Abstract
A concern has been raised that the persistent COVID-19 infection in an immunocompromised host can be the source of the SARS-CoV-2 variants. This is the case of a 61-year-old man in complete remission of a follicular lymphoma after six cycles of rituximab and bendamustine with additional two cycles of rituximab completed eight months prior to the episode of COVID-19 pneumonia. The patient's respiratory failure was long-lasting, and required mechanical ventilation until day 75. Acquired immunity tested negative throughout the observational period. The viral RNA was detectable until day 100 while the infectious virus was isolated until day 79. Seven haplotypes were identified and the non-synonymous mutations accumulated in the spike gene which included E484Q and S494P. In the management of COVID-19 cases with suppressed immune statuses, initial evaluation of existing immunity and monitoring for infectiousness throughout the clinical course including the convalescent stage may be necessary.
Keywords: COVID-19, Immunocompromised host, Haplotype, Infectivity, Viral kinetic
1. Introduction
Coronavirus disease 19 (COVID-19) swept across the globe within the first quarter of 2020 under intensive surveillance. Although the scientific community decided on the vaccination by the late 2020, the next issue encountered was the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants, which may reduce the effectiveness of health and social measures.
In general, coronaviruses (CoVs) have a positive-sense single-stranded RNA genome, which is approximately 30 kilobases in length. As a nature of RNA viruses, the fidelity of their own RNA-dependent RNA polymerase (RdRp) is lower than that of DNA-dependent DNA polymerase. To help CoVs to replicate large RNA genomes with minimum error, they possess a non-structural protein (Nsp) 14, which provides them with a proof-reading system. As a consequence, the mutation rate of CoVs is lower compared with that of other RNA viruses [1], and CoVs may have reduced chance to acquire fitness by selecting suitable variants that arise from genetic diversity. Thus, elucidating factors driving the genetic diversity of SARS-CoV-2 may be important for the containment of COVID-19.
As prolonged infection and virus shedding among immunocompromised hosts had been reported [[2], [3], [4]], a concern over persistent COVID-19 infection in immunocompromised host being a possible source of the SARS-CoV-2 variants [5].
Here, we report the case of COVID-19 positive immunocompromised patient who possessed seven haplotypes of SARS-CoV-2 during 79 days of infectious period.
2. Case report
This is the case of a 61-year-old man in complete remission of a follicular lymphoma after six cycles of rituximab and bendamustine with an additional two cycles of rituximab completed eight months prior to the COVID-19 diagnosis. He was notified as a close contact of a COVID-19 case in late December 2020 when Japan was at the height of the third wave of the pandemic. The initial real-time Reverse Transcriptase PCR (qRT-PCR) was negative (day −3). However, he became febrile (day 0), and the qRT-PCR done on day 4 was positive. Based on the medical history, he was admitted to a nearby hospital and treated with five-days courses of favipiravir (loading dose: 3600 mg, maintenance dose: 1600 mg/day) and oral administration of dexamethasone as indicated in the COVID-19 treatment guideline [6]. He became afebrile on day 10, and was discharged on day 13 according the discharge criteria [7]. However, he became febrile once again on day 14. Then, he was re-admitted on day 16 for observation, and treated with antibiotics and additional oral dexamethasone. On day 19, he developed dyspnea and was transferred to a tertiary hospital.
