Abstract
Cellular metabolism requires dissolved oxygen gas. Because evolutionary refinements have constrained mammalian dissolved oxygen levels, intracellular oxygen sensors are vital for optimizing the bioenergetic and biosynthetic use of dissolved oxygen. Prolyl hydroxylase domain (PHD) homologs 1–3 (PHD1/2/3) are molecular oxygen dependent non-heme dioxygenases whose enzymatic activity is regulated by the concentration of dissolved oxygen. PHD oxygen dependency has evolved into an important intracellular oxygen sensor. The most well studied mechanism of PHD oxygen-sensing is its regulation of the hypoxia-inducible factor (HIF) hypoxia signaling pathway. Heterodimeric HIF transcription factor activity is regulated post-translationally by selective PHD proline hydroxylation of its HIF1α subunit, accelerating HIF1α ubiquitination and proteasomal degradation, preventing HIF heterodimer assembly, nuclear accumulation, and activation of its target oxygen homeostasis genes. Phd2 has been shown to be the key isoform responsible for HIF1α subunit regulation in many cell types and accordingly disruption of the Phd2 gene results in embryonic lethality. In bone cells Phd2 is expressed in high abundance and tightly regulated. Conditional disruption of the Phd1, Phd2 and/or Phd3 gene in various bone cell types using different Cre drivers reveals a major role for PHD2 in skeletal growth and development. In this review, we will summarize the state of current knowledge on the role and mechanism of action of PHD2 as oxygen sensor in regulating bone metabolism.
Keywords: Bone, Prolyl hydroxylase domain, Hypoxia-inducible factor, Bone regeneration, Oxygen sensor, Epigenetic regulation
1. Introduction
It is estimated that 200 million people world-wide and 54 million people in the United States have osteoporosis. One in 2 women and 1 in 4 men after age 40 will have an osteoporosis-related fracture over their lifetime [1]. Osteoporosis is responsible for more than 2 million fractures year and this number continues to grow [2]. Two major causes are known to contribute to the pathogenesis of osteoporosis: 1) failure to achieve peak bone mass that typically occurs around the age of 30; and 2) menopause and age-induced excessive bone resorption that is not compensated by corresponding increase in bone formation. Bone loss occurs with age in part because the rate of bone resorption surpasses the rate of bone formation [3]. Many systemic and local growth factors regulate bone formation processes. At the molecular level, hypoxia signaling is identified as a key signaling pathway that plays a central role in the regulation of bone formation both during normal physiology and under disease conditions [[4], [5], [6]]. Hypoxia-inducible factor (HIF) activity, a heterodimeric (HIF1α/HIFβ) transcription factor for oxygen homeostasis genes, is largely regulated post-translationally by prolyl hydroxylase domain enzymes (PHD1/2/3). PHD1/2/3 were identified originally as the primary intracellular oxygen sensor [7]. PHDs, in the presence of normal dissolved oxygen levels, constitutively hydroxylate specific HIF1α subunit proline residues, accelerating HIF1α turnover through ubiquitination and proteasomal degradation, thereby preventing HIF-heterodimer assembly and its nuclear transcription activity. PHD2 has been shown to be the key isoform responsible for HIF1α regulation in many cell types and accordingly disruption of the Phd2 gene results in embryonic lethality [8,9]. Phd2 is found to be expressed in high abundance in bone cells and disruption of Phd2 gene in bone cells result in severe skeletal phenotype [10,11]. In this review article, we will discuss our current understanding of the role of PHD oxygen sensors in regulating bone metabolism.
2. Troposphere and dissolved oxygen
Cellular metabolism requires dissolved oxygen gas. Both evolutionary refinement of cellular dissolved oxygen utilization and species radiation occurred under oxygen deplete tropospheric conditions [[12], [13], [14], [15], [16], [17], [18], [19]] (Fig. 1). Consequently, these influences imbued mammalian species with unique physiological and cellular adaptions to lowered dissolved oxygen concentrations. Fig. 2 shows human dissolved oxygen concentrations differ considerably throughout the body, and their activities normally occur in dissolved oxygen environments markedly lower than atmospheric levels [20,21]. Similarly, hypoxia, a state of metabolically insufficient dissolved oxygen, differs by organ systems. To maximize intracellular dissolved oxygen utilization among widely varying levels of dissolved oxygen requirements and its availability, and to mitigate against consequential cellular hypoxia, multiple confluent intracellular systems have evolved to optimize cellular oxygen utilization; the pairing of the 2-oxoglutarate-dependent oxygenase, prolyl hydroxylase domain (PHD) enzyme, with transcription factor, hypoxia-inducible factor alpha (HIF1α) being one such system.
Fig. 1.
