., 2021; Gaebler et al., 2021; Rodda et al., 2021; Sokal et al., 2021a, 2021b). MBCs not only persist after infection but evolve and mature over several months by the progressive acquisition of somatic mutations in their variable region genes to improve affinity through an ongoing germinal center response (Gaebler et al., 2021; Rodda et al., 2021; Sokal et al., 2021a, 2021b). MBCs display a diverse repertoire, allowing for an adaptive response upon re-exposure to the pathogen, especially in the case of variants (Purtha et al., 2011; Weisel and Shlomchik, 2017). Upon restimulation, either in the context of natural infection or vaccinal boost, MBCs can rapidly differentiate into the plasma cell lineage, secreting the diverse array of high-affinity antibodies contained in their repertoire (Purtha et al., 2011; Weisel and Shlomchik, 2017). A deep analysis of the repertoire of vaccinated individuals has so far suggested that a sizable proportion of such MBCs is able to neutralize all VOCs up to the Beta variant (Sokal et al., 2021b; Wang et al., 2021). Recent reports demonstrate that SARS-CoV-2 Omicron escapes vaccine-elicited antibodies or antibodies from SARS-CoV-2 recovered sera to a large extent (Cameroni et al., 2022; Carreño et al., 2022; Dejnirattisai et al., 2022; Garcia-Beltran et al., 2022; Hoffmann et al., 2022; Muik et al., 2022; Planas et al., 2022). Yet, the intrinsic capacity of the MBC pool to recognize the Omicron variant remains largely unexplored.
Results
Omicron RBD evades recognition from a large proportion of MBC-derived monoclonal antibodies from mRNA-vaccinated individuals
We recently performed an in-depth characterization of over 400 single-cell sorted and cultured RBD-specific MBCs isolated from 8 vaccinated SARS-CoV-2 recovered and 3 vaccinated naive individuals (Sokal et al., 2021b). Our analyses included affinity measurements against VOCs and variants of interest (VOIs) (B.1.1.7, Alpha; B.1.351, Beta; P.1, Gamma; B.1.617.2, Delta; and B.1.617.1 Kappa) and neutralization potency against the D614G and Beta SARS-CoV-2 variants. Selected individuals were part of two longitudinal cohorts. 8 SARS-CoV-2-recovered COVID-19 patients (4 severe [S-CoV] and 4 mild [M-CoV] patients) were selected from the MEMO-COV-2 cohort (Sokal et al., 2021a), which is constituted of patients infected during the first wave in France and longitudinally followed up since, including after their vaccine boost. Additionally, 3 vaccinated SARS-CoV-2-naive subjects were selected from a cohort of healthcare workers, with no clinical history of COVID-19 and no serological evidence of previous SARS-CoV-2 infection. All subjects were vaccinated with the BNT162b2 vaccine as part of the French vaccination program. All the individuals were then sampled for circulating MBCs 7 days and 2 months after their vaccine boost (Sokal et al., 2021b). In all the individuals, we detected a sizable fraction of MBCs encoding antibodies with high affinity and neutralizing potential against all the tested VOCs, with the Beta variant showing the largest extent of immune escape at the time. This suggested that MBCs elicited by prior infection or vaccination would be able to provide an efficient secondary layer of protection in case of the waning of the serological protective antibodies or escape of a novel SARS-CoV-2 variant from this antibody pool.
To test whether such a conclusion still holds true in the context of the newly emerging B.1.1.529 (Omicron) variant, we generated a recombinant Omicron RBD harboring the 15 described mutations, including N501Y, K417T/N, E484K/Q/A, and T478K found in other VOCs and additional mutations G339D, S371L, S373P, S375F, N440K, G446S, S477N, Q493R, G496S, Q498R, and Y505H (Figures 1A and 1B). All the previously described naturally expressed monoclonal IgGs from single-cell culture supernatants of MBCs were assayed anew using biolayer interferometry (BLI) to assess their affinity against the Omicron RBD, and 313 monoclonal supernatants passed quality controls. As previously reported, monoclonal antibodies encoded by MBCs isolated from vaccinated COVID-19-recovered patients and vaccinated naive patients contained a vast majority of high-affinity binders against the ancestral Hu-1 RBD, among which ∼50% retained high affinity (KD <10−9 M) against the Beta and Delta RBD (160/312 and 170/306, respectively). This proportion further decreased against the Omicron RBD to 33% (102/313; Figures 1C and 1D). Importantly, ∼40% of the clones showed no detectable or low affinity (KD>10−8 M) to the Omicron RBD, in both vaccinated recovered and vaccinated naive individuals, which is in line with the prediction that Omicron accumulated numerous mutations associated with antibody evasion. Similar conclusions could be drawn when focusing only on the highly mutated antibodies found in the recovered individuals (Figures 2A and 2B). By contrast, a few clones (n = 14) displayed ultra-high affinity toward Omicron (KD < 10−10 M). Overall, this suggests that the result of the affinity maturation process, which takes place in the germinal centers of both infected and vaccinated individuals and selects over time for a narrow range of high-affinity clones (KD < 10−9 M) against the ancestral Hu-1 RBD (Figures 2A–2C), is greatly altered in the context of such a highly mutated variant like Omicron.
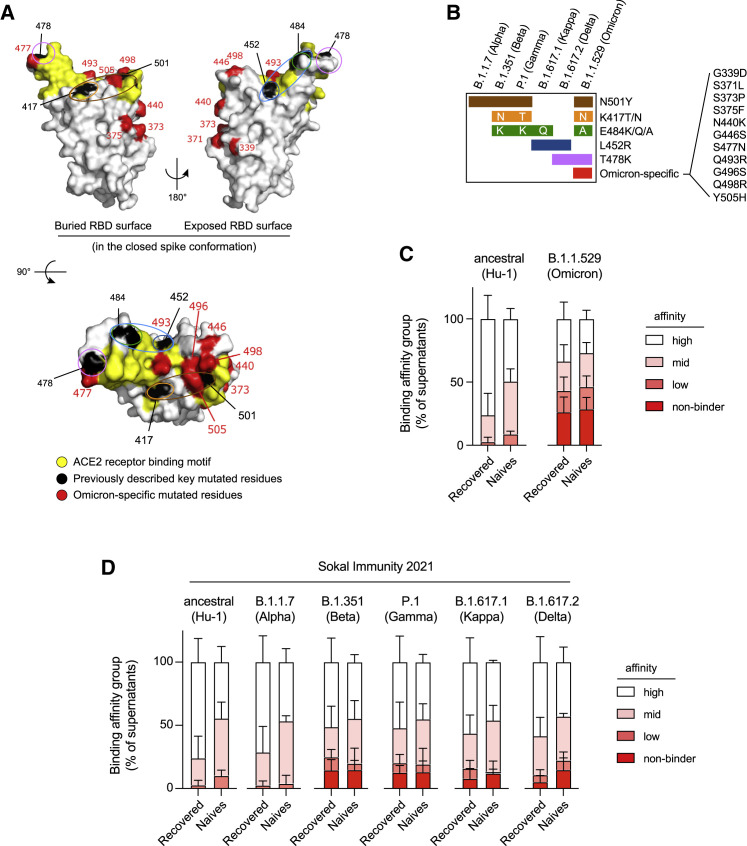
Figure 1.
