Abstract
Background
Fungal infections are common life-threatening diseases amongst immunodeficient individuals. Invasive fungal disease is commonly treated with an azole antifungal agent, resulting in selection pressure and the emergence of drug resistance. Antifungal resistance is associated with higher mortality rates and treatment failure, making the current clinical management of fungal disease very challenging. Clinical isolates from a variety of fungi have been shown to contain mutations in the MSH2 gene, encoding a component of the DNA mismatch repair pathway. Mutation of MSH2 results in an elevated mutation rate that can increase the opportunity for selectively advantageous mutations to occur, accelerating the development of antifungal resistance.
Objectives
To characterize the molecular mechanisms causing the microevolutionary emergence of antifungal resistance in msh2 mismatch repair mutants of Cryptococcus neoformans.
Methods
The mechanisms resulting in the emergence of antifungal resistance were investigated using WGS, characterization of deletion mutants and measuring ploidy changes.
Results
The genomes of resistant strains did not possess mutations in ERG11 or other genes of the ergosterol biosynthesis pathway. Antifungal resistance was due to small contributions from mutations in many genes. MSH2 does not directly affect ploidy changes.
Conclusions
This study provides evidence that resistance to fluconazole can evolve independently of ERG11 mutations. A common microevolutionary route to the emergence of antifungal resistance involves the accumulation of mutations that alter stress signalling, cellular efflux, membrane trafficking, epigenetic modification and aneuploidy. This complex pattern of microevolution highlights the significant challenges posed both to diagnosis and treatment of drug-resistant fungal pathogens.
Introduction
Cryptococcosis is a life-threatening human disease caused by invasive infection with fungi in the Cryptococcus neoformans or Cryptococcus gattii species complexes. Disseminated cryptococcosis or cryptococcal meningitis are treated initially with amphotericin B in combination with 5-flucytosine, then long-term with fluconazole. Mild-to-moderate cases of isolated pulmonary cryptococcal infections can be treated with fluconazole as first-line therapy. In recent years, many clinical studies have demonstrated an increase in resistance to azole antifungal agents, resulting in an emerging threat to the management of invasive fungal diseases in clinical practice.1–4 A recent review of drug resistance in Cryptococcus spp. revealed 10.6% of clinical isolates are resistant to fluconazole and this increases to 24.1% in patients with relapsed disease.3 The molecular mechanisms of how drug resistance emerges in C. neoformans remain poorly defined.5
The most common mechanism of amphotericin B resistance is alterations to the sterol composition of the membrane via mutations in ERG genes of the ergosterol biosynthesis pathway.6 Several azole resistance mechanisms have been identified; most involve point mutations in the ERG11 (cyp51A) gene, which alters the ability of the azole molecule to bind to the target lanosterol 14α-demethylase, or which occur in regulatory regions in the promoter.4,7–10 Overexpression of genes encoding drug efflux transporters, or the transcription factors that regulate their expression, can also result in drug resistance in Candida species.11,12 However, mutations located within ERG11 may not be the most predominant mode of azole drug resistance across all fungi. Only four ERG11 mutations have been associated with fluconazole resistance in C. neoformans and studies show 50%–70% of fluconazole-resistant clinical isolates lack mutations in ERG11.8,13–16 Another study investigating C. gattii clinical isolates from the Pacific Northwest epidemic concluded neither ERG11 overexpression nor coding variations were responsible for the increased fluconazole resistance.17 In addition, more than 50% of azole-resistant Aspergillus fumigatus clinical isolates contain no mutations in cyp51A or its promoter.12,18 A recent study that followed the process of microevolution of azole resistance using WGS, during persistent and recurrent aspergillosis, found that the microevolution of azole resistance was driven by both Cyp51A-independent and Cyp51A-dependent mechanisms.19
In addition to point mutations, resistance in fungi can be conferred upon exposure to azoles by aneuploidy, the gain or loss of chromosomes. Aneuploidy is a common strategy utilized by fungi to adapt to stress and drug-resistant aneuploids with permanent chromosomal duplications have been found in clinical isolates.20 Aneuploidy can also be transient in a process called heteroresistance, where one or more aneuploidies (chromosome duplications) develop that are subsequently lost when the azole is removed.21 In cryptococcal meningitis patients, transient aneuploidy of chr1 has been shown to be associated with increased fluconazole MIC and clinical relapse.22
Recent studies have uncovered a role for DNA mismatch repair (MMR) in the microevolution of antifungal drug resistance.23–26 Defective DNA repair mechanisms lead to an elevated mutation rate that provides an avenue for the rapid acquisition of beneficial mutations that contribute to drug resistance evolution.23–25,27–29 Non-synonymous variation in the MMR gene MSH2 has been found with varying prevalence in clinical populations of Candida glabrata, A. fumigatus, Cryptococcus deuterogattii and C. neoformans.23–27 The exact prevalence of msh2 mutators in clinical populations and their clinical relevance remains contentious. Studies in C. glabrata have found between 37%–77% of clinical strains possess non-synonymous variation in MSH2 (North America, 55%; India, 69%; France, 44%; South Korea, 65%; China, 77%; Spain, 44%; and Australia, 37%) however only some studies have shown a correlation with antifungal drug resistance (North America, 65%; South Korea, 69%; China, 43%).27,30–35 In addition, some C. glabrata clinical isolates with naturally occurring MSH2 alleles, previously called mutators, were subsequently shown not to possess a mutator phenotype.36 Studies have shown that 18.2% and 18.1% of A. fumigatus and C. neoformans clinical isolates, respectively, possess non-synonymous variation in MSH2, however the number of strains analysed was small.24,25 Deletion of MSH2 in C. deuterogattii or C. glabrata does not reduce virulence.26,27 Initially the A. fumigatus Δmsh2 mutant displayed reduced virulence, however, passaging allowed virulence to be rapidly regained.25 Deletion of MSH2 leads to the rapid emergence of azole resistance in all these species in vitro.24–27 Deletion of MSH2 in C. deuterogattii results in increased emergence of resistance to 5-flucytosine through point mutations in three different genes.28 The mutations accumulating during the emergence of azole resistance in an msh2Δ mutant remain undefined and are the focus of this study.
Materials and methods
Strains and growth conditions
C. neoformans strains used in this study are in Tables 1 and 2. Strains were stored as glycerols at −80°C and struck off glycerol prior to each experiment to minimize passaging. Strains were cultured on yeast extract-peptone dextrose (YPD) ±2% agar at 30°C or liquid at room temperature in a roller drum.
Table 1.
C. neoformans strains used in this study
| Strain name | Source/parent strain | Isolated on | MSH2 genotype | Elevated mutation ratea | ERG11 genotype | MIC (mg/L) | |
|---|---|---|---|---|---|---|---|
| FLCb | AMB | ||||||
| KBCN001 (KN99) | 63 | — | MSH2 + | — | ERG11+ | 8 | 2 |
| KBCN105 (AI187) | 64 | — | MSH2 +/MSH2+ | — | ERG11+/ERG11+ | ND | ND |
| msh2Δ (AISVCN195) | 24 | — | msh2Δ | Yes | ERG11+ | 8 | 2 |
| KBCN0137 | msh2Δ | FLC | msh2Δ | Yes | ERG11+ | 24 | 16 |
| KBCN0138 | msh2Δ | FLC | msh2Δ | Yes | ERG11+ | >256 | 4 |
| SACN00B1 | msh2Δ | FLC | msh2Δ | Yes | ERG11+ | >256 | 16 |
| SACN00B2 | msh2Δ | FLC | msh2Δ | Yes | ERG11+ | >256 | 8 |
| KBCN0140 | msh2Δ | AMB | msh2Δ | Yes | ERG11+ | 16 | 4 |
| KBCN0142 | msh2Δ | AMB | msh2Δ | Yes | ERG11+ | 12 | 4 |
| KBCN0134 | msh2Δ | FLC | msh2Δ | Yes | ERG11+ | >256 | 4 |
| KBCN0135 | msh2Δ | FLC | msh2Δ | Yes | ERG11+ | >256 | 8 |
| C23 | Clinical isolate40 | — | msh2 Δ379–397,V378N,S398L,K399E | Yes | ERG11+ | 32 | ND |
| C8 | Clinical isolate40 | — | MSH2 N900D,D904G | No | 6 SNPs in 5′ ERG11I99V | 16 | ND |
| A5-35-17 | Environmental isolate40 | — | MSH2 N900D,D904G | No | 6 SNPs in 5′ ERG11I99V | 24 | ND |
| C27 | Clinical isolate40 | — | MSH2 D512N,N900D,D904G | No | 6 SNPs in 5′ ERG11+ | 24 | ND |
| C36 | Clinical isolate40 | — | MSH2 + | No | ERG11+ | 64 | ND |
FLC, fluconazole; AMB, amphotericin B; ND, not determined.