Evolution of earth's atmospheric oxygen and animal species. Red and green lines represent the range of estimates of oxygen build-up in the earth's atmosphere while time is measured in billions of years ago. P02, partial pressure of oxygen.
Fig. 2.
Normal human dissolved oxygen partial pressures. Normal dissolved oxygen in water, human plasma as well as extracellular fluids in different organs are shown.
3. PHDs are oxygen sensors
PHD homologs (PHD1/2/3), and factor inhibiting HIF [FIH]) are non-heme dioxygenases which sense oxygen via direct enzymatic interaction with dissolved oxygen [[22], [23], [24]]. PHD1/2/3 are Fe2+ 2-oxoglutarate-dependent dioxygenases (2OGD) which represent the largest group of non-heme, Fe2+ bound, oxidizing enzymes. 2OGD enzymes are distributed throughout the bacterial, plant, animal phyla, and are engaged in a wide variety of metabolic processes including hydroxylation, demethylation, epoxidation, desaturation, and halogenation reactions. 2OGD substrates are diverse and found in proteins, nucleic acids, lipids, carbohydrates, and small molecules. For example, cytoplasmic procollagen's vitamin C-dependent post-translational proline and lysine hydroxylation, modifications required for its tertiary folding and quaternary assembly is dependent on 2OGD processing [24,25], and nuclear epigenetic Jumonji C domain containing histone lysine demethylation (KDM) enzyme and 10–11 translocation hydroxylates (TET1-3) enzyme, vital for regional context-dependent promotion or suppression of transcription, are as well 2OGD enzymes [23,24].
2OGD are non-equilibrium enzymes, favoring a forward enzymatic reaction velocity. 2OGD X-ray crystallography studies show a conserved catalytic double-stranded beta-helical core. Fe2+, possibly obtained from cytoplasmic iron chaperone poly-(26)-binding proteins (PCBP1/2) is held in a 3 amino acid histidine, aspartate/glutamate, and histidine coordinating motif, [23,27]. The oxidation state of Fe2+ increases during 2-oxoglutarate dependent catalytic conversion and returns to its ferrous state in the presence of soluble reducing agents such as vitamin C. Fe2+ also protects against intramolecular 2OGD cysteine oxidation (Fig. 3) [28].
Fig. 3.
Schematic review of PHD2 regulation of the HIF system. Regulation of hydroxylase activity by small molecules and protein factors are indicated. PHD proteins catalyze the hydroxylation of proline residues of target proteins including HIFs using oxygen (O2), iron (Fe2+), α-ketoglutarate (also known as 2-oxoglutarate, 2-OG) and ascorbic acid (vitamin C) as co-factors. Poly(rC)-binding protein (PCBP)1 and PCBP2 are iron chaperones that deliver ferrous iron via metal-mediated, protein-protein interaction. Tricarboxylic acid (TCA) cycle provides a source of α-ketoglutarate. Hydroxylation of prolyl residues within the oxygen-dependent degradation domain of HIFs in the presence of co-factors triggers recognition by the von Hippel-Lindau tumor suppressor protein (p-VHL), which is part of a E3 ubiquitin ligase complex (also containing elongin [Elo] C and B, cullin-2 [Cul-2) and RING-Box protein (Rbx) 1 to induce proteosomal degradation. In contrast, under hypoxia PHDs are unable to hydroxylase HIF1α subunits which accumulate and migrate to nucleus, subsequently forming an HIF-complex with other nuclear proteins to induce transcription of HIF1α targets.
2OGD regulates HIF transcription activity [7,23,24,[29], [30], [31]]. HIF is a heterodimer transcription cofactor composed of an 2OGD substrate targeted alpha-subunit isoform (HIF1α/826aa, HIF2α/870aa, HIF3α/667aa) and a constitutively expressed beta-subunit HIFβ. The HIFα isoforms share multiple functional domains. These domains include an oxygen-dependent degradation domain (ODD), terminal trans activation domains (NTAD, CTAD), nuclear localization signal domains (NLS), a transcription activation domain (TAD), an amino-terminal basic helix-loop-helix (bHLH) DNA binding domain, and protein interaction PER-ARNT-SIM domains (PASA, PASB). The HIF3α isoform also possess a leucine zipper transactivation domain (LZIP). A heterodimer of non-hydroxylated HIFα and HiF1β in complex with other transcription factors associates with cis-hypoxia response elements (HRE; 5′-G/ACGTG-3′) within transcription regulatory regions of target genes. A multitude of HRE genes have been identified [32]. HRE genes span a range of processes, developmental to pathophysiological. HIF1β was previously described as the aryl hydrocarbon nuclear translocator (ARNT) protein, a basic helix-loop-helix–Per-ARNT-Sim (bHLH–PAS) protein [33].