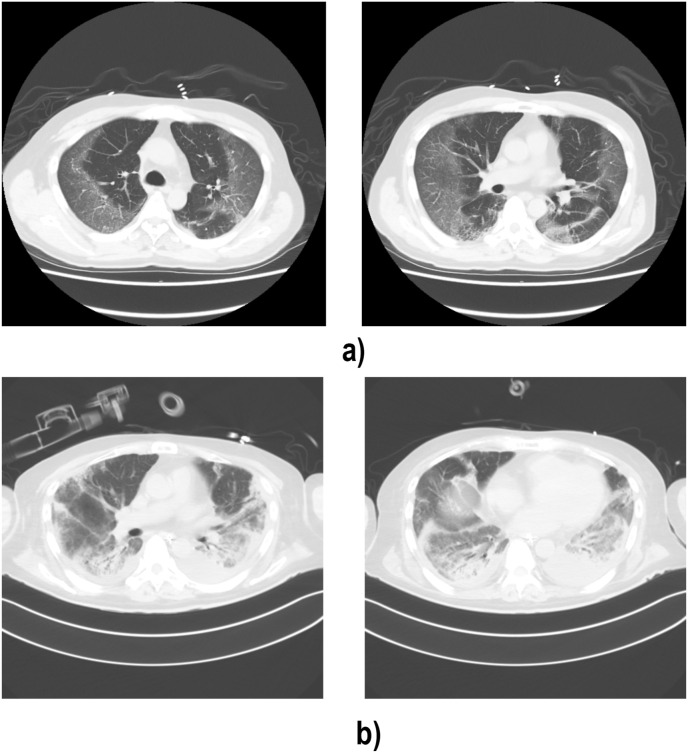
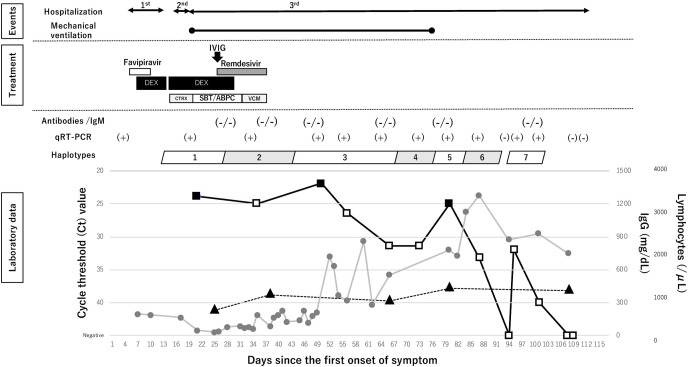
Upon presentation at the third admission (day 19), he required 2 L/min of supplemental oxygen, and a computed tomography (CT) of his chest revealed bilateral ground-glass opacity (Fig. 1 a). Due to the rapid increase in the patient's demand for oxygen, he was intubated and underwent mechanical ventilation on the 2nd day of the third admission (day 20). Under the empirical antibiotic administration, he was managed as the recurrence of COVID-19 infection. He received a 10-day course of additional intravenous dexamethasone (6.6 mg/day) from day 20 and a 10-day course of remdesivir (loading dose: 200 mg, maintenance dose: 100 mg/day) from day 27. He showed hypogammaglobulinemia (IgG: 229 mg/dL) on day 24 and 30,000 mg of intravenous immunoglobulin (IVIG) was administered on day 27 (Fig. 2 ). He also had leukocytopenia (lymphocyte count: 116/μL) from day 20, which persisted until day 48. The respiratory failure was long-lasting due to active viral pneumonia (Fig. 1b), and required mechanical ventilation until day 75. On day 116, he no longer required oxygen and was transferred to another hospital for rehabilitation.
Fig. 1.
Chest computed tomography (CT). a Bilateral ground-glass opacity with a pronounced peripheral distribution, which is typical of COVID-19 pneumonia (Day 30). b Dense consolidation are also confirmed, which indicates progression of COVID-19 pneumonia (Day 66).
Fig. 2.
Timeline of treatment and diagnostic tests. Treatments are shown in above and key laboratory findings are shown in below. Viral kinetics are shown as the cycle threshold (Ct) value in the box. The black boxes indicate the Ct value of a viral isolation positive sample; while the white boxes indicate the Ct value of the viral isolation negative sample; triangles indicate IgG; circles indicate lymphocytes count. IVIG, intravenous immunoglobulin; DEX, dexamethasone, CTRX, ceftriaxone; SBT/ABPC, sulbactam/ampicillin; VCM, vancomycin; qRT-PCR, real-time reverse transcriptase PCR.
To evaluate acquired immunity against SARS-CoV-2, antibodies against the nucleocapsid (N) protein and IgM against spike (S) protein were quantified in serum samples collected on days 26, 37, 48, 65, 79, and 100 using Elecsys Anti-SARS-CoV-2 (Roche Diagnostics) and ARCHITECT SARS-CoV-2 IgM (Abbott), respectively. All six samples tested were negative for both antibodies (Fig. 2).
To monitor the viral kinetics, nasopharyngeal swabs (NPSs) that were periodically collected during the third admission (day 20 to day 108) underwent q-RT-PCR [8], and the cycle threshold (Ct) value was used as a proxy indicator of the viral load (Fig. 2). A total of 13 NPSs underwent qRT-PCR, of which ten were positive. The viral load gradually decreased from day 55 to day 72, but it suddenly increased on day 79. During the interval between days 72 and 79, there was no sign of clinical deterioration, and the case was extubated on day 75. The viral genome was once negative on day 94, but remained detectable until day 100. He had been under isolation until the confirmation of two consecutive negative qRT-PCR tests on day 107 and 108.