Mutation rate was qualitatively assessed by measuring the frequency of resistant 5-fluoroorotic acid-resistant colonies.24
MICs from at least two biological repeats. If not indicated in the table, the standards errors of the mean were ±0.
Table 2.
Fluconazole MICs of C. neoformans deletion mutants
| Biological process | Predicted role | Strain name | FLC MIC (mg/L)a |
|---|---|---|---|
| Membrane trafficking | Trafficking from ER to the Golgi | KBCN0303 trs130Δ | 8 |
| Trafficking through the Golgi | KBCN0298 kes1Δ | 12 | |
| Trafficking from ER to the vacuole directly via the ALP pathway | KBCN0300 apl3Δ | 16 | |
| Trafficking from the ER to the vacuole indirectly via the endosomes and VPS pathway | KBCN0301 vps10Δ | 8 | |
| Multivesicular sorting in the late endosome (ESCRT-0) | KBCN0302 hse1Δ | 8 | |
| Multivesicular sorting in the late endosome (ESCRT-III) | KBCN0205 snf7Δ | 4 | |
| Fusion of vesicles and guanyl-nucleotide exchange factor for the GTPase controlling TOR signalling at the vacuole membrane | KBCN0210 vps39Δ | 6 | |
| Chromatin remodelling | Component of CAF chromatin assembly complex | KBCN0226 rfl2Δ | 16 |
| Chromodomain helicase predicted to play a role in chromatin remodelling | KBCN0299 CNAG_01306Δ | 20 ± 6 | |
| Histone acetyltransferase | NuA4 complex | KBCN0182 yaf9Δ | 16 |
| KBCN0188 eaf6Δ | 24 | ||
| KBCN0190 eaf1Δ | 28 ± 6 | ||
| KBCN0180 swc4Δ | 32 |
FLC, fluconazole.
MICs from at least two biological repeats. If not indicated in the table, the standards errors of the mean were ±0.
The frequency of antifungal drug resistance was assessed by measuring the frequency of colony formation of WT, msh2Δ and an msh2Δ MSH2+ on fluconazole or amphotericin B. The WT strain used for comparison was KN99 (KNCN001), which is the parent strain to msh2Δ. Three separate YPD cultures were inoculated per strain with 1 × 105 cells from overnight YPD cultures. After growth for 48 h, 1 × 106 cells of each culture were plated onto YPD ±72 mg/L fluconazole (16× MIC) or ±4.8 mg/L amphotericin B (32× MIC) and plates incubated at 30°C for 7 days. These concentrations were chosen to enable comparison between similar experiments.23–27 Experiments were performed in triplicate. Frequencies were calculated as number of colonies on the drug divided by the total cfu plated. Frequency averages and standard errors of the mean were calculated using Prism 4.0c. Two-tailed Student’s t-tests were performed to determine statistical significance.
To isolate msh2Δ antifungal drug-resistant strains, 1 × 105 cells from 27 independent YPD msh2Δ cultures (strain AISVCN195) were grown for 48 h and 1 × 105 cells from 15 of the cultures were plated onto YPD +54 mg/L fluconazole (12× MIC) and from the remaining 12 cultures onto YPD +4.8 mg/L amphotericin B (32× MIC) plates. Plates were incubated at 30°C for 7 days and an individual colony was picked from each plate and streak purified. Colonies from three of the fluconazole plates were also patched onto amphotericin B to identify MDR strains. To assess antifungal resistance of each of the 27 isolates, individual strains were cultured overnight in YPD, 10-fold serially diluted, plated onto YPD ±54 mg/L fluconazole (12× MIC) or 2.4 mg/L (16× MIC) amphotericin B and incubated at 28°C for 2 days. Fluconazole and amphotericin B MICs were determined using Etest strips on yeast nitrogen base pH 7 or the broth dilution method, respectively.37 Etests were from Integrated Sciences Pty Ltd with a concentration range of 0.16–256 mg/L. MICs were replicated twice and mean and standard errors of the mean were calculated using Prism 4.0c.
Deletion mutants were obtained from the Madhani collection (http://www.fgsc.net/crypto/crypto.htm). Gene deletion was confirmed using PCR with primers (Table S1, available as Supplementary data at JAC-AMR Online) designed outside and within the deleted region (product present in WT and absent in the deletion) and also with a primer outside the deleted region and within the NAT selectable marker (product absent in WT and present in the deletion if NAT is integrated at the specific gene locus). The parent strain for these deletion mutants is KN99 (KNCN001). This reference was used as a control in all experiments. Twenty deletion strains were screened for changes to fluconazole MIC. The CNAG_06356Δ, CNAG_05838Δ, CNAG_05998Δ, CNAG_00372Δ, CNAG_00051Δ, CNAG_06276Δ and CNAG_06373Δ strains did not showing any significant difference in MIC and are not shown.
Relative fitness of the msh2Δ, SACN00B1, SACN00B2 and a nourseothricin-resistant control compared with WT was determined in three independent experiments by making 50:50 ratios of cells from YPD overnight cultures, incubating for 2 days and plating 1 × 102 cells onto YPD. After incubation at 28°C for 4 days, 100 random colonies were re-struck onto YPD ± nourseothricin (Jena Bioscience) (100 mg/L) and scored as NAT− (WT) or NAT+ (msh2Δ::NAT, SACN00B1 or SACN00B2). Two-tailed Student’s t-tests were performed to determine statistical significance using GraphPad Prism software. All data were expressed as means with a CI of 95% with P values at ≤0.05 considered significant.
Genome sequencing and bioinformatic analysis
Genomic DNA was extracted using a CTAB buffer [100 mM Tris-HCl pH 7.5, 0.7 M NaCl, 10 mM EDTA, 1% β-mercaptoethanol and 1% CTAB (alkyltrimethyl ammonium bromide, Sigma)]. HiSeq paired-end 125 bp Illumina sequencing was performed on shotgun libraries by the Australian Genome Research Facility and Victorian Clinical Genetics Services. Samples had between 16.3 and 22.6 million reads (∼215–300× coverage). Sequencing reads were aligned to the KN99 reference sequence from FungiDB in Geneious version 11.0.2.32–38 Aneuploidy of each strain was determined by comparison of read coverage every 100 bp across the genome to the control strain, calculating the log2 fold difference which was graphed using Prism 8.4.1. Sequence variants were detected using the criteria of a minimum coverage of 10 reads, minimum frequency of 95%, and the maximum P value of 10−6 (0.0001% to observe by chance). Chromosomal position of mutations was performed in Prism 8.4.1. Sequencing reads were deposited to the GenBank database as accession PRJNA664881. Gene Ontology (GO) enrichment was performed with Fisher’s exact test with the background defined as all genes from C. neoformans and a P value cut-off of 0.05 at FungiDB.39
Flow cytometry to assess ploidy
Between 1 × 105 and 1 × 106 cells from overnight YPD cultures of WT (haploid and diploid controls), msh2Δ, SACN00B1 and SACN00B2 were used to inoculate YPD ± fluconazole (two duplicates). The cultures were stained with NucRed Live 647 DNA stain (Thermofisher) at room temperature for 30 min in the dark and non-stained controls were included. Cells were washed in PME buffer [50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.7), 5 mM magnesium sulphate (MgSO4) in 500 mL dH2O, 25 mM ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA) (pH 8.0)]. The samples were analysed in a fluorescence stimulated cell sorting Becton Dickinson FACSCanto II with a 633 nm commotion laser. Signals were captured from a minimum of 10 000 cells per sample in the APC channel (red, excitation 633 nm) at a medium flow rate of 20 000 cells/sec. The FACS Diva (8.0.1) acquisition software was used to measure forward and side scatter on a four-decade logarithmic scale and red fluorescence (APCA) on a linear scale using FlowJo software (version 10.6.1). Experiments were repeated in triplicate.