HIFα isoforms are constitutively 2OGD hydroxylated at one or both of their N- terminal ODDs (NODD, HIF1α/402aa, HIF2α(EPAS1)/405aa, HIF3α/465aa), and C-terminal ODDs (CODD, HIF1α/564aa, HIF2α/531aa, HIF3α/568aa) [23,29]. Selective HIFα 2OGD hydroxylation increases binding affinity to von-Hippel Lindau tumor suppressor protein (pVHL), the E3 ubiquitin ligase substrate recognition subunit [29,[33], [34], [35]]. Complexed with Ring-box 1 (Rbx1), cullin 2 (Cul2), and elongin B and C, HIFa is polyubiquitinated at lysine residues, directing it to ubiquitin-dependent proteasome degradation. Constitutive 2OGD HIFα hydroxylation decreases HIFa half-life, reducing HIF1α/HIF1b complex concentrations, and ineffective HIF1α/HIF1β HRE directed transcription activity (Fig. 3). Conversely, inhibition of 2OGD activity leads to elevated HIF1α/HIF1β complex concentrations and robust HRE targeted transcription. HRE gene elements are also located in the GLUT 1 glucose transporter, many of the glycolytic enzymes (hexose kinase, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4, aldolase, phosphoglycerate kinase, enolase, and lactate dehydrogenase A), and mitochondrial COX4-1, together capable of increasing glucose entry, glycolytic processing, and respiration [36].
Four 2OGD isoforms, prolyl hydroxylase domain-containing proteins PHD1/2/3 and factor inhibiting HIF (FIH) modify HIF homologs. PHD1/2/3 were previously designated as Egl nine homologs 1–3, and hypoxia-inducible factor prolyl hydroxylase homologs 1–3 in different numerical ordering (PHD1/EGLN2/HPH3, PHD2/EGLN1/HPH2, PHD3/EGLN3/HPH1) [8,37]. PHD2 is also characterized as a Jumonji-C domain (JmjC) containing histone demethylase transcription factor [38]. These 2OGDs utilize cofactors Fe2+, vitamin C, and co-substrates oxygen and 2-oxoglutarate to hydroxylate specific HIFα proline residues in its ODD domain (PHD1/2/3) or in the case of FIH hydroxylate asparagine residues in the HIF CTAD domain. PHD1/2/3-mediated HIFα hydroxylation increases HIFa turnover. FIH asparaginyl hydroxylation disrupts HIFa C-terminal transactivation domain (C-TAD) association with the acetyltransferase transcriptional coactivator CBP/p300 [39].
In addition to vitamin C, PHD activity requires several cofactors including alpha-ketoglutarate, an intermediate metabolite of the mitochondrial inner membrane tricarboxylic cycle (TCA, Krebs Cycle, Citric Acid Cycle) [23,36,40] Alpha-ketoglutarate is derived from isocitrate, by isocitrate dehydrogenase oxidative decarboxylation [28]. Alpha-ketoglutarate accumulates in the cytoplasm through 2 sequential mitochondrial export process; oxoglutarate carrier (OGC, a mal99ate-a-ketoglutarate antiporter) transport across inner mitochondrial membranes, and subsequent cytosol gradient dependent diffusion through outer mitochondrial membrane channels. Accordingly, alpha-ketoglutarate is also a required co-substrate for the epigenetic 2OGD Jumonji-C domain lysine demethylases and 2OGD 10–11 translocation hydroxylases [23,41,42]. Concurrently, alpha-ketoglutarate analogues and as well other mitochondrial metabolites inhibit PHD activity, such as L, 2-hydroxyglutaric acid (L-2HGA), fumarate, succinate, and reactive oxygen species (ROS) [41]. Succinate activity has also been implicated in inflammation, immune signaling, ischemia and reperfusion injury processes, while mutations in isocitrate dehydrogenase (IDH), fumarate hydratase (FH), and succinate dehydrogenase (SDH) genes, enzymatic steps leading to alpha-ketoglutarate and analogues, are linked to cancer [32,[43], [44], [45], [46]]. Other distinguishing 2OGD PHD features are a catalytic core with a slower active-site Fe2+ - O2 binding and reaction time than other 2OGDs [23]. Among the PHDs, PHD2 is the most conserved isoform [23]. Importantly, PHD2's oxygen Km is higher than measured tissue concentrations (Fig. 2), imparting its catalytic activity responsive to changes in intracellular dissolved oxygen concentrations [[47], [48], [49]]. These cytosolic metabolite cofactors along with the variable cytoplasmic dissolved oxygen concentrations add significant regulatory refinement to the catalytic activity of PHDs. One vital biological process dependent upon 2OGD activity is PHD2 regulation of bone development and repair.