The memory B cell pool of vaccinated individuals contains a reduced frequency of high-affinity clones against the Omicron RBD
(A) RBD (extracted from the PDB:6XR8 spike protein trimer structure) shown in three orthogonal views with the ACE2-receptor-binding motif highlighted in yellow and the residues found mutated in at least one of the Alpha, Beta, Gamma, Kappa, or Delta variants (L452, K417, T478, E484, and N501) highlighted in black. Residues specifically mutated in Omicron (G339, S371, S373, N440K, G446, S477, Q493, G496, Q498, and Y505H) are highlighted in red. Single or groups of mutations predicted as key binding residues for particular antibodies are further highlighted by colored ovals according to the color scheme used in (B) and Figure 2B.
(B) The distribution of known mutations in the RBD domain between B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.1.529 (Omicron) SARS-CoV-2 variants.
(C) Histograms showing the binding affinity distribution of monoclonal antibodies from single-cell culture supernatants of RBD-specific MBCs isolated from vaccinated SARS-CoV-2 recovered (n = 225) and vaccinated naive donors (n = 88) against the ancestral (Hu-1) RBD and the B.1.1.529 (Omicron) RBD variant, defined as follows: high (KD < 10−9 M), mid (10−9 ≤ KD < 10−8 M), and low (10−8 ≤ KD < 10−7).
(D) Histograms showing the binding affinity distribution of monoclonal antibodies from the same single-cell culture supernatants against the ancestral (Hu-1) RBD and the B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Kappa), B.1.617.2 (Delta), and B.1.1.529 (Omicron) RBD variants, defined as follows: high (KD < 10–9 M), mid (10−9 ≤ KD < 10–8 M), and low (10−8 ≤ KD < 10–7). The clones with KD ≥ 10−7 reflecting an undetectable binding using the BLI were defined as “non-binders.” All the data from (D) come from previously published affinity measurements (Sokal et al., 2021b), whereas all the data in (C), including the Hu-1 RBD measurement, represent new measurements on these supernatants. (C and D) Bars indicate mean ± SEM. See also Table S1.
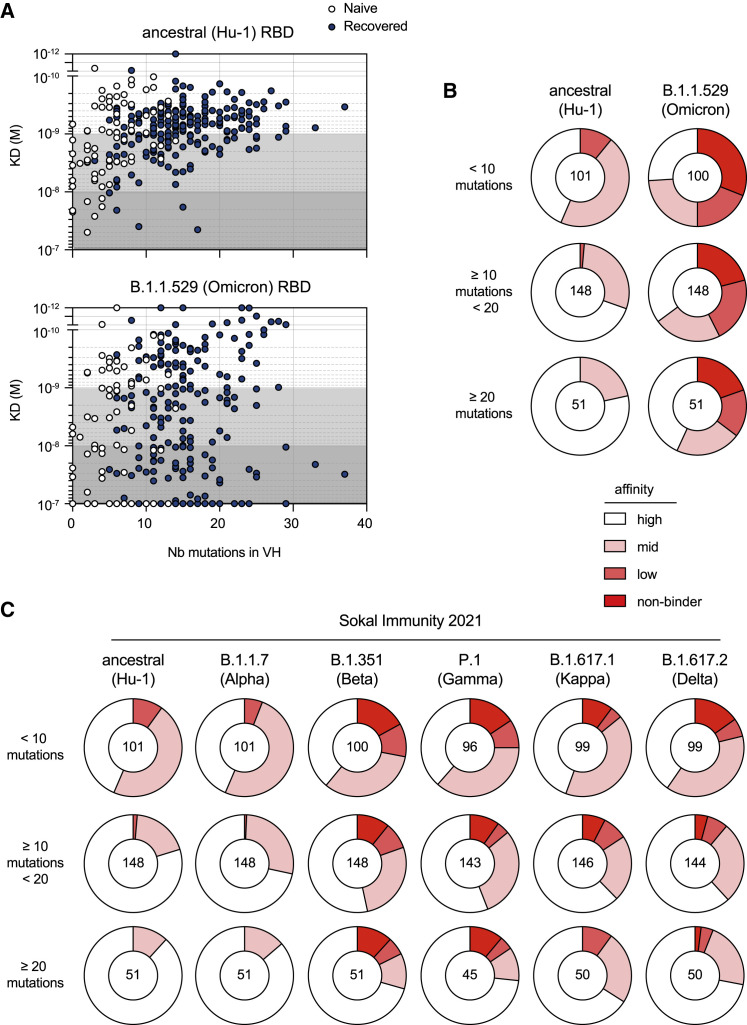
Figure 2.
Omicron RBD evades a large share of Hu-1 selected hypermutated and high-affinity memory B cell-derived antibodies
(A) The measured KD (M) against the ancestral (Hu-1) (top panel) or B.1.1.529 (Omicron) RBD (bottom panel) versus the number of VH mutations for all the tested monoclonal antibodies from the single-cell culture supernatants of the RBD-specific MBCs isolated from the vaccinated SARS-CoV-2 recovered (n = 225) and vaccinated naive donors (n = 75) with available VH sequence from the SARS-CoV-2 recovered (dark blue) and naive (white) donors (the Spearman correlations for all sequences are as follow: VH mutation/Hu-1 KD: r = 0.3791, p < 0.0001; VH mutation/B.1.1.529 KD: r = 0.1597, p = 0.0058).
(B and C) (B) Pie charts showing the binding affinity distribution of all tested monoclonal antibodies against the ancestral (Hu-1) RBD and B.1.1.529 (Omicron) RBD variants, and (C) pie charts showing the binding affinity distribution of the same supernatants against the ancestral (Hu-1), B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Gamma), and B.1.617.2 (Delta) RBD variants, when available. In both cases, the monoclonal antibodies are grouped according to their overall number of mutations as follows: low (<10 mutations, upper panel), intermediate (<20 and ≥10, mid panel), or high VH mutation numbers (≥20, lower panel). All the data from (C) come from previous affinity measurements (Sokal et al., 2021b), whereas all the data in (B), including the Hu-1 RBD measurement, represent new measurements on these supernatants.
(B and C) The affinity groups are defined in Figure 1C and the numbers at the center of each pie chart indicate the total number of tested monoclonal antibodies in each group. See also Table S1.