Results
The elevated mutation rate of an msh2Δ mutant enables rapid isolation of antifungal drug-resistant strains
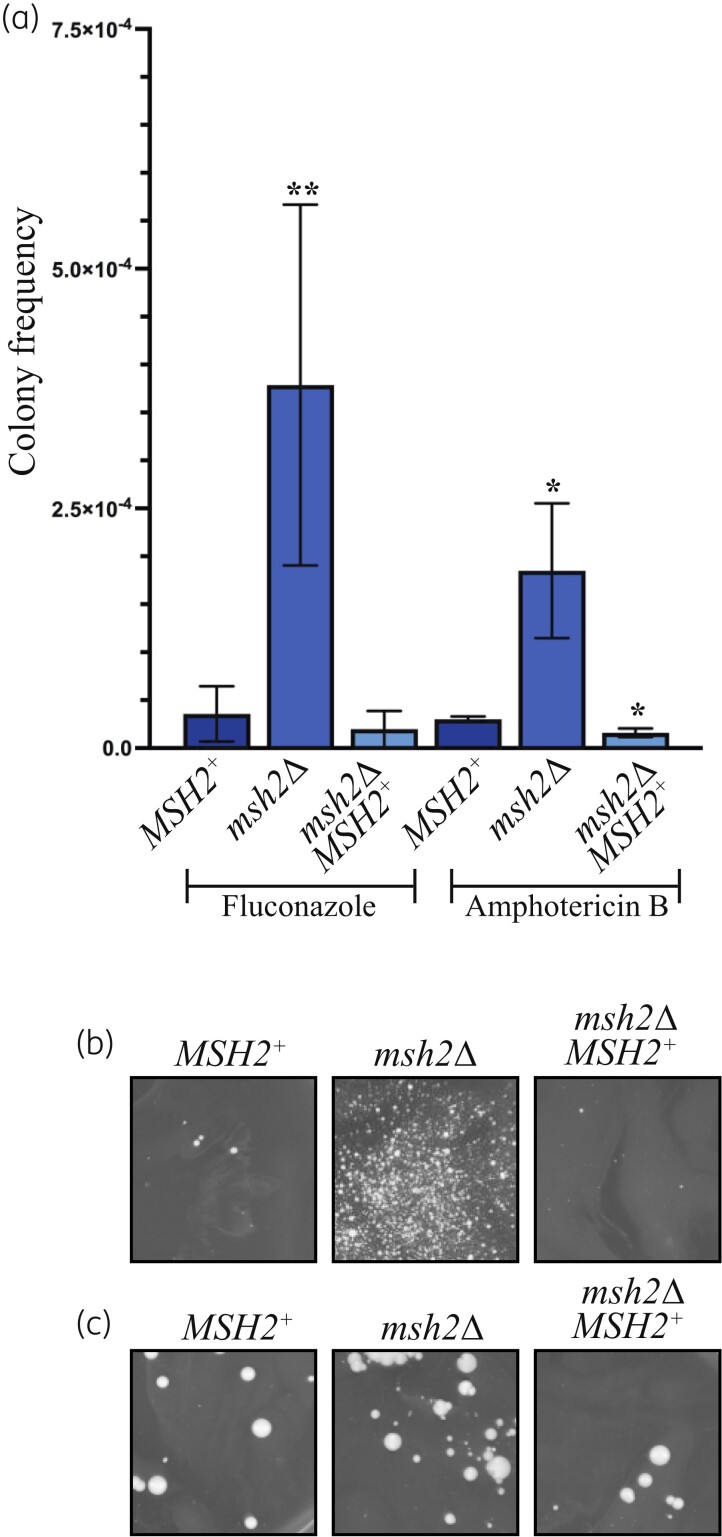
A WT control, msh2Δ mutant and an msh2Δ MSH2+ control strain were inoculated into liquid cultures and incubated to allow mutations to accumulate during mitotic division. Cells of each culture were plated onto either fluconazole (16× MIC) or amphotericin B (32× MIC) and the frequency of antifungal resistance calculated. These concentrations were chosen to enable comparison between similar experiments in other studies.23–27 Compared with the WT and the msh2Δ MSH2+ control, the msh2Δ mutant displayed an increase in the frequency of colonies resistant to fluconazole and amphotericin B (Figure 1). Therefore, mutation of MSH2 can promote the rapid emergence of resistance to antifungal drugs due an elevated mutation rate.
Figure 1.
The frequent emergence of antifungal drug resistance in the msh2Δ mutant enables rapid isolation of antifungal drug-resistant strains. (a) Compared with the WT (MSH2+) and msh2Δ MSH2+ controls, the msh2Δ exhibits an elevated frequency of fluconazole-resistant and amphotericin B-resistant colonies on media containing 72 mg/L fluconazole (16× MIC) or 4.8 mg/L amphotericin B (32× MIC). Asterisks indicate statistical significance using a two-tailed Student’s t-test; *P < 0.05, **P < 0.005. (b) An example of one of the independent cultures of msh2Δ grown in liquid culture to allow mutations to accumulate and plated on plates containing fluconazole (12× MIC) or (c) amphotericin B (32× MIC). WT (MSH2+) and msh2Δ MSH2+ controls are included for comparison.
To uncover the molecular mechanisms causing the emergence of antifungal resistance, 27 independently isolated fluconazole- or amphotericin B-resistant strains were isolated. From these, four fluconazole, two amphotericin B and two MDR strains possessing a range of resistance levels were chosen for further analysis. Each strain represents an independent antifungal drug resistance microevolution experiment. The fluconazole and amphotericin B MIC was determined for each strain (Table 1). Compared with WT and msh2Δ, strains isolated on fluconazole displayed high levels of resistance, with greater than 32× the fluconazole MIC of WT and the original msh2Δ susceptible strains. In addition, these strains also exhibited an elevated amphotericin B MIC. MDR strains exhibited greater than 32× the fluconazole MIC and 2–4× the amphotericin B MIC of WT and msh2Δ. The strains isolated directly on amphotericin B showed a 2× increase in the amphotericin B MIC and a small elevation in the fluconazole MIC (Table 1).
Antifungal drug-resistant msh2Δ strains do not possess mutations in ERG11 or components of the ergosterol biosynthesis pathway
To investigate the genetic changes underpinning the emergence of antifungal drug resistance, the genomes of the original msh2Δ strain (susceptible to antifungals) and the msh2Δ antifungal-resistant strains were analysed by WGS. Reads were aligned to the C. neoformans KN99 WT genome sequence (susceptible to antifungals) in which the msh2Δ strain was generated. Sequence variants were identified that differed between genomes of the KN99 and msh2Δ strains to identify mutations that had accumulated in msh2Δ during laboratory passaging in the absence of antifungal selection. Although passaging was minimized, 284 sequence variants were identified in the original msh2Δ strain prior to antifungal selection. GO term enrichment analysis of the mutated genes showed significant enrichment in the biological processes of hyperosmotic response, peroxisome fission, negative regulation of melanin biosynthetic process, biological process involved in interaction with host, carbohydrate metabolic process, fructose metabolic process and fructose 2,6-bisphosphate metabolic process (Table S2).
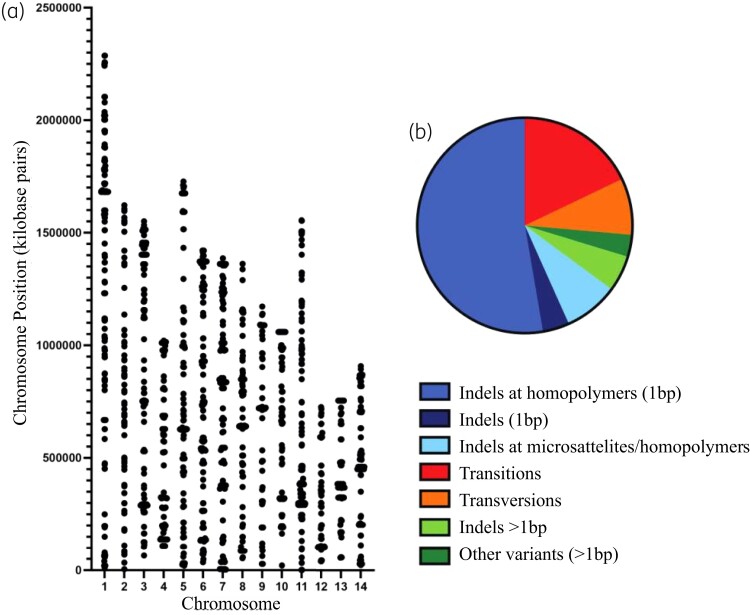
Sequence variants that differed between the genomes of the WT and msh2Δ strains (both antifungal susceptible) and the msh2Δ antifungal drug-resistant strains were also identified to find variants responsible for the emergence of antifungal resistance. A total of 1783 sequence variants were detected in the antifungal-resistant strains that were not present in KN99 or msh2Δ. These variants were distributed across all chromosomes and were predominately single base pair mutations (83.1%); transitions (17.9%), transversions (8.5%) and single base indels at homopolymers (52.7%) and also included larger indels at microsatellites and homopolymers (8.2%) (Figure 2).