To assess the infectiousness, all ten qRT-PCR positive NPSs underwent the viral isolation using the VeroE6/TMPRSS2 cell line [8]. NPSs collected on days 20, 49, and 79 showed cytopathic effects, and the viral propagation was confirmed by qRT-PCR (Fig. 2).
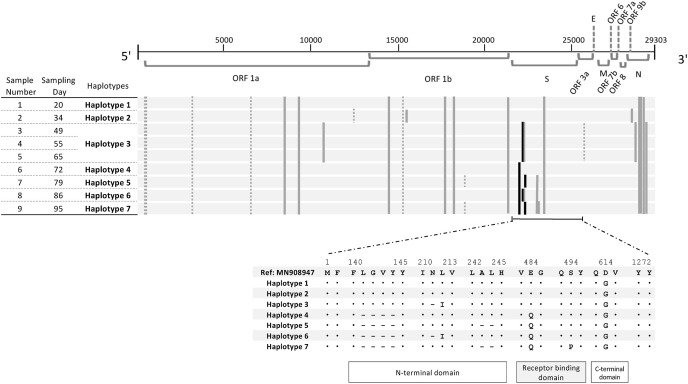
To assess the genetic trait of the SARS-CoV-2 in a course of infection, near-complete genomes of nine out of ten qRT-PCR positive NPSs were amplified based on a protocol published by the ARTIC network using the V3 multiplex primer scheme and cDNA library was sequenced using Illumina NovaSeq platform (2 × 151 cycles; paired-end). All seven haplotypes were classified in 20B in Nextstrain clade or B.1.1.214 in Pangolin Lineage V.3.1.11, which were the dominant circulating strains in Japan in December 2020 [9]. When haplotype 1 was compared with Wuhan-Hu-1/2019 (GenBank accession number; MN908947), it had ten non-synonymous (ORF1a:Q2702H and S2981F, ORF1b:P314L, T1404 M, P1567L, and R2684I, S gene:D614G, N gene:R203K, G204R, and M234I) and five synonymous mutations (Fig. 3 ). Haplotype 2 acquired two non-synonymous mutations in two different open reading frames (ORFs) (ORF1b and ORF9b) and one synonymous mutation. One of the non-synonymous mutations, V658I, was located in nsp12, which encodes RdRp. Haplotype 3 acquired additional four non-synonymous mutations (ORF1a, S gene, and N gene), a single deletion (S gene), and a synonymous mutation; however, it lost two previous non-synonymous mutations including V658I. From haplotype 4 through haplotype 7, diversity was observed mainly in the S gene, and two of them (E484Q and S494P) were additional mutations in the receptor-binding motif of the receptor-binding domain (RBD) [10].
Fig. 3.
Mutations observed in haplotypes. Nucleotide mutations in seven haplotypes of nine nasopharyngeal swab samples are shown in above and amino acid mutations of the spike proteins are shown in below. The SARS-CoV-2 isolate, Wuhan-Hu-1(MN908947.3), was used as a reference genome. The black bar indicates deletion, the gray bar indicates a non-synonymous mutation, and the dashed bar indicates a synonymous mutation. ORF, open reading frame; S, spike; E, envelope; M, membrane; N, nucleocapsid.
3. Discussion
We experienced a COVID-19 case with long-term infectiousness with a series of haplotype switches, which provided another insight for patient management for COVID-19 in immunocompromised hosts.
Unlike SARS in 2003, the emergence of the variants became a serious issue once COVID-19 had sustained the chain of transmission across the globe. Considering the nature of the RNA virus, SARS-CoV-2 accumulates mutation as it reiterates replication among the susceptible population. Especially, the propagation among immunocompromised hosts may favor viral replication since it is an infection under zero or weakened immune pressure for extended periods. Indeed, the pooled analysis revealed that the rate of sequence change over time was faster in immunocompromised patients than in the general population [11].
Our case had undergone B-cell depleting therapies with rituximab for the treatment of follicular lymphoma eight months prior to the current COVID-19 episode. In general, the administration of rituximab suppresses the production of antibodies and induces hypoglobulinemia for 5–11 months [12]. Since our case had hypoglobulinemia and did not acquire immunity against SARS-CoV-2 throughout the clinical course, his humoral immunity should have been under suppression. On the other hand, he had lymphocytopenia from the third admission, and the lymphocyte count recovered gradually. Lymphocytopenia is one of the common laboratory findings in severe COVID-19. However, our case had a total of 18 days of an extended course of the dexamethasone treatment. The administration of dexamethasone is generally known to cause immunosuppression by depleting both cellular and humoral immunity [13]. Thus, extra caution may be required for a series of dexamethasone treatments when administered in immunocompromised hosts. Taken together, our case had suppressed humoral immunity alongside temporary cellular immunosuppression. Unfortunately, the IgG levels and CD4 count were not measured before SARS-CoV-2 infection but, if measured, they might have been decreased.
