Abstract
A Tn1546-related element with IS1216V at position 8839 underwent a structural change after storage of the host strain of Enterococcus faecium at 4°C. The element acquired IS1542 at position 3932, nucleotides 8732 to 8831 were deleted, and the first 3417 nucleotides were lost and replaced by an inverted copy of the IS1216V–vanY–vanZ-inverted-repeat block from the 3′ end. Insertion sequence movement is likely to play a key role in the evolution of VanA resistance elements.
Elements that mediate VanA glycopeptide resistance in enterococci are diverse, although all retain the vanRSHAX gene cluster of the prototype element, Tn1546 (6, 9). The presence of insertion sequences (IS elements) accounts for much of the heterogeneity. IS1216V is widespread among VanA enterococci of diverse geographical origins (6), whereas other elements appear to have a more restricted distribution (2, 3, 5). The presence of IS1216V at diverse positions within Tn1546 (4, 6, 8), including intragenic positions within the vanRSHAX cluster (A. Darini, M.-F. I. Palepou, D. James, and N. Woodford, Letter, Antimicrob. Agents Chemother. 43:995–996, 1999), provides indirect evidence that these IS elements are mobile. IS element movement has probably played a key role in the diversification of VanA elements (8).
Isolates of Enterococcus faecium representing a single clone, defined by pulsed-field gel electrophoresis (PFGE), may contain distinct VanA elements (7, 9), suggesting that these elements change more rapidly than other molecular markers, such as the distribution of SmaI digestion sites. Nevertheless, we are unaware of any reports of structural alterations of a VanA element during a period of study. For the present report, we characterized the changes observed in a VanA element following storage of the host enterococcus at 4°C.
Twenty-four strains of E. faecium harboring VanA elements of groups A to X (9) were stored at 4°C in a cold room on Columbia blood agar plates for 2 to 3 months. Strains were subcultured once onto fresh agar plates, and the VanA elements were then characterized using overlapping PCR (1) and long PCR (L-PCR) restriction fragment length polymorphism (RFLP) analysis with primer LP1 (located in the terminal inverted repeats of Tn1546), followed by ClaI digestion of the amplicons (6). In addition, L-PCR with primers LP1 (forward) and LP4 (reverse; 5′-GGC AAG GTC AAT CTC AGA CTT GTC T-3′) was performed on BamHI-digested genomic DNA to amplify only the 5′ ends of VanA elements, upstream of position 7294 within vanA. To study the stability of VanA elements, two strains with VanA elements of groups A (Tn1546) and B were passaged in L broth for 30 days; they were subcultured in fresh broth daily. In parallel experiments, the strains were passaged for 30 days in L broth containing vancomycin (Sigma, Poole, United Kingdom) at 10 μg/ml. On days 10, 20, and 30, samples from each broth were checked for purity and vancomycin resistance and stored at −70°C. PFGE (10) was performed to confirm that no contamination of strains occurred during the experiment. IS1216V and IS1542 were sought within the genomes of enterococci using previously described primers and amplification conditions (4, 9). The sequences of selected regions of VanA elements were determined and analyzed as described previously (2, 9). Most sequencing was performed on purified PCR amplicons with a Hybaid Recovery Kit (Hybaid, Teddington, United Kingdom), although the termini of element b (see below) were sequenced using vector-based primers M13R and M13-20F from the LP1-LP1 L-PCR product cloned into pCR2.1-TOPO (Invitrogen, Groningen, The Netherlands).
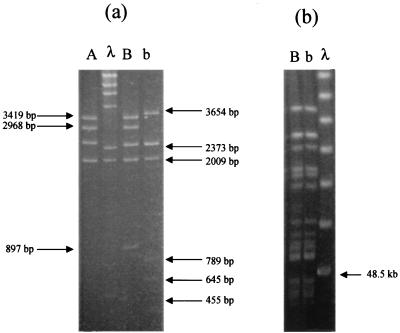
After recovery from prolonged storage at 4°C, 23 VanA elements gave overlapping PCR patterns indistinguishable from those reported previously (9). The recovered group B element gave a distinct pattern. This profile was unlike those of elements A and C to X, and the novel element was designated b (Table 1). L-PCR RFLP analysis with primer LP1 and ClaI revealed that bands of 3,419 and 2,968 bp which were present in B and also in Tn1546 were absent in b, being replaced by a single larger band of ca. 3.6 kb. There were additional changes in smaller bands (<1 kb; Fig. 1a); in contrast to B, the 897-bp band was absent from b and three new small bands appeared. These data indicated that the main structural changes were located upstream of the vanRSHAX cluster. Isolates containing VanA elements B and b were indistinguishable after PFGE of SmaI-digested genomic DNA (Fig. 1b), indicating that the strain had not become contaminated during the experiment.