Figure 2.
Mutations accumulate in the msh2Δ antifungal drug-resistant strains in response to exposure to antifungal drugs. (a) The position of mutations in msh2Δ antifungal drug-resistant strains not present in the original msh2Δ before antifungal selection across the 14 C. neoformans chromosomes. (b) The mutational spectrum of sequence variants in the genomes of msh2Δ antifungal drug-resistant strains: red, transitions; orange, transversions; medium blue, single base indels at homopolymers; dark blue, single base indels; light blue, larger indels at microsatellites and homopolymers; light green, large indels; dark green, other variants.
The fluconazole-resistant strains showed only 12 mutations that were absent in the amphotericin B-resistant strains. Likewise, the amphotericin B resistant strains showed 37 mutations that were absent in the fluconazole-resistant strains. In addition, these mutations were either in non-coding regions or were located in genes that also contained numerous additional mutations common to all antifungal drug-resistant strains. This suggests that the mutations are arising due to a general stress response, rather than to a specific antifungal drug.
Most of the genetic causes of resistance to azole antifungal agents described to date involve point mutations that affect either the expression or activity of the ERG11 (cyp51A) gene of the ergosterol biosynthesis pathway.4,7–10 None of the msh2Δ antifungal drug-resistant strains possessed mutations in ERG11 or other ergosterol biosynthesis genes (Table S3).
To investigate whether clinical isolates also exhibit azole resistance caused by mutations in genes other than ERG11, the genomes of fluconazole-resistant clinical and environmental isolates were also analysed by WGS. These isolates are the same molecular type (VNI) as the msh2Δ and exhibit varying degrees of fluconazole resistance (Table 1).40 One of these (C23) is an msh2 mutant with elevated mutation rates.24 None of these clinical isolates were aneuploids (Figure S1). The isolates with relatively lower levels of fluconazole resistance (16–24 mg/L) possessed an ERG11I99V conservative missense mutation (C8 and A5-35-17) and six identical SNPs in the 5′ regulatory region, which is unlikely to lead to disruption in function but which could result in changes in the levels of ERG11 expression (C8, C27 and A5-35-17) (Table 1). However, clinical isolates with high resistance (C23, 32 mg/L and C36, 64 mg/L) had no detectable sequence variation in ERG11 (Table 1).
Antifungal drug resistance is due to small contributions from mutations in many genes
The genomes of the msh2Δ antifungal-resistant strains possessed variants located in 601 different genes suggesting that drug resistance emerges due to small contributions from many genes rather than due to large contributions from a few genes (Table S4). GO term enrichment analysis of these genes showed significant enrichment in the biological processes: reproduction, vesicle docking involved in exocytosis, regulation of chromosome organization, signalling, signal transduction and response to stimulus, biological regulation, carbohydrate catabolic process, monocarboxylic acid metabolic processes, small molecule metabolic process, phosphorus metabolic process, N-terminal protein amino acid modification and protein containing complex subunit organization (translation initiation) (Table S5). The mutated genes were also significantly enriched for the cellular component GO terms related to translation initiation (Table S5).
The genomes of the msh2Δ antifungal-resistant strains were compared with those of the antifungal resistant C23, C8, C27 and C36 clinical isolates and A5-35-17 environmental isolate to investigate the clinical relevance of variants identified in the in vitro generated msh2Δ antifungal-resistant strains. In total, 28 sequence variants were identified common to all in vitro generated, clinical and environmental strains, either located in intragenic regions or within seven different genes (CNAG_00843 salicylate hydrolase predicted to have a role in drug metabolism, CNAG_05394 membrane transporter, CNAG_02980 membrane dipeptidase, CNAG_04087 hypothetical protein, CNAG_01537 helicase Dhh1, CNAG_06487 chitin synthase and CNAG_05395 guanyl-nucleotide exchange factor Vps39 involved in TOR signalling at the vacuole membrane and membrane trafficking). To compare the sequence variants specifically in msh2 mutants, common sequence variants were identified in the genomes of the msh2 C23 clinical isolate and the msh2Δ antifungal-resistant strains. In total, 103 common sequence variants were identified and GO term enrichment analysis was performed on genes containing common variants. In addition to enrichment in the biological processes of interaction with the host, interspecies interaction and peroxisome fission, there was enrichment in biological processes previously shown to be enriched in the in vitro-generated msh2Δ antifungal-resistant strains including vesicle-mediated transport, organic substance metabolic processes, response to stimulus and regulation of translation.
The genomes of drug-resistant strains contain mutations in genes encoding stress-activated signalling pathways and drug efflux transporters
Mutations in the in vitro-generated msh2Δ antifungal-resistant strains were found in genes encoding components of each of the four major stress-activated signalling pathways in C. neoformans: the protein kinase A (PKA)/Ras signalling pathways, calcium signalling, mitogen activated protein kinase (MAPK) signalling and the high osmolarity glycerol (HOG) response (Table S6).41 However, the mutations in HOG1 occurred in msh2Δ prior to selection on antifungal drugs. A recent study using quantitative trait locus mapping and WGS to identify the genetic basis of antifungal drug sensitivity has shown complex epistatic interactions occur within these signalling pathways to regulate antifungal drug resistance in Cryptococcus.42 Deletion of HOG1 and STE7 have previously been shown to result in increased fluconazole resistance, and inhibition of the HOG pathway increases expression of ergosterol biosynthesis genes and cellular ergosterol content.43,44 Mutations were also located in many genes encoding drug efflux transporters (Table S6).
Defects in membrane trafficking affect the susceptibility of C. neoformans to fluconazole
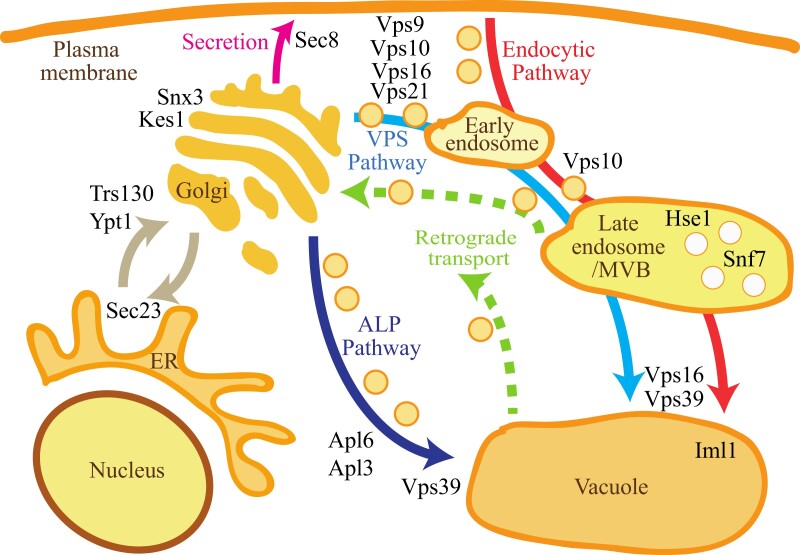
Shared mutations in genes encoding components required for membrane trafficking were also found in the msh2Δ antifungal-resistant strains (Figure 3, Table S6). Mutations in the gene encoding the kinase subunit of TOR complex, TOR1, was also present. In addition to the role in vesicle fusion, Vps39 acts as a guanyl-nucleotide exchange factor for the GTPase controlling TOR signalling at the vacuole membrane.45 There was significant GO term enrichment in the biological processes of vesicle docking involved in exocytosis and small (Rho) GTPase mediated signal transduction which, in addition to being involved in signal transduction, is directly involved in controlling intracellular membrane sorting and trafficking (Table S5).46
Figure 3.