The determination of infectiousness is essential in controlling infectious diseases. In Japan, the discharge criteria for patient with symptomatic COVID-19 are either ten days after the onset and clinical improvement in the last 72 hours, or two consecutive negative results for SARS-CoV-2 that are at least 24 hours apart after improvement of symptoms in the last 24 hours [7]. The former criterion, which required only clinical judgment, was based on an early study that reported that the viable virus could not be isolated beyond ten days after the onset of the disease [14], which cannot be extrapolated for immunocompromised population, as we experienced. Our case was not only infectious beyond such a designated isolation period, but also experienced a resurgence of the infectiousness late in the clinical course. Interestingly, he had a stable condition upon such resurgence, which suggests that the symptom-based monitoring may not be the appropriate indicator for infectiousness [15]. In addition, the measurement of the antibody titers may not be informative since the case may overcome COVID-19 without presence of the humoral immunity [16], as observed in our case. Viral isolation is the best way to assess infectiousness; however, it is generally not applicable in hospital laboratory. However, the measurement of the viral RNA titers by qRT-PCR can be used as the proxy indicator for the viral load. To prevent the secondary infection of immunocompromised COVID-19 cases, we suggest following up of the patient's infectiousness, at least monitoring viral kinetics, beyond the designated isolation period. According to Centers for Disease Control and Prevention (CDC) guidance, some conditions other than immunosuppressive chemotherapy may lead to longer infectiousness in COVID-19 patients, such as solid organ transplantation, acquired immunodeficiency syndrome (AIDS), and primary immunodeficiency [17]. Patients under these conditions are also required to be closely followed up in convalescent phase.
The decrease in viral titers on day 55 coincided with the recovery of lymphocytopenia. An animal study showed that cellular immunity plays an important role in protection when the humoral response is suboptimal [16]. So, the recovery of the cellular immunity might have facilitated the viral clearance in our case. To evaluate existing humoral and cellular immunity and predict the clinical course of COVID-19 pneumonia, it is advisable to measure the number of B-cells, IgG levels and CD4 count at the early phase of infection.
From a genetic perspective, we detected seven haplotypes during the course of the infection. This longitudinal genetic diversity was in line with past reports [2,3], which illustrated that the virus may interchange even within a single host when the infection persisted in immunocompromised hosts. Although our case had depleted humoral immunity, his recovered cellular immunity single-handedly put pressure on the virus to accelerate its intra-host evolution. Apparently, the switches to haplotypes 4 through 7, which had mutations in the S gene, are distinct from the switches from haplotypes 1 to 2, 2 to 3, and from 3 to 4, which consisted of concurrent mutations in different ORFs. Interestingly, the mutations in RBD have accumulated from haplotype 4 to haplotype 7, which consisted of two mutations in the receptor-binding motif of RBD. Haplotypes 4 to 7 acquired an additional mutation, E484Q, which is a one of the signature mutations in the kappa variant (B.1.617.1). E484 is a hot-spot for an escape mutation from the RBD specific monoclonal antibody [18]. In addition, haplotype 7 acquired S494Q. Both E484Q and S494Q are common mutations in isolates of RBD escape mutants [18]. Thus, we speculated that the immune pressure from the host results in the emergence of escape mutants and induces a fluctuation in viral titers.
Our case underwent antiviral therapy with favipiravir and remdesivir, which may also trigger emergence of resistant virus [19]. Regarding RdRp, we observed two mutations in nsp12: P314L and V658I. P314L (P323L) had been one of the common mutations in PANGON lineage B [20], and all seven haplotypes in our study possessed the mutation as the background. On the other hand, V658I was observed during the administration of remdesivir (haplotype 2), but disappeared thereafter. Such transient observation of the haplotype may suggest the emergence of resistant virus, but there had been no report on V658I. Further studies on V658I under drug pressure among immunocompromised host are needed.
As a part of the case management of COVID-19 patient with a suppressed immune status, we propose evaluating both humoral and cellular immunity at the early phase of infection, and monitoring infectiousness throughout the clinical course and even in the convalescent stage in order to minimize the secondary transmission of mutants.