Omicron-specific RBD mutations expand their overall escape of memory B cell-derived antibodies
A direct comparison of binding affinities toward the ancestral Hu-1 RBD and the Omicron RBD variant for all antibodies extensively characterized against other variants showed that ∼50% (153/310) of the monoclonal antibodies had reduced binding to the Omicron RBD compared with Hu-1 RBD (Omicron/Hu-1 RBD equilibrium dissociation constant [KD] ratio > 2; Figure 3A). Additional two-by-two comparisons of the binding affinities toward the Hu-1 and the previous RBD variants for these Omicron-affected antibodies further suggested that a large fraction of them is uniquely affected by the Omicron variant (Figure 3A). The distribution of mutations in the previous RBD variants (Figures 1A and 1B) had allowed us to predict the identity of key binding amino acid residues within the RBD for 116 MBC-derived monoclonal antibodies among the 139 affected by at least one of the previous RBD variants in the 310 antibodies included in this analysis (Figure 3B; Sokal et al., 2021b). 98 of these 139 MBC-derived monoclonal antibodies, including most of the ones recognizing the E484, K417, and N501 receptor-binding motif (RBM) residues, appeared to be also affected in their recognition of the Omicron variant. In addition, 55 out of the 171 previously unaffected antibodies appeared to be selectively affected by the new mutated residues uniquely displayed by Omicron (Omicron-specific; Figure 3B), though we cannot predict the role of each individual mutation in this context of multiple co-selected mutations in the Omicron RBD. The clones recognizing the Omicron-specific residues were not enriched for the recurrent and convergent antibody rearrangements observed in convalescent and vaccinated individuals (IGHV1-2, IGHV1-69, IGHV3-53, and IGHV3-66; Figure 3C) (Barnes et al., 2020). Therefore, Omicron-specific mutations appear to affect a more diverse range of VH recognizing the RBD than other VOCs and VOIs do. A few clones predicted to bind the L452 residue based on their B.1.617.1 and B.1.617.2 recognition profiles were also affected in their binding to B.1.1.529, despite the absence of this mutation in this variant. This could suggest the recognition of multiple residues (as seen for the E484/L452 and N501/K417 residues) or more complex structural changes resulting from the large number of mutations included in the Omicron’s RBD. Finally, when analyzed in the context of our previous knowledge of the neutralization potency of 163 out of the 313 monoclonal antibodies tested by BLI (Sokal et al., 2021b), the loss of affinity against Omicron RBD appeared to be mostly restricted to the potent neutralizers of the D614G SARS-CoV-2 (Figure 3D). As previously described for the Beta variants, this result highlights the selective pressure imposed on SARS-CoV-2 by such neutralizing antibodies. Similarly, the antibodies that still displayed a potent neutralization potency against the Beta SARS-CoV-2 appeared to be selectively targeted for additional loss of affinity by the Omicron-specific mutations not included in the Beta variant (Figure 3E).
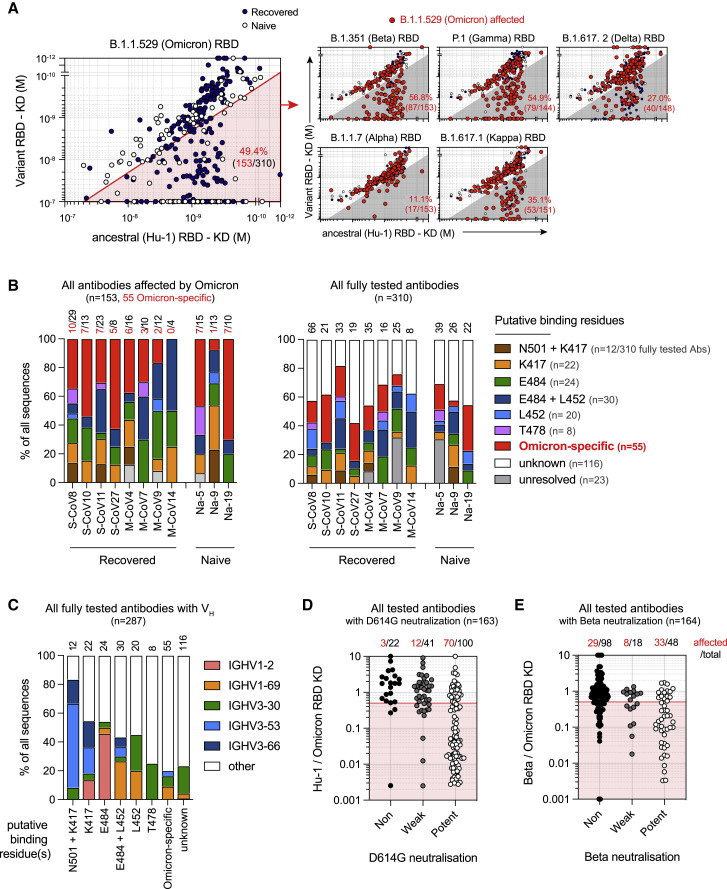
Figure 3.
Omicron-specific RBD mutations expand their overall escape of memory B cell-derived antibodies
(A) Dot plot representing the KDs for B.1.1.529 (Omicron) RBD versus ancestral (Hu-1) RBD for all tested monoclonal antibodies from the SARS-CoV-2 recovered (dark blue dots) and naive donors (white dots). The red shaded area indicates monoclonal antibodies with at least two-fold increased KD for B.1.1.529 than for Hu-1 (termed “B.1.1529-affected antibodies” herein) (left panel). Dot plots representing KDs against the B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Kappa), and B.1.617.2 (Delta) RBD versus the ancestral (Hu-1) RBD for all tested antibodies (right panels). B.1.1.529-affected antibodies are highlighted as larger size red dots (corresponding to clones present in the red sector in the left panel). The percentages indicate the proportion of B.1.1.529-affected monoclonal antibodies also affected by the indicated RBD variant (gray zone).
(B) The frequencies of antibodies targeting one of the predicted essential binding residue groups in the Omicron-affected antibodies (left panel) and in all the tested antibodies (right panel), as defined by RBD variants recognition profile in BLI, among all tested antibodies for each of the 11 individuals from whom memory B cells were assayed. The numbers of monoclonal antibodies for each donor are indicated on top of each histogram in black. The number of antibodies affected by Omicron-specific mutations is detailed in red. S-CoV, patients who recovered from severe COVID-19; M-CoV, patients who recovered from mild COVID-19; Na, patients naive from COVID-19.
(C) The proportion of IGHV1-2, IGHV1-69, IGHV3-30, IGHV3-53, and IGHV3-66 usage among all the tested monoclonal antibodies with available VH sequence and grouped based on their predicted essential binding residues, as defined in (B). The numbers of tested monoclonal antibodies from all donors are indicated on top of each histogram.
(D and E) The ratio of Hu-1 over B.1.1.529 (Omicron) RBD KD (D) or B.1.351 (Beta) over B.1.1.529 (Omicron) KD for all the monoclonal antibodies tested, grouped based on their neutralization potency against D614G (D) or B.1.351 SARS-CoV-2 (E) (refer to Sokal et al., 2021b). The numbers on top indicate the numbers of monoclonal antibodies with a [KD for Hu-1 RBD / KD for B.1.1.529 RBD] ratio <0.5 (D) or a [KD for B.1.351 / KD for B.1.1.529 RBD] ratio <0.5 (E). Values above 10 or below 0.001 were plotted on the axis. See also Table S1.
Omicron SARS-CoV-2 evades neutralization from most but not all MBC-derived monoclonal antibodies from mRNA-vaccinated individuals
To better characterize the remaining neutralizing potential of MBC-derived antibodies against the Omicron variant, we next tested 253 supernatants in an in vitro focus reduction neutralization assay against authentic D614G and B.1.1.529 (Omicron) SARS-CoV-2 viruses. As previously described, a majority of RBD-specific MBC-derived antibodies displayed some neutralization potency against the D614G SARS-CoV-2 strain in all the donor groups (Figures 4A and 4B) and all the tested individuals (Figure 4C), except in one of the 3 naive donors. These numbers were strongly reduced in all the individuals when looking at Omicron SARS-CoV-2 neutralization (Figures 4A and 4B), with notably less than 20% of potent or weak neutralizing antibodies in most vaccinated naive donors and M-CoV patients (Figure 4C). Nonetheless, we could detect potent neutralizing antibodies in 10 out of the 11 tested donors (Table S1) and neutralizing antibodies in all of them (Figure 4C). Additionally, 3 out of the 4 S-CoV patients harbored more than 20% of potent neutralizers against Omicron SARS-CoV-2. Cross-examining our neutralization data in the light of our RBD affinity measurements further confirmed a strong escape by Omicron SARS-CoV-2 from most antibodies targeting the core RBM epitopes mutated in Omicron (N501, K417, and E484) (Figure 4D). However, in line with the absence of the L452 mutation in Omicron, 6 out the 7 identified potent neutralizing antibodies with predicted binding to this residue retained their potency against Omicron. Additionally, about one-third of the antibodies identified as binding Omicron-specific residues did retain potent neutralization despite clear affinity loss (Figures 4E and 4F). Conversely, it is important to note that none of the non or weak D614G neutralizers acquired potent neutralization potency against Omicron. Five of the 14 ultra-high Omicron binders (KD < 10−10 M for Omicron; Figure 2A) were included in this assay. Two out of these five were non-neutralizers of D614G SARS-CoV-2 to begin with and did not acquire any neutralization activity against Omicron SARS-CoV-2. The other three fell into a group of antibodies (n = 29) that showed the loss of neutralization potency without any detectable reduction in RBD affinity (Figures 4E and 4F). These results are in line with our previous results of neutralization against B.1.351 (Beta) SARS-CoV-2 and most likely result from allosteric changes at the level of the whole spike imposed by mutations outside of the RBD (Sokal et al., 2021b).