Genes encoding components of membrane trafficking pathways mutated in the msh2Δ antifungal drug-resistant strains. Schematic of the membrane trafficking pathways in a fungal cell (adapted from Feyder et al.45) showing the role of proteins encoded by genes mutated in the msh2Δ antifungal drug-resistant strains (black text). Transporting newly synthesized proteins from the endoplasmic reticulum (ER) to the Golgi requires formation of COPII vesicles (involves Sec23) and the fusion of vesicles tethered to the Golgi membrane that is dependent on the GTPase Ypt1 and Trs130, a component of the transport protein particle (TRAPP) complex, which acts as a multimeric guanine nucleotide exchange factor for Ypt1 (brown arrow). Snx3 is a late-Golgi sorting nexin and Kes1 is required for negative regulation of Golgi secretory functions. Secretion from the Golgi to the plasma membrane (pink arrow) requires the exocyst complex, containing Sec8, to tether post-Golgi secretory vesicles to sites of exocytosis. There are two pathways from the Golgi to the vacuole: the direct ALP pathway (dark blue arrow) and the indirect VPS pathway via endosomes (light blue arrow). In the ALP pathway (dark blue arrow), ALP is packaged into vesicles through the AP-3 adapter complex, containing Apl6 and Apl3, which also recruits clathrin. In the VPS pathway (light blue arrow), soluble carboxypeptidase Y pro-protease (CPY) binds to its receptor Vps10 in the Golgi lumen and is transported from the trans-Golgi network via AP-1 adapter and clathrin-coated vesicles to endosomes. The Rab GTPase Vps21 and guanine exchange factor for Rab GTPases Vps9 are required for Golgi-endosome trafficking. Vps16 and Vps39 are part of the HOPS complex essential for docking and fusion of vesicles. Fusion of the late endosome (multivesicular body, MVB) to the vacuole also requires the HOPS complex. In the late endosome, proteins are sorted into vesicles that bud into the lumen in a process that requires the ESCRT complexes containing Hse1 (ESCRT-0) and Snf7 (ESCRT-III). The endocytic pathway (red) and retrograde transport (green dashed arrows) are also indicated.
To investigate the effect of defects in membrane trafficking on antifungal susceptibility, the fluconazole MIC was determined for deletion strains of TRS130, KES1, VPS10, APL3, VPS39, HSE1 and SNF7. The trs130Δ, vps10Δ and hse1Δ mutants showed no significant difference in MIC compared with WT. Compared with WT, the vps39Δ and snf7Δ displayed increased susceptibility to fluconazole (P < 0.05 and P < 0.0005, respectively). The kes1Δ and apl3Δ mutants resulted in a significant increase in resistance to fluconazole compared with WT (Table 2) (P < 0.0005 and P < 0.0001, respectively).
Mutations in genes encoding proteins required for epigenetic control, DNA repair and bypassing replication stress accumulate in antifungal-resistant strains
The genomes of the msh2Δ antifungal-resistant strains also contain shared mutations in genes encoding proteins required for histone modification, specifically acetylation (Table S6). The two main histone acetyltransferases (HATs) in fungi are components of the Spt-Ada-Gcn5 acetyltransferase (SAGA) and nucleosome acetyltransferase of histone H4 (NuA4) complexes.47 The msh2Δ antifungal-resistant strains contain mutations in genes from both the SAGA and NuA4 complexes and four additional HAT genes (Table S6). Deletion of ADA2 (SAGA complex) has previously been shown to result in increased fluconazole resistance.48 Chromatin remodellers (NSR1, SWR1, INO80 and RFL2) and components of the RSC chromatin remodelling complex (RSC1 and RSC7) were also mutated. Genes involved in histone and chromatin modification have been shown to also play a role in DNA repair and bypassing replication stress and additional genes involved in these processes were also mutated; replication stress (DHH1 and RAD53), double-stranded break repair (RAD51, RDH54, RAD57 and SCC2), single-stranded DNA repair (RAD2), MMR (POL3) and cell cycle checkpoints (SWI6, REV7, BUB1 and BUB3).49–51
The fluconazole MIC was determined for deletions of genes encoding NuA4 complex components; EAF1, EAF6, YAF9 and SWC4 and the chromatin remodellers RLF2 and CNAG_01306. Compared with WT, all deletion mutants displayed a significant increase in fluconazole resistance (Table 2) (CNAG_01306Δ and eaf1Δ, P < 0.05 and rfl2Δ, yaf9Δ, eaf6Δ and swc4Δ, P < 0.005).
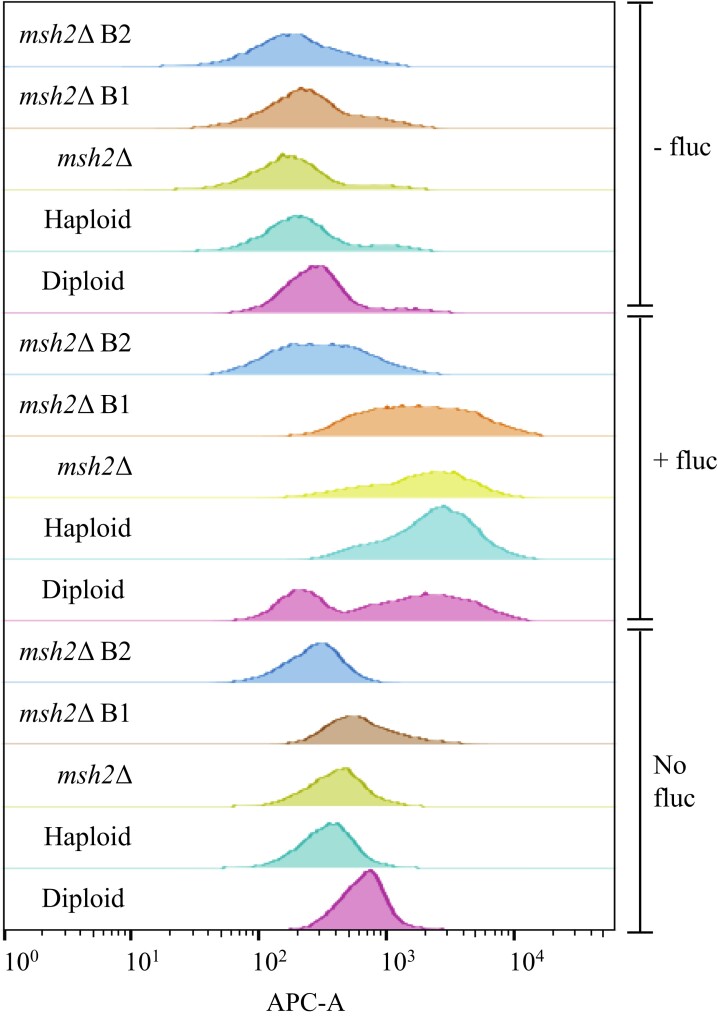
Deletion of MSH2 does not directly affect heteroresistance but can result in aneuploids
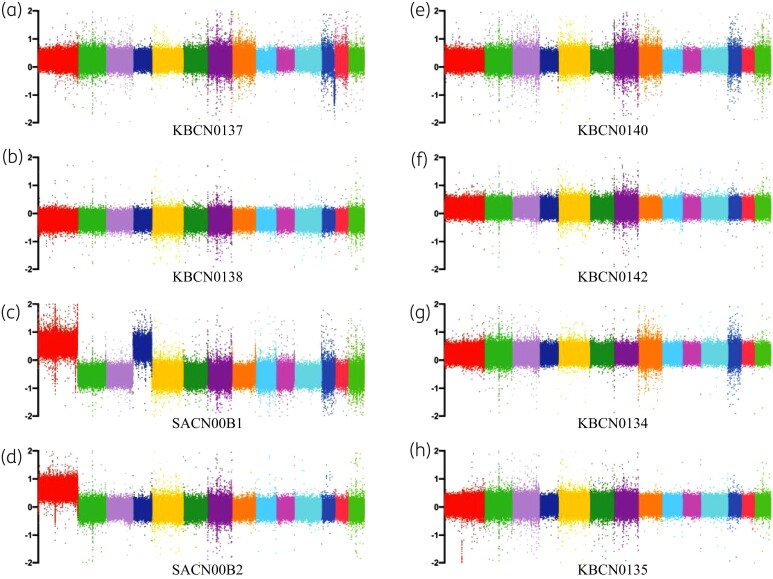
Changes in ploidy can also result in resistance to azoles, although this is usually transient.21,52 During heteroresistance, in the presence of azoles, chr1, chr4, chr10 and chr14 are successively duplicated to produce aneuploids but normal ploidy is re-established when the azole is removed.21 Analysis of ploidy using WGS read coverage showed that SACN00B1 and SACN00B2 are stable aneuploids, possessing duplications of chr1 and chr4 (SACN00B1) and chr1 (SACN00B2) (Figure 4).