TABLE 1.
Overlapping PCR for Tn1546 and VanA elements B and b
| VanA element | Amplificationa with PCR primer (annealing sites):
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p1p2 (22–1330) | p3p4 (1222–2353) | p5p6 (2227–3525) | p7p8 (2769–4042) | p9p10 (3569–4793) | p11p12 (4675–6353) | p13p14 (6229–8021) | p15p16 (6979–8920) | p17p18 (8889–10473) | p19p1 (10403–10830) | |
| A (Tn1546)b | + | + | + | + | + | + | + | + | + | + |
| B | + | + | + | + | + | + | + | ++ | + | + |
| b | − | − | − | − | ++ | + | + | ++ | + | + |
−, no amplification; +, amplification; ++, amplicon larger than that of Tn1546.
Group A served as the control.
FIG. 1.
(a) L-PCR RFLP patterns (ClaI digests) of VanA resistance elements B and b. Lane A shows the pattern of the prototype VanA element, Tn1546. A phage λ DNA HindIII digest is shown as a size standard. (b) PFGE patterns of SmaI-digested genomic DNA from E. faecium isolates containing VanA elements B and b. A concatemer of phage λ DNA (48.5 kb) is shown as a size standard.
The stability of VanA elements of groups A (Tn1546) and B was studied prospectively. After 30 days of passage in broth containing no vancomycin, strains containing group A and B elements remained glycopeptide resistant. The group A element was structurally unchanged, but the group B element had become indistinguishable, by L-PCR RFLP analysis, from b. No changes were observed in either the group A or B elements following passage in broth containing vancomycin.
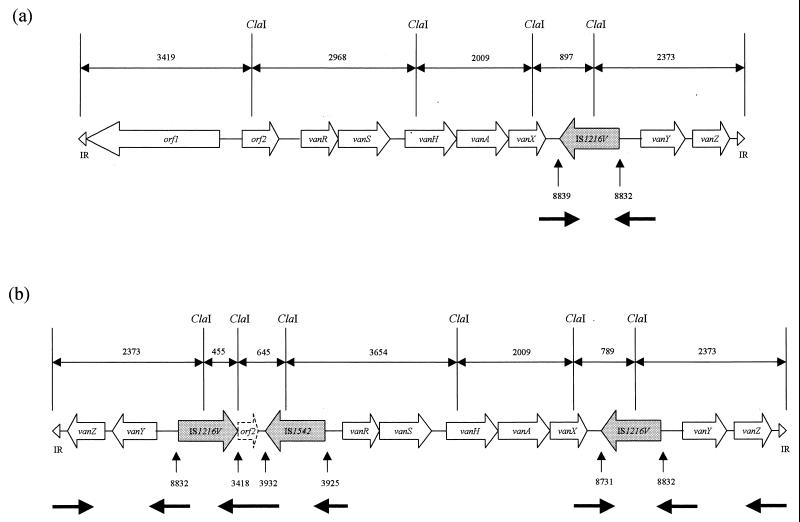
Element B had a copy of IS1216V at position 8839 and corresponded to transposon type B1 (8) (Fig. 2a). PCR indicated that IS1542 was present within the genome of the host strain, although not within the VanA element. Element b acquired IS1542 at position 3932 (Fig. 2b), as reported for other VanA elements (2, 9). Element b had lost nucleotides 8732 to 8831 but gained an additional copy of IS1216V immediately before position 3418. IS1216V has been documented at this position in VanA elements with deletions of the first 3,417 nucleotides (8). Sequencing confirmed loss of orf1 and partial loss of orf2 from b; the 5′ end comprised an inverted duplication of the IS1216V–vanY–vanZ–inverted-repeat block from the 3′ end (Fig. 2b). The structures of B and b (Fig. 2a and b) were consistent with their L-PCR RFLP profiles (Fig. 1a). The presence of vanY and vanZ at the 3′ end of element B but at both the 5′ and 3′ ends of element b was confirmed by hybridization of BamHI-digested LP1 L-PCR products with specific intragenic probes under stringent conditions (data not shown). The changes in b required at least two genetic events, transposition of IS1542 and duplication of the gene block.