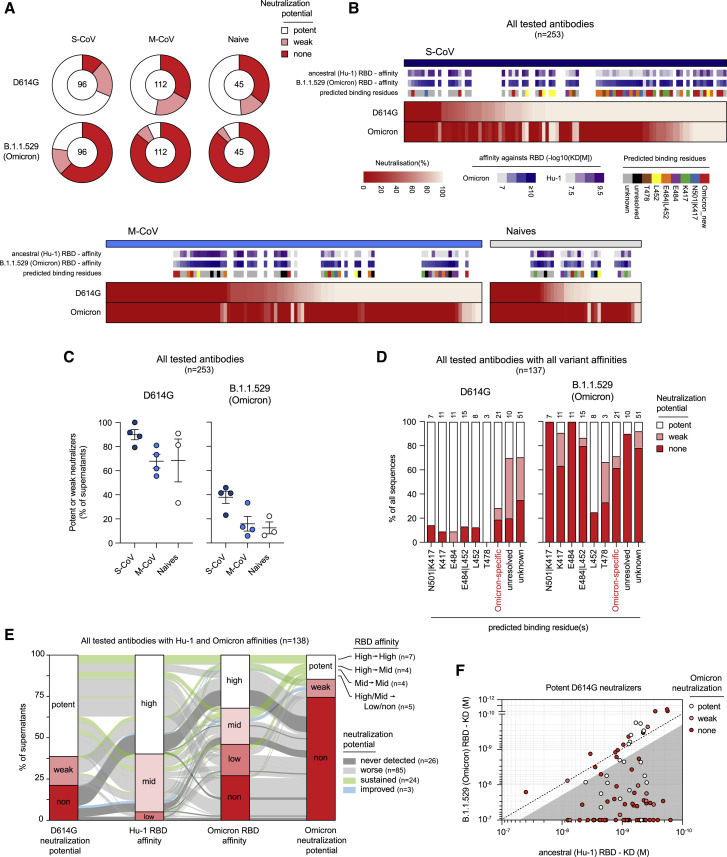
Figure 4.
The Omicron VOC evades neutralization from a large proportion of memory B cell-derived monoclonal antibodies
(A) Pie charts showing the proportion of single-cell culture supernatants of RBD-specific MBCs isolated from SARS-CoV-2 recovered (S-CoV, n = 96; M-CoV, n = 112) and naive donors (n = 45) displaying potent, weak, or no neutralization potency (none) against D614G SARS-CoV-2 and B.1.1.529 (Omicron) SARS-CoV-2 variant. Potent neutralizers are defined as >80% neutralization at 16 nM and weak neutralizers as neutralization between 25% and 80% at 16 nM. None neutralizers are defined as neutralization <25% at 16 nM.
(B) A heatmap showing the in vitro neutralization of D614G SARS-CoV-2 and Omicron variant at 16 nM for all cultured supernatants tested. KD (M) against the ancestral (Hu-1) and B.1.1.529 (Omicron) RBD for tested monoclonal antibodies are represented on top along with predicted binding residues.
(C) The percentage of potent neutralizers against SARS-CoV-2 D614G or variant B.1.1.529 (Omicron) viruses among monoclonal antibodies analyzed for each donor in each group.
(D) The proportion of potent, weak, or non-D614G or B.1.1.529 (Omicron) SARS-CoV-2 neutralizers among all tested monoclonal antibodies, grouped based on their predicted binding residues, as defined in Figure 3B.
(E) A river plot connecting affinity for the ancestral (Hu-1) and B.1.1.529 (Omicron) RBD with neutralization potency for D614G and B.1.1.529 (Omicron) SARS-CoV-2. The binding and neutralizing affinity groups are defined in Figures 1C and 4A. Clones are connected with colored lines. The line colors indicate the evolution of the neutralization potency toward B.1.1.529 (Omicron) SARS-CoV-2 as follows: green indicates potent or weak clones remaining respectively potent or weak; light gray indicates clones with diminished neutralization potency (potent to weak, none, or weak to none); dark gray indicates clones that do not neutralize any of the 2 viruses; and blue indicates clones with improved neutralization toward B.1.1.529 (only none to weak).
(F) KD (M) for the B.1.1.529 (Omicron) RBD versus the ancestral (Hu-1) RBD for all D614G SARS-CoV-2 potent neutralizers monoclonal antibodies. The dot color indicates the neutralization potency against B.1.1.529 SARS-CoV-2 variant. The gray-shaded area highlights binding-impaired clones against the B.1.1.529 RBD variant as defined in Figure 3A. See also Table S1.
Discussion
Previous analyses of the MBC repertoire elicited after mRNA vaccination in COVID-19 recovered and naive individuals had demonstrated the presence of a sizable pool of high-affinity neutralizing clones against all VOCs prior to Omicron (Cho et al., 2021; Goel et al., 2021; Sokal et al., 2021b). These data suggested that vaccine recalls would be effective at reconstituting protective levels of serum antibodies to prevent infection by VOCs. Our results suggest a strong but incomplete immune escape by the Omicron variant from specific MBCs elicited by natural infection with the D614G SARS-CoV-2 and/or vaccination with mRNA vaccine encoding the ancestral Hu-1 spike. This is consistent with the recent studies on sera collected from individuals who had received three mRNA vaccine doses (Carreño et al., 2022; Dejnirattisai et al., 2022; Garcia-Beltran et al., 2022; Gruell et al., 2022; Hoffmann et al., 2022; Planas et al., 2022), as well as with one recent study of single-cell MBCs from pooled vaccinated SARS-CoV-2-naive individuals (Kotaki et al., 2022). The reduction in the range of 4- to 18-fold in overall serum neutralizing activity against the Omicron variant as compared with the ancestral Hu-1 strain was reported in these studies, although the overall binding potential of such serum antibodies appeared less drastically reduced (Carreño et al., 2022).