Figure 4.
Two of the msh2Δ fluconazole-resistant strains are aneuploids. Log2 fold changes in read coverage (every 100 bp) across the 14 chromosomes in drug-resistant strains isolated on fluconazole: KBCN0137 (a), KBCN0138 (b), SACN00B1 (c), SACN00B2 (d); on amphotericin B: KBCN0140 (e) and KBCN0142 (f); and on fluconazole and amphotericin B: KBCN0134 (g) and KBCN0135 (h). Two of the eight strains, SACN00B1 (c) and SACN00B2 (d), are aneuploids, possessing duplications of chr1 (shown in red) and chr4 (shown in dark blue).
These results suggest that MSH2 deletion may result in increased aneuploidy in response to fluconazole. To investigate this further, changes in ploidy and the ability to undergo heteroresistance were analysed by flow cytometry after culturing strains in the absence or presence of fluconazole and after subsequent removal of fluconazole. Total DNA of the WT haploid increases in the presence of fluconazole, indicating changes in chromosome ploidy (Figure 5). Changes in ploidy can also be observed in the diploid in the presence of fluconazole although the cells appear to have a larger range of changes to ploidy than the haploid (Figure 5). The increases in ploidy are transient, as the original ploidy of the haploid and diploid is re-established when the fluconazole is removed (Figure 5). The changes in ploidy in the msh2Δ are indistinguishable from the haploid, indicating that this strain can undergo heteroresistance and deletion of mshA does not directly affect ploidy (Figure 5). This result suggests that the stable chromosomal aneuploidy in the msh2Δ strains is likely caused by a mutation in a gene affecting chromosomal stability. Deletion of mshA in A. fumigatus also does not directly affect ploidy.25
Figure 5.
Deletion of MSH2 does not directly affect heteroresistance. Populations of cells from the WT haploid control strain KBCN001 (Haploid), a diploid control strain KBCN105 (Diploid), the msh2Δ mutant strain AISVCN195 (msh2Δ) and the fluconazole-resistant aneuploid strains SACN00B1 (msh2Δ B1) and SACN00B2 (msh2Δ B2) were analysed by flow cytometry to assess changes in DNA content as the ploidy changes in response to the absence of fluconazole (No fluc), the presence of fluconazole (+fluc) and 1 day after subsequent removal of fluconazole (−fluc). The diploid control has twice the DNA content of the haploid and msh2Δ in the absence of fluconazole. The chromosome aneuploidy of strain msh2Δ B1 (chr1 and chr4) can be observed as an increase in DNA content compared with the haploid control; however, the increased aneuploidy of strain msh2Δ B2 (chr1) cannot be detected. In response to fluconazole, the ploidy of the haploid, diploid, msh2Δ and msh2Δ B1 increases. Some cells of the diploid do not increase ploidy resulting in two fluorescent peaks. No increase in ploidy observed in the msh2Δ B2 strain indicating it cannot undergo heteroresistance. When the fluconazole is removed, the haploid, diploid, msh2Δ and msh2Δ B1 strains revert back to their original ploidy.
Although strain SACN00B2 is an aneuploid of chr1, this cannot be detected using this method (Figure 5). The total DNA of SACN00B1, which is an aneuploid of both chr1 and chr4, is slightly increased compared with the haploid but less than the diploid as expected (Figure 5). SACN00B1 can undergo heteroresistance, indicated by increased ploidy in the presence of fluconazole and subsequent loss of chromosomes (Figure 5). However, SACN00B2 does not display an increase in ploidy in the presence of fluconazole suggesting it cannot undergo heteroresistance (Figure 5).
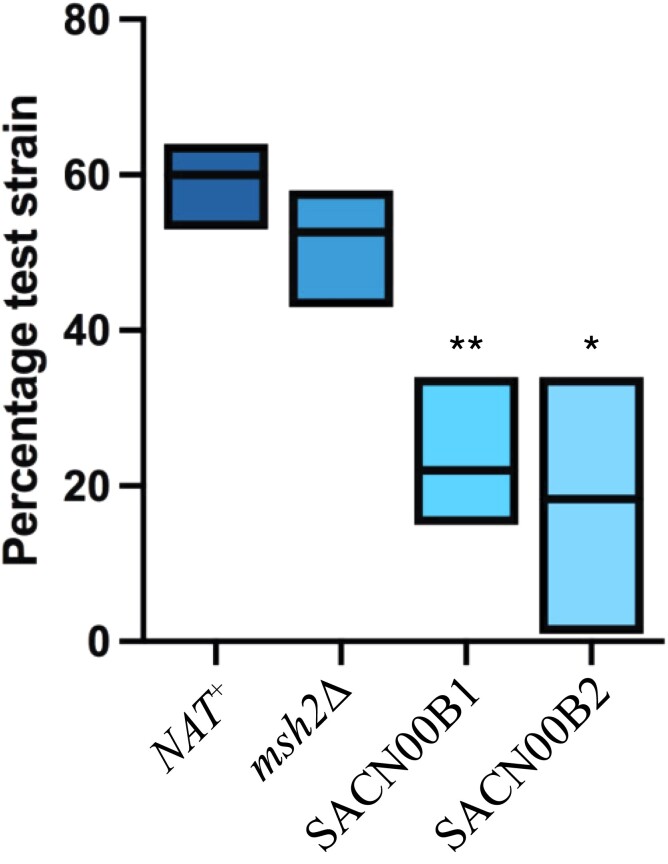
During heteroresistance, aneuploidies are subsequently lost when the azole is removed as they reduce fitness.21 To assess the relative fitness of the aneuploid strains, competition assays were performed with WT. The msh2Δ mutant did not show a statistically significant difference in growth to WT. In contrast, SACN00B1 or SACN00B2 displayed reduced relative fitness in competition with WT in the absence of fluconazole suggesting that aneuploidy reduces fitness (Figure 6).
Figure 6.
The msh2Δ fluconazole-resistant aneuploids show reduced relative fitness compared with WT in the absence of fluconazole. Competition assays with WT and the nourseothricin-resistant control (NAT+), msh2Δ::NAT, SACN00B1 or SACN00B2 cells showing the percentage of colonies derived from each original strain. Aneuploid strains SACN00B1 or SACN00B2 show decreased growth in competition with WT. Asterisks indicate statistical significance using a two-tailed Student’s t-test; *P < 0.05, **P < 0.005.
Discussion
The mutation of MSH2 provides pathogenic fungi with an avenue for the rapid acquisition of mutations, which can contribute to the emergence of favourable phenotypes without negatively impacting virulence or fitness in the short term. This study has shown that antifungal drug resistance emerges rapidly in C. neoformans msh2 mutants in vitro due to single base pair mutations, predominately indels at homopolymers, a mutational profile that matches previous observations in a range of fungi.26,53 To date, most of the genetic causes of resistance to azole antifungal agents involve point mutations that affect ERG11 (cyp51A).4,7–10 Interestingly, none of the msh2Δ antifungal drug-resistant strains possessed mutations in ERG11, providing evidence that antifungal resistance can evolve independently of ERG11 mutations in C. neoformans. This finding is supported by other studies, which show that >50% of azole-resistant clinical isolates of C. neoformans and A. fumigatus lack mutations in ERG11 and that the microevolution of azole resistance can be driven during infection by Cyp51A-independent mechanisms.8,12–16,18,19 Another interesting result from this study was that the genomes of the C. neoformans msh2Δ antifungal drug-resistant strains contained a large number of mutations in many genes suggesting that drug resistance can evolve due to the small contributions of mutations in many genes rather than due to large contributions from a few genes. None of the single gene deletion mutants tested in this study displayed MICs as high as the antifungal drug-resistant strains, also supporting the idea of a multi-gene effect. The results suggest a common microevolutionary route to the emergence of antifungal resistance that involves the accumulation of many mutations that alter stress signalling, cellular efflux, membrane trafficking, epigenetic modification and aneuploidy.