Authorship statement
All authors meet the ICMJE authorship criteria; Akira Suzuki and Kosuke Shoji were responsible for the conception of the work, interpretation of data, and draft the work. Michiko Okamoto, Michio Kobayashi, and Masaru Yanai was responsible for the acquisition, analysis, and interpretation of the data. Emmanuel Kagning Tsinda, Naoko Sugawara, and Mie Sasaki were responsible for the acquisition and analysis of the data. Hitoshi Oshitani was responsible for the analysis and interpretation of the data. Yoshihiko Nogami was responsible for the design of the work. All authors revised this work critically and contributed to the writing of the final manuscript. All authors agreed to be accountable for all aspects of the work. Kosuke Shoji and Akira Suzuki equally contributed to this study.
Declaration of competing interest
No reported conflicts of interest.
Acknowledgements
The authors would like to thank the medical staffs for taking part in the patient care and the public health staffs for conducting contact tracing. The authors would like to thank Enago (www,enago.jp) for the English language review.
This work was supported (in part) by the Ministry of Health, Labour, and Welfare [grant number JPMH20HA2007] and by AMED [grant number JP21wm0125001].
References
- 1.Nie Q., Li X., Chen W., Liu D., Chen Y., Li H., et al. Phylogenetic and phylodynamic analyses of SARS-CoV-2. Virus Res. 2020 doi: 10.1016/j.virusres.2020.198098:198098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183:1901–19012 e9. doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi B., Choudhary M.C., Regan J., Sparks J.A., Padera R.F., Qiu X., et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aydillo T., Gonzalez-Reiche A.S., Aslam S., van de Guchte A., Khan Z., Obla A., et al. Shedding of viable SARS-CoV-2 after immunosuppressive therapy for cancer. N Engl J Med. 2020;383:2586–2588. doi: 10.1056/NEJMc2031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corey L., Beyrer C., Cohen M.S., Michael N.L., Bedford T., Rolland M. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med. 2021;385:562–566. doi: 10.1056/NEJMsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ministry of Health, Labour and Welfare, Japan Clinical guildeline for COIVD-19. 2020. https://www.mhlw.go.jp/content/000788513.pdf fourth ed.
- 7.Ministry of Health, Labour and Welfare, Japan Re-notification, discharge criteria for COVID-19 cases act on infectious disease. 2020. https://www.mhlw.go.jp/content/000698210.pdf
- 8.National Institute for Infectious Diseases J SARS-CoV-2 genomic detection and virus isolation manual Ver2.9. 2021. https://www.niid.go.jp/niid/images/lab-manual/SARS-CoV-2_gene_detect_and_isolation_manual_Ver1_1.pdf
- 9.Takashi S., Kentaro I., Koji Y., Rina T., Satsuki E., Risa S., et al. Molecular analysis of SARS-CoV-2 (2021.1.14) Infectious Agents Surveillance Reports. 2011;42:61–64. [Google Scholar]
- 10.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary M.C., Crain C.R., Qiu X., Hanage W., Li J.Z. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America; 2021. SARS-CoV-2 sequence characteristics of COVID-19 persistence and reinfection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacco K.A., Abraham R.S. Consequences of B-cell-depleting therapy: hypogammaglobulinemia and impaired B-cell reconstitution. Immunotherapy. 2018;10:713–728. doi: 10.2217/imt-2017-0178. [DOI] [PubMed] [Google Scholar]
- 13.Olnes M.J., Kotliarov Y., Biancotto Al, Cheung F., Chen J., Shi R., et al. Effects of systemically administered hydrocortisone on the human immunome. Sci Rep. 2016;6 doi: 10.1038/srep23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones T.C., Biele G., Muhlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., et al. Science; New York, NY): 2021. Estimating infectiousness throughout SARS-CoV-2 infection course. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention Ending isolation and precautions for people with COVID-19: interim guidance. 2022. https://www.cdc.gov/coronavirus/2019-ncov/hcp/duration-isolation.html
- 18.Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gandhi S., Klein J., Robertson A., Pena-Hernandez M.A., Lin M.J., Roychoudhury P., et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. medRxiv. 2021 doi: 10.1101/2021.11.08.21266069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alam A., Islam O.K., Hasan M.S., Islam M.R., Mahmud S., Al-Emran H.M., et al. Dominant clade-featured SARS-CoV-2 co-occurring mutations reveal plausible epistasis: an in silico based hypothetical model. J Med Virol. 2021 doi: 10.1002/jmv.27416. [DOI] [PMC free article] [PubMed] [Google Scholar]