Looking at the single MBC level, the unique accumulation of mutations in key amino acid residues within the RBD of the Omicron variant, either shared with previous variants or unique, resulted in a loss of affinity for close to 50% of all the MBCs analyzed in our study. The link between affinity loss and neutralization potency, however, is not as straightforward. On the one hand, a few clones with reduced binding maintained their neutralization potency and represented a large share of the remaining potent Omicron neutralizers (11/20). On the other hand, non-affected binders were enriched in non-neutralizing antibodies against D614G SARS-CoV-2, which remained non-neutralizers against Omicron SARS-CoV-2. Additionally, many non-affected binders with weak or potent D614G neutralization potency did not display any detectable Omicron-neutralization potency, suggesting more complex allosteric effects at the level of the spike (Cerutti et al., 2022; Sokal et al., 2021b; Yin et al., 2022). Overall, of all the antibodies tested for both neutralization and affinity in our study, over 80% of the high- to mid-binder of the Hu-1 RBD displayed some neutralization potency against the D614G SARS-CoV-2 (105/131), but less than 35% of the high- to mid-binder of the Omicron RBD displayed neutralization potency against the Omicron SARS-CoV-2 (24/74). Nonetheless, Omicron-neutralizing antibodies still represented more than 10% of the anti-RBD MBC repertoire for 8 out of the 11 donors in our study and could be detected in all donors (5.3 ± 3.4-fold reduction in the proportion of Omicron neutralizers as compared with D614G). Unlike plasma cells producing serum antibodies, MBCs are endowed with great proliferative potential and, at a rate of one division every 10–12 h, could theoretically compensate for an up to an 8-fold loss in protective clone frequency in less than 2 days. The low frequencies of the available protective MBC clones detected in our study could, therefore, still be sufficient to avoid severe forms of COVID-19, as observed in clinical reports (Nemet et al., 2022).
An important next step will be to characterize the long-term remodeling of the MBC repertoire after an Omicron breakthrough infection in vaccinated individuals and to address whether an Omicron-specific B cell response can be achieved, with the recruitment of naive B cells or the maturation of pre-existing low-affinity MBCs. Answering these questions will provide crucial information regarding the available immune protection against the Omicron or subsequent variants and allow an informed decision as to whether a vaccine boost specifically targeted against VOCs would be of interest in the near future.
Limitations of the study
Potential limitations of our work include a limited number of subjects included in this study for in-depth MBC characterization (n = 11). As such, the observed differences between the donor groups should be interpreted with caution, notably the slightly higher frequency of potent neutralizing antibodies detected in the severe-COVID-19 patient group. Additionally, the samples analyzed were collected early after the first vaccine boost in all individuals. The MBC repertoire of COVID-19-naive vaccinated individuals has been shown to evolve up to 6 months after the boost (Cho et al., 2021; Goel et al., 2021) and it remains to be assessed how the breadth of MBC repertoire will further evolve after an additional vaccine boost with a third dose of the ancestral Hu-1 spike mRNA vaccine (Wang et al., 2022a). Finally, our study was focused on the RBD domain of the SARS-CoV-2 spike protein, which represents the major target of neutralizing antibodies (Tong et al., 2021; Vanshylla et al., 2022). Broadly neutralizing antibodies against other domains of the trimeric spike have been described, notably against the conserved epitopes of the NTD (Wang et al., 2022b). A similar point could be made for memory T cell responses, which so far appear to be less affected by mutations selected by SARS-CoV-2 variants (Goel et al., 2021; Tarke et al., 2022).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibody | ||
| Anti-Human Fc Capture Biosensors | Sartorius | Cat#18-5060 |
| Biological samples | ||
| Monoclonal antibodies from single cell cultured RBD-specific memory B cells | INSERM U1151 – Generated from Sokal et al. 2021b | N/A |
| D614G SARS-CoV-2 virus (hCoV-19/France/GE1973/2020) | Institut Pasteur, CNR Respiratory Viruses (S.Van der Werf) | N/A |
| B.1.1.529 SARS-CoV-2 virus | Institut Pasteur (Olivier Schwartz) | N/A |
| Chemical, peptides, and recombinant proteins | ||
| Ancestral (Hu-1) SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| B.1.1.7 SARS-CoV-2 (Alpha) RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| B.1.351 (Beta) SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| P.1 (Gamma) SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| B.1.617.1 (Kappa) SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| B.1.617.2 (Delta) SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| B.1.1.529 (Omicron) SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| Deposited data | ||
| Clinical data, affinity measurements, neutralization values, VDJ sequencing | Mendeley | https://doi.org/10.17632/wwrxgn65h2.1 (https://data.mendeley.com/datasets/wwrxgn65h2/2) |
| VDJ sequencing | GenBank | BioProject PRJNA819082 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA819082) |
| Software and algorithms | ||
| GraphPad Prism v9 | GraphPad | https://www.graphpad.com |
| R v4.0.2 | R Foundation | https://www.r-project.org |
| RStudio v1.3.1056 | RStudio | https://rstudio.com |
| HT Data analysis software 11.1 | ForteBio | https://www.sartorius.com |
| Adobe Illustrator (CS6) | Adobe | https://www.adobe.com |
| PyMOL Molecular Graphics System, v2.1 | Schrödinger, LLC | https://pymol.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthieu Mahévas (matthieu.mahevas@aphp.fr).
Materials availability
No unique materials were generated for this study.
Experimental model and subject details
Study participants
In this study, we included 11 subjects whose memory B cells had been analyzed as part of two larger cohorts described in Sokal et al. 2021b (4 patients with severe COVID-19 (S-CoV), hospitalized and requiring oxygen, 4 patients with mild COVID-19 (M-CoV), mainly healthcare workers with ambulatory form, and 3 SARS-CoV-2 naive donors (Naive) (Sokal et al. 2021b)). All subjects received the BNT162b2 mRNA vaccine. Naive subject received two doses of vaccine as part of the French vaccination campaign, while SARS-CoV-2 recovered patients received only one dose, in line with French guidelines. All subjects were followed longitudinally after the last vaccine dose (boost) and all time points in this study refer to the date of this boost. Detailed information on the individuals included in this study, including gender and health status, can be found in Table S1.
SARS-CoV-2 infection was defined as confirmed reverse transcriptase polymerase chain reaction (RT-PCR) on nasal swab or clinical presentation associated with typical aspect on CT-scan and/or serological evidence. Naive patients were healthcare workers who had no history of COVID-19 and negative IgG anti-nucleocapsid (and/or Spike) (“Collection Vaccin” [2018-A01610-55, CPP EST-III]). Patients were recruited at the Henri Mondor University Hospital (AP-HP), between March and April 2021. MEMO-COV-2 study (NCT04402892) was approved by the ethical committee Ile-de-France VI (Number: 40-20 HPS), and was performed in accordance with the French law. Written informed consent was obtained from all participants.
Virus strains
The reference D614G strain (hCoV-19/France/GE1973/2020) was supplied by the National Reference Centre for Respiratory Viruses hosted by Institut Pasteur and headed by Sylvie van der Werf. The Omicron strain (B.1.1.529 GISAID ID: EPI_ISL_6794907) was a generous gift from Olivier Schwartz, Institut Pasteur, and was generated as described in (Planas et al., 2022).
Method details
Recombinant protein purification
Construct design
The SARS-CoV-2 Receptor Binding Domain (RBD) was cloned in pcDNA3.1(+) encompassing the Spike (S) residues 331-528, and it was flanked by an N-terminal IgK signal peptide and a C-terminal Thrombin cleavage site followed by Hisx8, Strep and Avi tags. The mutations present on the B.1.1.7 (Alpha, N501Y), B.1.351 (Beta, K417N, E484K, N501Y), P.1 (Gamma, K417T, E484K, N501Y), B.1.617.1 (Kappa, L452R, E484Q), and B.1.617.2 (Delta, L452R, T478K) variants as compared to the ancestral (Hu-1) strain were introduced by PCR mutagenesis using standard methods. B.1.1.529 (omicron variant) RBD plasmid was specifically designed with its mutations (N501Y, K417T/N, E484K/Q/A, T478K, G339D, S371L, S373P, S375F, N440K, G446S, S477N, Q493R, G496S, Q498R and Y505H).