This study showed that the genomes of the C. neoformans msh2Δ antifungal-resistant strains contained mutations in TOR1 and VPS39, encoding the kinase subunit of TOR complex and the guanyl-nucleotide exchange factor (GEF) for the GTPase controlling TOR signalling at the vacuole membrane (Gtr1p), which also plays a role in membrane trafficking.54,55 This finding supports the proposed role of TOR signalling at the vacuole membrane in the response to antifungal drugs. TOR signalling in Cryptococcus has recently been shown to modulate the expression of genes required to aid pathogen survival in macrophages during infection.56 As well as controlling TOR signalling at the vacuole membrane, Vps39 is part of the homotypic fusion and protein sorting (HOPS) complex required for the fusion of vesicles either derived directly from the Golgi, via the alkaline phosphatase (ALP) pathway, or indirectly from the late endosome, via the indirect vacuolar protein sorting (VPS) pathway, to the vacuole.45 This study has shown that deletion of VPS39 in C. neoformans results in increased susceptibility to fluconazole, suggesting an essential role of membrane trafficking to the vacuole in the tolerance to antifungal drugs. The antifungal-resistant environmental and clinical isolates contained a sequence variant in VPS39 in common with the in vitro generated strains, suggesting mutations in VPS39 could be relevant to antifungal susceptibility in the clinic. Recent characterization of the role of this gene in C. neoformans by Fan and Liu (2021)57 has shown this gene is also required for capsule and melanin formation, response to membrane stress and pathogenicity in a mouse model of cryptococcosis. This study has also shown that deletion of vps10 in C. neoformans, which affects trafficking through the VPS pathway and the endocytic pathway, does not affect susceptibility to fluconazole. This is similar to what is observed in C. albicans, where deletion of the gene encoding the Rab GTPase required for endocytosis and the VPS pathway, vps21, does not affect the susceptibility to fluconazole.58 In C. albicans the vps21 mutant exhibits more growth than WT under standard antifungal susceptibility testing conditions and is more susceptible to antifungal drugs that target alternative steps of the ergosterol biosynthesis pathway, a finding which has led to the proposal that endosomal trafficking through the late endosomal pre-vacuolar compartment (PVC) leads to the redistribution of toxic sterol intermediates that accumulate in response to inhibition of the enzyme lanosterol 14-demethylase by azoles, in order to allow limited tolerance to the antifungal drug.58 The present study and previous studies show that deletion of snf7 in C. neoformans, encoding a subunit of the endosomal sorting complexes required for transport (ESCRT) complex, which is required for the additional sorting step at the endosome for delivery to the vacuolar lumen, also results in increased susceptibility to fluconazole.59,60 Consistent with this, mutations in msh2Δ antifungal drug-resistant strains were located in the 5′ regulatory region only. ESCRT mutants in C. albicans are also sensitive to fluconazole.61 In addition, we have also shown that mutation of genes required for membrane trafficking directly from the Golgi to the vacuole via the ALP pathway, kes1 and apl3, results in increased resistance to fluconazole in C. neoformans.
Interestingly, the msh2Δ mutant accumulated mutations in genes involved in regulation of chromatin organization and histone modification prior to selection on antifungals and continued to accumulate these during antifungal selection, in addition to within genes encoding proteins required for DNA repair and bypassing replication stress. Both histone deactelylases and alterations in chromatin structure are associated with the response to DNA damage in yeast.49–51,62 Deletion of some of these genes (EAF1, EAF6, YAF9, SWC4, RLF2 and CNAG_01306) resulted in a significant increase in resistance to fluconazole. This raises the interesting possibility that in msh2 mutants there is selection for mutations occurring in genes required for DNA repair and bypassing replication stress in the absence of antifungal selection, and these mutations also confer drug resistance. This possible link between the genotoxic stress experienced within the host during infection and the emergence of antifungal resistance even in the absence of selection is intriguing and warrants further investigation.
What is clear from this study is that the microevolutionary process resulting in the emergence of antifungal resistance is extremely complex and will pose significant challenges both to the diagnosis and treatment of fungal pathogens.
Supplementary Material
Acknowledgements
Strains were kindly provided by Alexander Idnurm, Mark Bleackley and James McKenna who obtained them from the Madhani laboratory collection. Strains from the Madhani laboratory collection have been made freely available ahead of publication to the scientific community (supported by NIH funding project R01AI100272).
Funding
This study was supported by internal funding. S. H. I. Albehaijani is sponsored by Qassim University, represented in Australia by the Saudi Arabian Cultural Mission (SACM).
Transparency declarations
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary data
Figure S1 and Tables S1 to S6 are available as Supplementary data at JAC-AMR Online.
References
- 1. Gamaletsou MN, Walsh TJ, Sipsas NV. Invasive fungal infections in patients with hematological malignancies: emergence of resistant pathogens and new antifungal therapies. Turk J Haematol 2018; 35: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sanguinetti M, Posteraro B, Lass-Flörl C. Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 2015; 58: 2–13. [DOI] [PubMed] [Google Scholar]
- 3. Bongomin F, Gago S, Oladele ROet al. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel) 2017; 3: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meis JF, Chowdhary A, Rhodes JLet al. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos Trans R Soc Lond B Biol Sci 2016; 371: 20150460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Robbins N, Caplan T, Cowen LE. Molecular evolution of antifungal drug resistance. Annu Rev Microbiol 2017; 71: 753–75. [DOI] [PubMed] [Google Scholar]
- 6. Carolus H, Pierson S, Lagrou Ket al. Amphotericin B and other polyenes-discovery, clinical use, mode of action and drug resistance. J Fungi (Basel) 2020; 6: 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morio F, Loge C, Besse Bet al. Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: new substitutions and a review of the literature. Diagn Microbiol Infect Dis 2010; 66: 373–84. [DOI] [PubMed] [Google Scholar]
- 8. Rodero L, Mellado E, Rodriguez ACet al. G484S amino acid substitution in lanosterol 14-α demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrob Agents Chemother 2003; 47: 3653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whaley SG, Rogers PD. Azole resistance in Candida glabrata. Curr Infect Dis Rep 2016; 18: 41. [DOI] [PubMed] [Google Scholar]
- 10. Bidaud AL, Chowdhary A, Dannaoui E. Candida auris: an emerging drug resistant yeast - a mini-review. J Mycol Med 2018; 28: 568–73. [DOI] [PubMed] [Google Scholar]
- 11. Beardsley J, Halliday CL, Chen SCet al. Responding to the emergence of antifungal drug resistance: perspectives from the bench and the bedside. Future Microbiol 2018; 13: 1175–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fraczek MG, Bromley M, Buied Aet al. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 2013; 68: 1486–96. [DOI] [PubMed] [Google Scholar]
- 13. Sionov E, Chang YC, Garraffo HMet al. Identification of a Cryptococcus neoformans cytochrome P450 lanosterol 14α-demethylase (Erg11) residue critical for differential susceptibility between fluconazole/voriconazole and itraconazole/posaconazole. Antimicrob Agents Chemother 2012; 56: 1162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Selb R, Fuchs V, Graf Bet al. Molecular typing and in vitro resistance of Cryptococcus neoformans clinical isolates obtained in Germany between 2011 and 2017. Int J Med Microbiol 2019; 309: 151336. [DOI] [PubMed] [Google Scholar]
- 15. Bosco-Borgeat ME, Mazza M, Taverna CGet al. Amino acid substitution in Cryptococcus neoformans lanosterol 14-alpha-demethylase involved in fluconazole resistance in clinical isolates. Rev Argent Microbiol 2016; 48: 137–42. [DOI] [PubMed] [Google Scholar]
- 16. Gago S, Serrano C, Alastruey-Izquierdo Aet al. Molecular identification, antifungal resistance and virulence of Cryptococcus neoformans and Cryptococcus deneoformans isolated in Seville, Spain. Mycoses 2017; 60: 40–50. [DOI] [PubMed] [Google Scholar]
