Abstract
VanD-type Enterococcus faecium BM4416 was constitutively resistant to vancomycin and to teicoplanin by synthesis of peptidoglycan precursors ending in d-alanyl–d-lactate. Like E. faecium BM4339, the only VanD-type strain described so far, BM4416 produced an impaired d-alanine:d-alanine ligase. Unlike for BM4339, which had a 5-bp insertion in ddl, inactivation of the gene in BM4416 was due to insertion of IS19.
Acquired glycopeptide resistance in enterococci is due to the synthesis of peptidoglycan precursors ending in the depsipeptide d-alanyl–d-lactate (d-Ala–d-Lac) instead of the dipeptide d-alanyl–d-alanine (d-Ala–d-Ala) (5). The former considerably reduces binding affinity of glycopeptides for their peptidoglycan targets (2). Inducible high-level resistance to both vancomycin and teicoplanin and inducible variable levels of resistance to vancomycin alone are due to acquisition of the vanA and vanB operons, respectively (1, 2). Recently, a new glycopeptide resistance gene cluster, vanD, has been characterized in Enterococcus faecium BM4339, which is resistant to intermediate levels of vancomycin and to low levels of teicoplanin (6, 10). The organization of the vanA, vanB, and vanD operons is similar. Three proteins are required for glycopeptide resistance: a ligase (VanA, VanB, or VanD) to form the depsipeptide d-Ala–d-Lac, a dehydrogenase (VanH, VanHB, or VanHD) to convert pyruvate into d-Lac (5), and a d,d-dipeptidase (VanX, VanXB, or VanXD) to hydrolyze the dipeptide d-Ala–d-Ala synthesized by the chromosomal d-Ala:d-Ala Ddl ligase (4, 6, 13, 14). A d,d-carboxypeptidase (VanY, VanYB, or VanYD), not essential for resistance in VanA strains (3), hydrolyzes the C-terminal d-Ala of the remaining pentapeptide. VanYD is distinct from VanY and VanYB since it displays higher identity with some penicillin-binding proteins than with VanY and VanYB and contains the motifs predicted to define the active sites of penicillin-binding proteins (6). A two-component regulatory system, encoded by the vanR or vanRB and vanS or vanSB genes, controls the expression level of the resistance genes. In BM4439, the first VanD-type strain that has been described, resistance is constitutively expressed despite the presence of the vanSD and vanRD genes (6). In this strain, a 5-bp insertion in the 5′ part of the ddl gene is responsible for inactivation of Ddl and accounts for the lack of precursors terminating in d-Ala–d-Ala (6).
E. faecium BM4416 was isolated from a stool specimen of a 59-year-old male treated with vancomycin after a liver transplant for alcoholic cirrhosis. This strain was resistant to vancomycin (MIC = 128 μg/ml) and to teicoplanin (MIC = 64 μg/ml); it was also resistant to erythromycin, penicillin G, and high levels of streptomycin.
BM4416 was identified as E. faecium by a PCR assay using primers specific for genes encoding d-Ala:d-Ala ligases in enterococci (7). A PCR product was obtained only with the primer pair specific for the E. faecium ddl gene, but the length of the amplified fragment, approximately 2,100 bp, exceeded the expected length of 1,077 bp. Species identification was confirmed by PCR using two pairs of primers (8FPL-806R and 515FPL-13B) located in consensus sequences of rrs coding for 16S rRNA (11, 12). Nucleotide sequences of the fragments amplified from BM4416 total DNA were identical to those from a control strain, E. faecium BM4107. PCR primers specific for resistance genes vanA, vanB, vanC-1, vanC-2, and vanD (7, 10) indicated that BM4416 was of glycopeptide resistance genotype vanD.
The cytoplasmic peptidoglycan precursors of E. faecium BM4416 grown with (4 μg/ml) or without vancomycin were analyzed as described previously (13) (Table 1). Strain BM4416 mainly produced UDP-MurNAc-pentadepsipeptide (69%) terminating in d-Ala–d-Lac, UDP-MurNAc-tetrapeptide (24%), and UDP-MurNAc-tripeptide (7%). No significant amounts of UDP-MurNAc-pentapeptide were found (<1%). There were no differences between the peptidoglycan precursors produced from uninduced and induced cells. The precursors synthesized by BM4416 were qualitatively and quantitatively similar to those of E. faecium BM4339. As in E. faecium BM4339, glycopeptide resistance was constitutively expressed.
TABLE 1.