Protein expression and purification
The plasmids coding for recombinant proteins were transiently transfected in Expi293F™ cells (Thermo Fischer) using FectoPRO® DNA transfection reagent (Polyplus), according to the manufacturer’s instructions. The cells were incubated at 37 °C for 5 days and then the culture was centrifuged and the supernatant was concentrated. The proteins were purified from the supernatant by affinity chromatography using His-Trap™ Excel columns (Cytiva) (SARS-CoV-2 RBD). A final step of size-exclusion chromatography (SEC) in PBS was also performed, using a Superdex200 10/300 (Cytiva) for the SARS-CoV-2 RBD.
Single-cell culture
All culture supernatants from single-cell cultured memory B cells used in this study were generated and reported as part as a previous study (see Sokal et al., 2021b). Briefly, single B cells were sorted in 96-well plates containing MS40Llo cells expressing CD40L (kind gift from G. Kelsoe, Crickx et al., 2021; Luo et al., 2009). Cells were co-cultured at 37°C with 5% CO2 during 21 or 25 days in RPMI-1640 (Invitrogen) supplemented with 10% HyClone FBS (Thermo Scientific), 55 μM 2-mercaptoethanol, 10 mM HEPES, 1 mM sodium pyruvate, 100 units/mL penicillin, 100 μg/mL streptomycin, and MEM non-essential amino acids (all Invitrogen), with the addition of recombinant human BAFF (10 ng/ml), IL2 (50 ng/ml), IL4 (10 ng/ml), and IL21 (10 ng/ml; all Peprotech). Part of the supernatant was carefully removed at days 4, 8, 12, 15 and 18 and the same amount of fresh medium with cytokines was added to the cultures. After 21 days of single cell culture, supernatants were harvested and stored at -20°C. Cell pellets were placed on ice and gently washed with PBS (Gibco) before being resuspended in 50 μL of RLT buffer (Qiagen) supplemented with 1% 2-mercaptoethanol and subsequently stored at -80°C until further processing.
ELISA
Total IgG from culture supernatants were detected by home-made ELISA. Briefly, 96 well ELISA plates (Thermo Fisher) were coated with goat anti-human Ig (10 μg/ml, Invitrogen) in sodium carbonate during 1h at 37°C. After plate blocking, cell culture supernatants were added for 1hr, then ELISA were developed using HRP-goat anti-human IgG (1 μg/ml, Immunotech) and TMB substrate (Eurobio). OD450 and OD620 were measured and Ab-reactivity was calculated after subtraction of blank wells.
Single-cell IgH sequencing
Clones whose culture had proven successful (IgG concentration ≥ 1 μg/mL at day 21-25) were selected and extracted using the NucleoSpin96 RNA extraction kit (Macherey-Nagel) according to the manufacturer’s instruction. A reverse transcription step was then performed using the SuperScript IV enzyme (ThermoFisher) in a 14 μl final volume (42°C 10 min, 25°C 10 min, 50°C 60 min, 94°C 5 min) with 4 μl of RNA and random hexamers (Thermofisher scientific). A PCR was further performed based on the protocol established by Tiller et al (Tiller et al., 2008). Briefly, 3.5 μl of cDNA was used as template and amplified in a total volume of 40 μl with a mix of forward L-VH primers (Table S1) and reverse Cγ primer and using the HotStar® Taq DNA polymerase (Qiagen) and 50 cycles of PCR (94°C 30 s, 58°C 30 s, 72°C 60 s). PCR products were sequenced with the reverse primer CHG-D1 and read on ABI PRISM 3130XL genetic analyzer (Applied Biosystems). Sequence quality was verified with the CodonCode Aligner software (CodonCode Corporation).
Computational analyses of VDJ sequences
Processed FASTA sequences from cultured single-cell VH sequencing were annotated using Igblast v1.16.0 against the human IMGT reference database. Clonal cluster assignment (DefineClones.py) and germline reconstruction (CreateGermlines.py) was performed using the Immcantation/Change-O toolkit (Gupta et al., 2015) on all heavy chain V sequences. Sequences that had the same V-gene, same J-gene, including ambiguous assignments, and same CDR3 length with maximal length-normalized nucleotide hamming distance of 0.15 were considered as potentially belonging to the same clonal group. Further clonal analyses on all productively rearranged sequences were implemented in R. Mutation frequencies in V genes were calculated using the calcObservedMutations() function from the Immcantation/SHazaM v1.0.2 R package. VH repartition was calculated using the countGenes() function from the Immcantation/alakazam v1.1.0 R package. Further analyses were implemented in R. Graphics were obtained using the ggplot2 v3.3.5, pheatmap v1.0.12 and ggalluvial v0.12.3 packages.
3D representation of known mutations to the RBD surface
Panel A in Figure 1 was prepared with The PyMOL Molecular Graphics System, Version 2.1 Schrödinger, LLC. The atomic model used for the RBD was extracted from the cryo-EM structure of the SARS-CoV-2 spike trimer (PDB:6XR8; (Cai et al., 2020)).
Affinity measurement using biolayer interferometry (Octet)
All affinity measurements were done using biolayer interferometry assays on the Octet HTX instrument (ForteBio). This high-throughput kinetic screening of supernatants using single antigen concentration has recently been extensively tested and demonstrated excellent correlation with multiple antigen concentration measurements (Lad et al., 2015). Briefly, anti-Human Fc Capture (AHC) biosensors (18-5060) were immersed in supernatants from single-cell memory B cell culture (or control monoclonal antibody) at 25°C for 1800 seconds. Biosensors were equilibrated for 10 minutes in 10x PBS buffer with surfactant Tween 20 (Xantec B PBST10-500) diluted 1x in sterile water with 0.1% BSA added (PBS-BT) prior to measurement. Association was performed for 600 s in PBS-BT with ancestral (Hu-1) or variant RBD at 100nM followed by dissociation for 600s in PBS-BT. Biosensor regeneration was performed by alternating 30s cycles of regeneration buffer (glycine HCl, 10 mM, pH 2.0) and 30s of PBS-BT for 3 cycles. Traces were reference sensor subtracted and curve fitting was performed using a local 1:1 binding model in the HT Data analysis software 11.1 (ForteBio). Sensors with response values (maximum RBD association) below 0.1nm were defined as “non-binders”. For variant RBD non-binding mAbs, sensor-associated data (mAb loading and response) were manually checked to ensure that this was not the result of poor mAb loading. For binding clones, only those with full R2>0.8 were retained for KD reporting and initial prediction of key binding residues. mAbs were automatically defined as affected against a given variant RBD if the ratio of calculated KD value against that RBD variant and the Hu-1 RBD was superior to two. RBD binding groups were defined as: high (KD <10-9 M), mid (10-9 ≤ KD <10-8 M) and low (10-8≤ KD <10-7). Clones with KD ≥10-7 reflecting an undetectable binding using the BLI were defined as “non-binders”.
A first set of affinity measurements against the ancestral (Hu-1), B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.1 (Kappa) and B.1.617.2 (Delta) RBD were performed and reported as part of a previous study (see Sokal et al., 2021b). Affinity measurement against the B.1.1.529 Omicron RBD was performed in the same way, using new biosensors. As an internal quality control step to ensure the correct conservation of the supernatants a new measurement of the Hu-1 RBD affinity was also done and correspond to the Hu-1 RBD affinities reported in all main figures (referred to as Hu-1 RBD-2 in Table S1). Only monoclonal antibodies displaying correct loading (>0.4nm) and binding (>0.1nm) in our latest Hu-1 RBD affinity measurement (313/414) were retained in the analysis presented in this report.