- 17. Gast CE, Basso LR Jr, Bruzual Iet al. Azole resistance in Cryptococcus gattii from the Pacific Northwest: investigation of the role of ERG11. Antimicrob Agents Chemother 2013; 57: 5478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denning DW, Park S, Lass-Florl Cet al. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis 2011; 52: 1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ballard E, Melchers WJG, Zoll Jet al. In-host microevolution of Aspergillus fumigatus: a phenotypic and genotypic analysis. Fungal Genet Biol 2018; 113: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai HJ, Nelliat A. A double-edged sword: aneuploidy is a prevalent strategy in fungal adaptation. Genes (Basel) 2019; 10: 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sionov E, Lee H, Chang YCet al. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathogens 2010; 6: e1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stone NRH, Rhodes J, Fisher MCet al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J Clin Invest 2019; 129: 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rhodes J, Beale MA, Vanhove Met al. A population genomics approach to assessing the genetic basis of within-host microevolution underlying recurrent cryptococcal meningitis infection. G3 (Bethesda) 2017; 7: 1165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boyce KJ, Wang Y, Verma Set al. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio 2017; 8: e00595-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. dos Reis TF, Silva LP, de Castro PAet al. The Aspergillus fumigatus mismatch repair MSH2 homolog is important for virulence and azole resistance. mSphere 2019; 4: e00416-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Billmyre RB, Clancey SA, Heitman J. Natural mismatch repair mutations mediate phenotypic diversity and drug resistance in Cryptococcus deuterogattii. Elife 2017; 6: e28802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Healey KR, Zhao Y, Perez WBet al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun 2016; 7: 11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Billmyre RB, Applen Clancey S, Li LXet al. 5-Fluorocytosine resistance is associated with hypermutation and alterations in capsule biosynthesis in Cryptococcus. Nat Commun 2020; 11: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyce KJ, Cao C, Xue Cet al. A spontaneous mutation in DNA polymerase POL3 during in vitro passaging causes a hypermutator phenotype in Cryptococcus species. DNA Repair (Amst) 2020; 86: 102751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh A, Healey KR, Yadav Pet al. Absence of azole or echinocandin resistance in Candida glabrata isolates in India despite background prevalence of strains with defects in the DNA mismatch repair pathway. Antimicrob Agents Chemother 2018; 62: e00195-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dellière S, Healey K, Gits-Muselli Met al. Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a French cohort of patients harboring low rates of resistance. Front Microbiol 2016; 7: 2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Byun SA, Won EJ, Kim MNet al. Multilocus sequence typing (MLST) genotypes of Candida glabrata bloodstream isolates in Korea: association with antifungal resistance, mutations in mismatch repair gene (Msh2), and clinical outcomes. Front Microbiol 2018; 9: 1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hou X, Xiao M, Wang Het al. Profiling of PDR1 and MSH2 in Candida glabrata bloodstream isolates from a multicenter study in China. Antimicrob Agents Chemother 2018; 62: e00153-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bordallo-Cardona MA, Agnelli C, Gomez-Nunez Aet al. MSH2 gene point mutations are not antifungal resistance markers in Candida glabrata. Antimicrob Agents Chemother 2019; 63: e01876-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Biswas C, Marcelino VR, Van Hal Set al. Whole genome sequencing of Australian Candida glabrata isolates reveals genetic diversity and novel sequence types. Front Microbiol 2018; 9: 2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shor E, Schuyler J, Perlin DS. A novel, drug resistance-independent, fluorescence-based approach to measure mutation rates in microbial pathogens. mBio 2019; 10: e00120-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mahabeer Y, Chang CC, Naidu Det al. Comparison of Etests and Vitek 2 ® to broth microdilution for the susceptibility testing of Cryptococcus neoformans. Diagn Microbiol Infect Dis 2014; 80: 294–8. [DOI] [PubMed] [Google Scholar]
- 38. Loftus BJ, Fung E, Roncaglia Pet al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 2005; 307: 1321–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Basenko EY, Pulman JA, Shanmugasundram Aet al. FungiDB: an integrated bioinformatic resource for fungi and oomycetes. J Fungi (Basel) 2018; 4: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Litvintseva AP, Mitchell TG. Most environmental isolates of Cryptococcus neoformans var. grubii (serotype A) are not lethal for mice. Infect Immun 2009; 77: 3188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jung KW, Bahn YS. The stress-activated signaling (SAS) pathways of a human fungal pathogen, Cryptococcus neoformans. Mycobiology 2009; 37: 161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roth C, Murray D, Scott Aet al. Pleiotropy and epistasis within and between signaling pathways defines the genetic architecture of fungal virulence. PLoS Genet 2021; 17: e1009313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ko YJ, Yu YM, Kim GBet al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell 2009; 8: 1197–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bahn YS, Kojima K, Cox GMet al. Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Mol Biol Cell 2005; 16: 2285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feyder S, De Craene JO, Bär Set al. Membrane trafficking in the yeast Saccharomyces cerevisiae model. Int J Mol Sci 2015; 16: 1509–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Olayioye MA, Noll B, Hausser A.. Spatiotemporal control of intracellular membrane trafficking by Rho GTPases. Cells 2019; 8: 1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bruzzone MJ, Grünberg S, Kubik Set al. Distinct patterns of histone acetyltransferase and mediator deployment at yeast protein-coding genes. Genes Dev 2018; 32: 1252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jung KW, Yang DH, Maeng Set al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun 2015; 6: 6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim JA, Haber JE. Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Proc Natl Acad Sci U S A 2009; 106: 1151–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Selth L, Svejstrup JQ. Vps75, a new yeast member of the NAP histone chaperone family. J Biol Chem 2007; 282: 12358–62. [DOI] [PubMed] [Google Scholar]
- 51. Wakatsuki T, Sasaki M, Kobayashi T. Defects in the NuA4 acetyltransferase complex increase stability of the ribosomal RNA gene and extend replicative lifespan. Genes Genet Syst 2019; 94: 197–206. [DOI] [PubMed] [Google Scholar]
- 52. Sionov E, Chang YC, Garraffo HMet al. Heteroresistance to fluconazole in Cryptococcus neoformans is intrinsic and associated with virulence. Antimicrob Agents Chemother 2009; 53: 2804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lang GI, Parsons L, Gammie AE. Mutation rates, spectra, and genome-wide distribution of spontaneous mutations in mismatch repair deficient yeast. G3 (Bethesda) 2013; 3: 1453–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Binda M, Péli-Gulli MP, Bonfils Get al. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol Cell 2009; 35: 563–73. [DOI] [PubMed] [Google Scholar]
- 55. Nakamura N, Hirata A, Ohsumi Yet al. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J Biol Chem 1997; 272: 11344–9. [DOI] [PubMed] [Google Scholar]
- 56. Piffer AC, dos Santos FM, Thomé MPet al. Transcriptomic analysis reveals that mTOR pathway can be modulated in macrophage cells by the presence of cryptococcal cells. Genet Mol Biol 2021; 44: e20200390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fan CL, Liu TB. The vacuolar morphogenesis protein Vam6-like protein Vlp1 is required for pathogenicity of Cryptococcus neoformans. J Fungi (Basel) 2021; 7: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luna-Tapia A, Kerns ME, Eberle KEet al. Trafficking through the late endosome significantly impacts Candida albicans tolerance of the azole antifungals. Antimicrob Agents Chemother 2015; 59: 2410–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tu J, Vallier LG, Carlson M. Molecular and genetic analysis of the SNF7 gene in Saccharomyces cerevisiae. Genetics 1993; 135: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hu G, Caza M, Cadieux Bet al. The endosomal sorting complex required for transport machinery influences haem uptake and capsule elaboration in Cryptococcus neoformans. Mol Microbiol 2015; 96: 973–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cornet M, Gaillardin C, Richard ML. Deletions of the endocytic components VPS28 and VPS32 in Candida albicans lead to echinocandin and azole hypersensitivity. Antimicrob Agents Chemother 2006; 50: 3492–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tkach JM, Yimit A, Lee AYet al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 2012; 14: 966–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nielsen K, Cox GM, Wang Pet al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and α isolates. Infect Immun 2003; 71: 4831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Idnurm A. A tetrad analysis of the basidiomycete fungus Cryptococcus neoformans. Genetics 2010; 185: 153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.