Cytoplasmic peptidoglycan precursors synthesized by E. faecium BM4416a
| Culture | % of peptidoglycan precursors
|
|||
|---|---|---|---|---|
| UDP-MurNAc-tripeptide | UDP-MurNAc-tetrapeptide | UDP-MurNAc-pentapeptide | UDP-MurNAc-pentadepsipeptide | |
| Uninduced | 7 | 24 | <1 | 69 |
| Induced (vancomycin, 4 μg/ml) | 11 | 24 | <1 | 65 |
Peptidoglycan synthesis was inhibited by addition of ramoplanin to the cultures for 20 min. The individual precursors were characterized by amino acid analysis and mass spectroscopy.
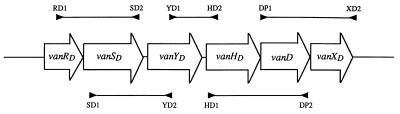
Organization of the vanD operon in BM4416 was determined by PCR mapping (Fig. 1). Several pairs of primers, specific for each gene of the BM4339 vanD operon, were used in PCR experiments (Table 2). The PCRs gave fragments with the expected size, indicating that all the genes constituting the vanD operon were present and in the same order as in BM4339.
FIG. 1.
PCR mapping of the vanD operon. Open arrows represent coding genes. Horizontal bars depict the PCR products. Arrowheads indicate direction of DNA synthesis. The sequences of the primers are given in Table 2.
TABLE 2.
Oligodeoxynucleotides used for gene mapping
| Primer | Nucleotide sequence | Positiona | Product size (bp) |
|---|---|---|---|
| RD1 | 5′-CCGTTTAACCCGCTGGAA | 304–321 | 1,269 |
| SD2 | 5′-CGAATGGTGGTATTCTC | 1556–1572 | |
| SD1 | 5′-CTATCATGATCGGGATG | 865–881 | 1,346 |
| YD2 | 5′-CTCTGGAACTGAGGGTA | 2194–2210 | |
| YD1 | 5′-GATTCGTCAACCGCATG | 2433–2449 | 746 |
| HD2 | 5′-ATCCAGCATTGCTTTCTG | 3161–3178 | |
| HD1 | 5′-CGTAAGCCATAAAGCGGA | 3204–3221 | 1,783 |
| DP2 | 5′-AAACCGGCTGCTGTCATCATG | 4966–4986 | |
| DP1 | 5′-AATCACAAAATCCGGCG | 4132–4148 | 1,499 |
| XD2 | 5′-TATGTATCCGGGTATGG | 5614–5630 |
Nucleotide numbering begins at the first base of the vanRD gene. Oligodeoxynucleotides were synthesized in the Unité de Chimie Organique, Institut Pasteur, Paris, France.
The vanD gene clusters were assigned to a chromosomal location in BM4416 and in BM4339 by contour-clamped homogeneous electric field gel electrophoresis. Agarose plugs were prepared and digested according to the manufacturer's recommendations (37°C, 3 h, 0.01 U) with I-CeuI, an intron-encoded endonuclease specific for rRNA genes (9). Fragments were separated on a 0.8% agarose gel using a CHEF-DRIII system (Bio-Rad Laboratories, Hercules, Calif.) under the following conditions: total migration, 24 h; initial pulse, 60 s; final pulse, 120 s; voltage, 6 V/cm; included angle, 120°; and temperature, 14°C. Fragments were transferred to a nitrocellulose sheet and hybridized (i) to an α-32P-labeled 16S rRNA (rrs) probe obtained by amplification of an internal portion of the rrs gene with primers RW01 and DG74 (8) and (ii) to a vanHDDXD probe obtained by PCR with primers HD1 and XD2 (Fig. 1 and Table 2). The rrs probe hybridized with five and four I-CeuI fragments from BM4416 and BM4339, respectively, and the vanHDDXD probe cohybridized with a 380-kb fragment from BM4416 and a 330-kb fragment from BM4339. These data indicate a chromosomal location for the vanD resistance operon in both strains and that the two isolates are distinct.
d,d-Peptidase activities in E. faecium BM4416 were assayed in enzyme extracts from E. faecium BM4416 prepared as described previously (4). Enzyme assays were performed with the S100 and C100 fractions of uninduced or induced (vancomycin concentration of 8, 16, or 64 μg/ml) cultures of E. faecium BM4416 (Table 3). Weak d,d-dipeptidase activity (VanXD) was found in the cytoplasmic extracts from vancomycin-induced or uninduced cells (Table 3). This is in contrast with strain BM4339, in which no VanX activity was detected (13). The difference in regulation of production of VanXD activity in these strains is currently under study. The amounts of d,d-carboxypeptidase (VanYD) activity found in membrane extracts of BM4416 and BM4339 were similar, and there were no differences between induced and uninduced cultures (Table 3) (10). As observed previously in BM4339, the VanYD activity of BM4416 was completely inhibited by penicillin G (10 mM).