Key binding residues predictions were extracted from our previous study (see Sokal et al.,2021b). Briefly, prediction were simply made based on mutations repartition in the different variants tested prior to the Omicron, for example mAbs affected only by B.1.351 and P.1 variants were predicted to bind to the K417 residue. Two exceptions to these simple rules were made: 1/ mAbs affected by the B.1.1.7, B.1.351 and P.1 variants were initially labeled as binding to the N501 residue but the ratios of KD values against the B.1.351 and P.1 RBD variants and the B.1.1.7 RBD variants were further calculated. All mAbs with ratio superior to two for these two combinations were labeled as binding both the N501 and K417 residues, as previously described for RBS-A type of anti-RBD mAbs (Yuan et al., 2021); 2/ mAbs affected by the B.1.351, P.1, B.1.617.1 and B.1.617.2 variants, but not the B.1.1.7 variant, were labeled as binding both the E484 and L452 residues based on reported data in the literature for RBS-B/C antibodies (Starr et al., 2021; Yuan et al., 2021). As part of this new analysis, Omicron-specific monoclonal antibodies that had not been previously defined as binding one of the residues shared by previous variants analyzed were defined as binding Omicron-specific mutated residues (Figure 2B). Sensors with missing values, KD ratio close to the cut-off or affinity profile not consistent with mutation distribution in tested variant RBD were manually inspected to resolve binding residues attribution, leaving 23 antibodies with an unresolved profile (including 19 previously predicted to bind the T478 residue based on the sole B.1.617.2 (Delta) RBD affinity measure but not affected in their binding to the B.1.1.529 (Omicron) RBD harboring the same mutation).
Virus neutralization assay
Virus neutralization against the D614G and B.1.351 (Beta) SARS-CoV-2 in Figures 2D and 2E were measured and reported as part of a previous study, and reanalyzed for the antibodies included in this study (respectively 163 and 164 out of the 313 antibodies with known affinities against Omicron, see Sokal et al., 2021b). For neutralization data reported in Figure 3, 253 monoclonal antibodies from memory B cells (including 137 with known affinities against various VOCs) were tested against B.1.1.529 (Omicron) SARS-CoV-2 and anew tested against D614G SARS-CoV-2 as an internal quality control (reported as D614G_2 in Table S1). Virus neutralization was evaluated by a focus reduction neutralization test (FRNT). Vero E6 cells were seeded at 2x104 cells/well in a 96-well plate 24h before the assay. Two-hundred focus-forming units (ffu) of each virus were pre-incubated with B-cell clone supernatants for 1hr at 37°C before infection of cells for 2hrs. The virus/antibody mix was then removed and foci were left to develop in presence of 1.5% methylcellulose for 2 (D614G) and 3 (Omicron) days. Cells were fixed with 4% formaldehyde and foci were revealed using a rabbit anti-SARS-CoV-2 N antibody (gift of Nicolas Escriou) and anti-rabbit secondary HRP-conjugated secondary antibody. Foci were visualized by diaminobenzidine (DAB) staining and counted using an Immunospot S6 Analyser (Cellular Technology Limited CTL). B-cell culture media and supernatants from RBD negative clones were used as negative control.
Percentage of virus neutralization was calculated as (100 - ((#foci sample / #foci control)∗100)). Culture supernatants were tested at 16 nM for each sample and each virus. Potent neutralizers were defined as >80% neutralization at 16 nM, weak neutralizer as neutralization between 25% and 80% at 16 nM. Clone with neutralization <25% at 16 nM were defined as non-neutralizing.
Quantification and statistical analysis
Statistical analyses were all performed using GraphPad Prism 9.0 (La Jolla, CA, USA).
Acknowledgments
We thank Garnett Kelsoe for providing the human cell culture system, together with invaluable advice; A. Boucharlat and the Chemogenomic and Biological Screening Core Facility headed by F. Agou, as well as P. England and the Molecular Biophysics Core Facility at the Institut Pasteur, Paris, France for their support during the course of this work; Sébastien Storck, Lucie Da Silva, and Sandra Weller for their advice and support; the physicians, Constance Guillaud, Raphael Lepeule, Frédéric Schlemmer, Elena Fois, Henri Guillet, Nicolas De Prost, and Pascal Lim, whose patients were included in this study; and Florence Guivel-Benhassine and Olivier Schwartz from the Institut Pasteur for providing authentic SARS-CoV-2 B.1.1.529 virus.
This work was initiated by a grant from the Agence Nationale de la Recherche and the Fondation pour la Recherche Médicale (ANR and MEMO-COV-2-FRM) and funded by the Fondation Princesse Grace, an ERC Advanced Investigator Grant (B-response), and a grant from the French Ministry of Health (Soutien Exceptionnel à la Recherche Clinique 2022, CAPNET [Comité ad-hoc de pilotage national des essais thérapeutiques et autres recherches]). Assistance Publique – Hôpitaux de Paris (AP-HP, Département de la Recherche Clinique et du Développement) was the promotor and the sponsor of MEMO-COV-2. Work in the Unit of Structural Virology was funded by Institut Pasteur, Urgence COVID-19 Fundraising Campaign of Institut Pasteur. A.S. was supported by a Poste d’accueil from INSERM, I.F. by a fellowship from the Agence Nationale de Recherches sur le Sida et les Hépatites Virales (ANRS), M.B. by a CIFRE fellowship from the Association Nationale de la Recherche et de la Technologie (ANRT), and A.D.L.S by a SNFMI fellowship. P.B. acknowledges funding from the French National Research Agency grant ANR-14-CE16-0011 project DROPmAbs, the Institut Carnot Pasteur Microbes et Santé (ANR 11 CARN 0017-01), the Institut Pasteur, and the Institut National de la Santé et de la Recherche Médicale (INSERM).
Author contributions
Conceptualization, P.C., A.S., J.-C.W., C.-A.R., and M. Mahévas; data curation, P.C., A.S., M.B., G.B.-S., and I.A.; formal analysis, A.S., P.C., M.B., G.B.-S., I.A., M.B.-A., A.D.L.S., A.V., S. Fourati, and I.F.; funding acquisition, S. Fillatreau, J.-C.W., C.-A.R., and M. Mahévas; investigation, A.S., P.C., A.D.L.S., I.A., and A.V.; methodology, A.S., J.-C.W., C.-A.R., P.C., and M. Mahévas; project administration, P.C., F.C.-P., J.-C.W., C.-A.R., and M. Mahévas; resources, J.-M.P., F.N.-P., S. Fourati, E.C., M. Michel, B.G., S.G., G.M., Y.N., V.Z., P.B., F.A.R., C.-A.R., and M. Mahévas; software, P.C.; supervision, P.C., J.-C.W., C.-A.R., P.B., F.A.R., and M. Mahévas; validation, A.S., I.A., A.V., A.D.L.S., M.B., C.-A.R., P.C., and M. Mahévas; visualization, P.C., A.S., I.A., A.D.L.S., M. Mahévas; writing – original draft, P.C., A.S., J.-C.W., C.-A.R., and M. Mahévas; writing – review & editing, all authors.