FIG. 2.
Schematic representation of VanA element B (a) and derivative b (b) showing the predicted ClaI cutting sites, fragment sizes, and positions of IS elements (indicating target site duplications where detected). The thick horizontal arrows indicate those regions sequenced. IR indicates a terminal inverted repeat.
In conclusion, we have reported a structurally novel VanA element with an inverted block of IS1216V and van genes at both the 5′ and 3′ ends. The frequent presence in VanA elements of IS1216V both within the vanX-vanY intergenic region and upstream of vanR (8) leads us to speculate that elements with similar structures may occur in some clinical isolates, and we have preliminary data to support this (C. H. Tremlett, D. F. J. Brown, and N. Woodford, unpublished data, 1999). The mechanism mediating the duplication requires further study, but a replicative transposition event involving a composite transposon carrying the vanY and vanZ genes and flanked by IS1216V elements is one possibility. Sequencing of the DNA flanking B and b will indicate the extent of any such composite element. IS element movement causes structural alterations in some VanA elements, including complete or partial loss of transposition genes orf1 and orf2. Such movement is likely to have played a key role in the evolution of these resistance elements and may modulate their future transmissibility.
Acknowledgments
We are grateful to Patricia Woodford (Imperial College of Science, Technology & Medicine at St. Mary's, London, United Kingdom) for processing samples on the automated sequencer and to Rob Willems (National Institute of Public Health and the Environment, Bilthoven, The Netherlands) for helpful discussion during the preparation of the manuscript.
A. L. C. Darini was funded by a grant from the Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP-Proc. 1998/00498-7), Brazil.
REFERENCES
- 1.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darini A, Palepou M-F I, Woodford N. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol Lett. 1999;173:341–346. doi: 10.1111/j.1574-6968.1999.tb13523.x. [DOI] [PubMed] [Google Scholar]
- 3.Handwerger S, Skoble J, Discotto L F, Pucci M J. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob Agents Chemother. 1995;39:362–368. doi: 10.1128/aac.39.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jensen L B, Ahrens P, Dons L, Jones R N, Hammerum A M, Aarestrup F M. Molecular analysis of Tn1546 in Enterococcus faecium isolated from animals and humans. J Clin Microbiol. 1998;36:437–442. doi: 10.1128/jcm.36.2.437-442.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacKinnon M G, Drebot M A, Tyrell G J. Identification and characterization of IS1476, an insertion sequence-like element that disrupts VanY function in a vancomycin-resistant Enterococcus faecium strain. Antimicrob Agents Chemother. 1997;41:1805–1807. doi: 10.1128/aac.41.8.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palepou M-F I, Adebiyi A-M A, Tremlett C H, Jensen L B, Woodford N. Molecular analysis of diverse elements mediating VanA glycopeptide resistance in enterococci. J Antimicrob Chemother. 1998;42:605–612. doi: 10.1093/jac/42.5.605. [DOI] [PubMed] [Google Scholar]
- 7.Tremlett C H, Brown D F J, Woodford N. Variation in structure and location of VanA glycopeptide resistance elements among enterococci from a single patient. J Clin Microbiol. 1999;37:818–820. doi: 10.1128/jcm.37.3.818-820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willems R J L, Top J, van den Braak N, van Belkum A, Mevius D J, Hendriks G, van Santen-Verheuvel M, van Embden J D A. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob Agents Chemother. 1999;43:483–491. doi: 10.1128/aac.43.3.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodford N, Adebiyi A-M A, Palepou M-F I, Cookson B D. Diversity of VanA glycopeptide resistance elements in enterococci from humans and nonhuman sources. Antimicrob Agents Chemother. 1998;42:502–508. doi: 10.1128/aac.42.3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woodford N, Morrison D, Johnson A P, Bateman A, Hastings J G M, Elliott T S J, Cookson B. Plasmid-mediated vanB glycopeptide resistance in enterococci. Microb Drug Resist. 1995;1:235–240. doi: 10.1089/mdr.1995.1.235. [DOI] [PubMed] [Google Scholar]