TABLE 3.
d,d-Peptidase activities in extracts from E. faecium BM4416
| Concn of vancomycin (μg/ml) | d,d-Dipeptidase activitya (nmol min−1 mg−1)b |
d,d-Carboxypeptidase activityc (nmol min−1 mg−1)b
|
|
|---|---|---|---|
| Without penicillin G | With penicillin G (10 mM) | ||
| 0 | 10.1 ± 2.1 | 14.9 ± 2.6 | <10−1 |
| 8 | 10.4 ± 1.7 | 13 ± 0.5 | <10−1 |
| 16 | 10.8 ± 1.6 | 12.4 ± 1.8 | <10−1 |
| 64 | 13.8 ± 4 | 13.1 ± 1.3 | <10−1 |
The activity was measured in the 100,000 × g supernatant of lysed bacteria.
Values are the means ± standard deviations for a minimum of three independent experiments.
The activity was measured in the resuspended pellet fraction after centrifugation of lysed bacteria at 100,000 × g for 45 min.
The unexpected length of the PCR product obtained with primers specific for the E. faecium ddl gene, the presence of UDP-MurNAc-tripeptide in the pool of peptidoglycan precursors, and the absence of pentapeptide were compatible with the fact that BM4416 produced an impaired d-Ala:d-Ala ligase. Amplification by PCR of the entire ddl gene with specific primers (Y. Gholizadeh et al., unpublished data) gave rise to a 2,100-bp fragment which was cloned into PCR2.1 (Invitrogen, Leek, The Netherlands) and sequenced. An insertion sequence, designated IS19, of 1,038 bp was found in the middle of the gene (Fig. 2). Insertion of the element generated a 9-bp (TATCAATAA) duplication of target DNA corresponding to nucleotides 762 to 770 of the ddl gene. IS19 was flanked by 21-bp perfect inverted repeats and contained an open reading frame which exhibited 39% identity with the structural gene for a putative transposase in IS982 from Lactococcus lactis (15). The presence of this genetic element accounts for the lack of synthesis of functional Ddl in E. faecium BM4416. As found for BM4339, the d-Ala:d-Ala ligase is not functional in BM4416. Constitutive expression of the resistance pathway, leading to production of modified precursors, allows growth of BM4416 in the absence of glycopeptides.
FIG. 2.
Sequence of IS19. The deduced amino acid sequence of the putative transposase is shown below the nucleotide sequence. The 21-bp perfect inverted repeats are indicated by arrows. The 9-bp duplication at the insertion site is indicated in bold characters. Putative −35 and −10 promoter regions and the putative ribosome-binding site (rbs) are indicated.
Resistance to glycopeptides in E. faecium BM4416 was due to synthesis of late peptidoglycan precursors ending in d-Ala–d-Lac. Constitutive resistance was encoded by a vanD operon closely related to that of E. faecium BM4339 and also located in the chromosome. Surprisingly, both VanD-type strains described so far produce an inactivated d-Ala:d-Ala ligase due to an insertion in the ddl gene. Differences in the levels of glycopeptide resistance of the two clinical isolates could be due to differences in the two-component regulatory systems or in individual resistance gene regulation.
Acknowledgments
We thank J. Conly for the gift of strain BM4416.
This work was supported in part by a Bristol-Myers Squibb Unrestricted Biomedical Research Grant in Infectious Diseases and by the “Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires” from the Ministère de l'Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Reynolds P E, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Depardieu F, Cabanié L, Reynolds P E, Courvalin P. Requirement of the VanY and VanX d,d-peptidases for glycopeptide resistance in enterococci. Mol Microbiol. 1998;30:819–830. doi: 10.1046/j.1365-2958.1998.01114.x. [DOI] [PubMed] [Google Scholar]
- 4.Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 5.Bugg T D H, Wright G D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 6.Casadewall B, Courvalin P. Characterization of the vanD glycopeptide resistance gene cluster from Enterococcus faecium BM4339. J Bacteriol. 1999;181:3644–3648. doi: 10.1128/jb.181.12.3644-3648.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greisen K, Loeffelholz M, Purohit A, Leong D. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J Clin Microbiol. 1994;32:335–351. doi: 10.1128/jcm.32.2.335-351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S L, Hessel A, Sanderson K E. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc Natl Acad Sci USA. 1993;90:6874–6878. doi: 10.1073/pnas.90.14.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perichon B, Reynolds P, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis—an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 12.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 13.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl–d-alanine. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu Z, Wright G D, Walsh C T. Overexpression, purification, and characterization of VanX, a d,d-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1995;34:2455–2463. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]
- 15.Yu Z, Mierau I, Mars A, Johnson E, Dunny G, McKay L L. Novel insertion sequence-like element IS982 in lactococci. Plasmid. 1995;33:218–225. doi: 10.1006/plas.1995.1023. [DOI] [PubMed] [Google Scholar]