Declaration of interests
Outside of the submitted work, M. Mahévas received research funds from GSK and personal fees from LFB and Amgen. J.-C.W. received consulting fees from Institut Mérieux. P.B. received consulting fees from Regeneron Pharmaceuticals. J.-M.P. received personal fees from Abbvie, Gilead, Merck, and Siemens Healthcare. F.A.R. is a member of the board of MELETIOS Therapeutics and of the Scientific Advisory Board of eureKARE.
Published: April 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.04.002.
Supplemental information
Data and code availability
-
•
Single-cell culture VDJ sequencing data were initially reported in Sokal et al., 2021b and all sequences used in this study are available as part of Table S1. All VDJ sequences were deposited as Targeted Locus Study projects at DDBJ/EMBL/GenBank under the accession numbers KFPV00000000-KFQZ00000000 (BioProject PRJNA819082, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA819082). The version described in this paper is the first version, KFPV01000000-KFQZ01000000. Affinity and neutralization data are also included in Table S1, which has been deposited on Mendeley Data (https://data.mendeley.com/datasets/wwrxgn65h2/2).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Liu L., Liu L., Zhang Z., Luo Y., Huang Y., Wang H.H., Ho D.D., Sheng Z., Shapiro L. Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep. 2022;38:110428. doi: 10.1016/j.celrep.2022.110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A., Muecksch F., Schaefer-Babajew D., Wang Z., Finkin S., Gaebler C., Ramos V., Cipolla M., Mendoza P., Agudelo M., et al. Anti-SARS-CoV-2 receptor-binding domain antibody evolution after mRNA vaccination. Nature. 2021;600:517–522. doi: 10.1038/s41586-021-04060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickx E., Chappert P., Sokal A., Weller S., Azzaoui I., Vandenberghe A., Bonnard G., Rossi G., Fadeev T., Storck S., et al. Rituximab-resistant splenic memory B cells and newly engaged naive B cells fuel relapses in patients with immune thrombocytopenia. Sci. Transl. Med. 2021;13:eabc3961. doi: 10.1126/scitranslmed.abc3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan H.L., Stamper C.T., Li L., Changrob S., Asby N.W., Halfmann P.J., Zheng N.Y., Huang M., Shaw D.G., Cobb M.S., et al. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity. 2021;54:1290–1303.e7. doi: 10.1016/j.immuni.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Cho A., Jankovic M., Schaefer-Babajew D., Oliveira T.Y., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466.e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., Kurth F., Sander L.E., Klein F. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat. Med. 2022;28:477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Krüger N., Schulz S., Cossmann A., Rocha C., Kempf A., Nehlmeier I., Graichen L., Moldenhauer A.S., Winkler M.S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456.e11. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaki R., Adachi Y., Moriyama S., Onodera T., Fukushi S., Nagakura T., Tonouchi K., Terahara K., Sun L., Takano T., et al. SARS-CoV-2 Omicron-neutralizing memory B-cells are elicited by two doses of BNT162b2 mRNA vaccine. Sci Immunol. 2022:eabn8590. doi: 10.1126/sciimmunol.abn8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad L., Clancy S., Kovalenko M., Liu C., Hui T., Smith V., Pagratis N. High-throughput kinetic screening of hybridomas to identify high-affinity antibodies using bio-layer interferometry. J. Biomol. Screen. 2015;20:498–507. doi: 10.1177/1087057114560123. [DOI] [PubMed] [Google Scholar]
- Luo X.M., Maarschalk E., O’Connell R.M., Wang P., Yang L., Baltimore D. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood. 2009;113:1422–1431. doi: 10.1182/blood-2008-09-177139. [DOI] [PubMed] [Google Scholar]
- Muik A., Lui B.G., Wallisch A.-K., Bacher M., Mühl J., Reinholz J., Ozhelvaci O., Beckmann N., Güimil Garcia R. de la C., Poran A., et al. Neutralization of SARS-CoV-2 Omicron by BNT162b2 mRNA vaccine-elicited human sera. Science. 2022;375:678–680. doi: 10.1126/science.abn7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemet I., Kliker L., Lustig Y., Zuckerman N., Erster O., Cohen C., Kreiss Y., Alroy-Preis S., Regev-Yochay G., Mendelson E., Mandelboim E. Third BNT162b2 vaccination neutralization of SARS-CoV-2 omicron infection. N. Engl. J. Med. 2022;386:492–494. doi: 10.1056/NEJMc2119358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Purtha W.E., Tedder T.F., Johnson S., Bhattacharya D., Diamond M.S. Memory B cells, but not long-lived plasma cells, possess antigen specificities for viral escape mutants. J. Exp. Med. 2011;208:2599–2606. doi: 10.1084/jem.20110740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2021;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Barba-Spaeth G., Fernández I., Broketa M., Azzaoui I., de La Selle A.de L., Vandenberghe A., Fourati S., Roeser A., Meola A., et al. mRNA vaccination of naive and COVID-19-recovered individuals elicits potent memory B cells that recognize SARS-CoV-2 variants. Immunity. 2021;54:2893–2907.e5. doi: 10.1016/j.immuni.2021.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong P., Gautam A., Windsor I.W., Travers M., Chen Y., Garcia N., Whiteman N.B., McKay L.G.A., Storm N., Malsick L.E., et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. Cell. 2021;184:4969–4980.e15. doi: 10.1016/j.cell.2021.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanshylla K., Fan C., Wunsch M., Poopalasingam N., Meijers M., Kreer C., Kleipass F., Ruchnewitz D., Ercanoglu M.S., Gruell H., et al. Discovery of ultrapotent broadly neutralizing antibodies from SARS-CoV-2 elite neutralizers. Cell Host Microbe. 2022;30:69–82.e10. doi: 10.1016/j.chom.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Jia Z., Bao L., Wang L., Cao L., Chi H., Hu Y., Li Q., Zhou Y., Jiang Y., et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature. 2022;603:919–925. doi: 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Muecksch F., Cho A., Gaebler C., Hoffmann H.H., Ramos V., Zong S., Cipolla M., Johnson B., Schmidt F., et al. Conserved neutralizing epitopes on the N-terminal domain of variant SARS-CoV-2 spike proteins. bioRxiv. 2022 doi: 10.1016/j.immuni.2022.04.003. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel F., Shlomchik M. Memory B cells of mice and humans. Annu. Rev. Immunol. 2017;35:255–284. doi: 10.1146/annurev-immunol-041015-055531. [DOI] [PubMed] [Google Scholar]
- Yin W., Xu Y., Xu P., Cao X., Wu C., Gu C., He X., Wang X., Huang S., Yuan Q., et al. Structures of the Omicron Spike trimer with ACE2 and an anti-Omicron antibody. Science. 2022;375:1048–1053. doi: 10.1126/science.abn8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.-C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahradník J., Marciano S., Shemesh M., Zoler E., Harari D., Chiaravalli J., Meyer B., Rudich Y., Li C., Marton I., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat. Microbiol. 2021;6:1188–1198. doi: 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell culture VDJ sequencing data were initially reported in Sokal et al., 2021b and all sequences used in this study are available as part of Table S1. All VDJ sequences were deposited as Targeted Locus Study projects at DDBJ/EMBL/GenBank under the accession numbers KFPV00000000-KFQZ00000000 (BioProject PRJNA819082, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA819082). The version described in this paper is the first version, KFPV01000000-KFQZ01000000. Affinity and neutralization data are also included in Table S1, which has been deposited on Mendeley Data (https://data.mendeley.com/datasets/wwrxgn65h2/2).
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.