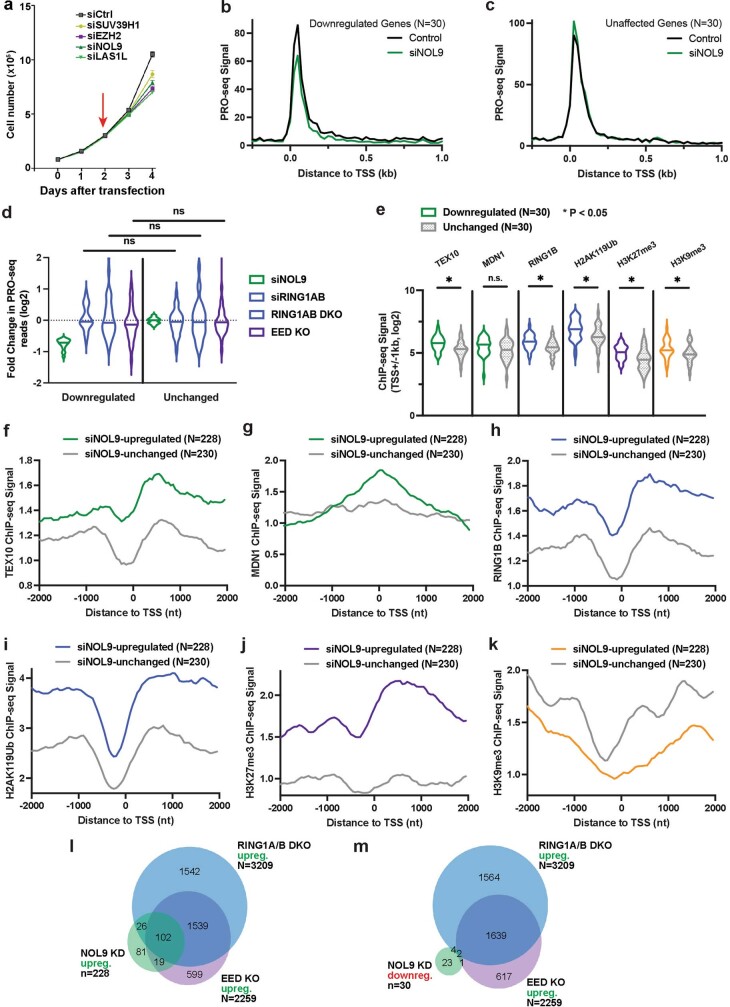
Extended Data Fig. 6. Rixosome subunits, H2AK119ub1, RING1B, and H3K27me3 are preferentially enriched at PRO-seq siNOL9-upregulated genes.
a, Growth curves show cell number changes at indicated time points after knockdowns with siCtrl, siSUV39H1, siEZH2, siNOL9, or siLAS1L in HEK293FT cells. Error bars represent standard deviation for three biological replicates. Data are presented as mean values +/− SEM. b, Average distribution of PRO-seq signal is shown at genes downregulated by siNOL9 (N = 30). Data are shown in 25-nt bins. c, Average distribution of PRO-seq signal is shown at a set of genes unaffected by siNOL9 (N = 30) which were expression matched for the downregulated genes in b. Data are shown in 25-nt bins. d, Violin plots depict the log2 (fold change) in PRO-seq for siNOL9 downregulated (N = 30) and unaffected (N = 30) genes in siNOL9, siRING1B, RING1AB DKO, and EED KO cells. Knockout cells were treated with control siRNA. p-values are from two-tailed Mann-Whitney test. P = 0.3581 for siRING1AB, P = 0.6438 for RING1AB DKO, P = 0.6228 for EED KO. e, Violin plots showing read counts for the indicated ChIP-seq experiments. Reads in were summed ± 1 kb from TSSs for the gene groups indicated. Violin plots depict the range of values, with median indicated by a line. p-values are from two-tailed Mann-Whitney test. P = 0.0034 for TEX10, P = 0.0648 for MDN1, P = 0.0058 for RING1B, P = 0.017 for H2AK119ub1, P = 0.0028 for H3K27me3, P = 0.0276 for H3K9me3. n.s., not significant. f–k, Average distribution of the indicated ChIP-seq reads at siNOL9-upregulated or siNOL9-unaffected genes. Read counts per gene were summed in 50-nt bins. l–m, Venn diagrams showing the overlap between siNOL9-upregulated (l) and siNOL9-downregulated (m) genes with genes upregulated in RING1AB DKO or EED KO cells in PRO-seq experiments.