4. Expression of PHDs in bone cells
The 3 PHD isoforms are known to be expressed differently in various tissues [50]. Both PHD1 and PHD2 are known to be expressed widely in multiple tissues with testis showing abundant PHD1 expression and adipose and skeletal muscle tissues displaying high levels of PHD2 expression. While PHD3 mRNA levels have been reported in several tissues, its expression level is much lower than that of PHD1 or PHD2. Studies on subcellular localization of the 3 PHD isoforms also show differences in their localization pattern with PHD1 and PHD2 being primarily localized in the nucleus and cytoplasm, respectively, and PHD3 being distributed in both compartments [51]. To determine which isoforms of PHDs are expressed in bone cells, we examined the transcripts of Phd1, Phd2 and Phd3 in primary calvarial osteoblasts and MC3T3-E1 osteoblast cell line by real time RT-PCR. Our data showed that Phd2 was expressed at higher levels than Phd1 and that ascorbic acid treatment significantly inhibited Phd2 but not Phd1 expression [52]. Phd3 expression was undetectable under the serum-free culture conditions used in our study. In another study, Irwin et al [53] allowed MC3T3-E1 cells to differentiate for 14 days in the presence of ascorbic acid under normoxic (21% oxygen) conditions and then evaluated the consequence of hypoxia (2%) on expression levels of all 3 PHD family members. While expression levels of Phd2 and Phd3 were increased by more than 10-fold under hypoxic conditions, Phd1 expression was not affected. In the human osteosarcoma cell line, U2OS, hypoxia-induced expression of Phd2 and Phd3 and selective suppression of HIF1α expression by RNA interference resulted in complete loss of hypoxic induction of PHD2 and PHD3 [54]. These data suggest that Phd2 appears to be the major form expressed in osteoblasts that is subject to regulation by both low oxygen and ascorbic acid treatments.
There are limited investigations evaluating expression levels of the 3 PHD proteins in various bone cell types that reside in bone during normal development and during metabolic perturbations, in vivo. Immunohistochemical staining of bone sections from 2-month-old C57BL/6J mice showed that both PHD1 and PHD2 were detected in osteoblasts that reside on the trabecular surfaces of femur. However, PHD3 expression was too weak to be detected by immunohistochemistry [55]. In our study, we evaluated the expression patterns of PHD2 and PHD3 by immunohistochemistry in the distal femoral articular cartilage in 2-week-old mice, when the articular cartilage formation is known to occur in mice [26]. While PHD2 protein was found to be expressed in high levels in the superficial zone of articular cartilage, its expression was low in the middle and deep zones. By contrast, PHD3 was undetectable in the superficial zone while it was highly expressed in the middle and deep zones where some of the chondrocytes were undergoing hypertrophy [26]. However, the issue of whether conditions that influence loss of bone or articular cartilage influence expression levels of PHDs remain to be determined.
5. Role of PHDs in skeletal development
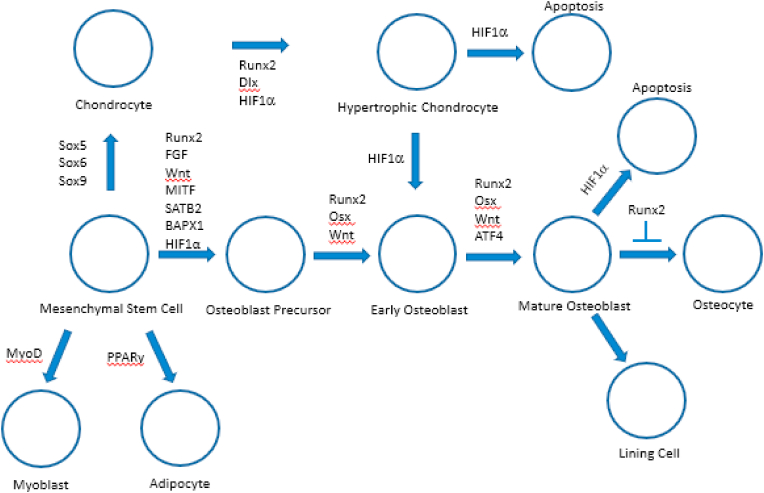
Bone development and repair occurs throughout life. It is a coordinated and ordered process. Axial skeletal development and mineralization began during the early Cambrian to mid-Ordovician period [56]. Bone is formed primarily via 2 routes known as endochondral and intramembranous bone ossification [57]. Both of these osteogenic pathways begin with a mesenchymal tissue precursor, but how it transforms into bone differs. During intramembranous ossification that occurs in the flat bones of the skull, clavicle, and most of the cranial bones, mesenchymal stem cells differentiate into osteoblasts to form bone. However, endochondral ossification occurring in the axial skeleton and the long bones involves a more complex route with mesenchymal tissue transforming into a cartilage intermediate which is subsequently replaced by bone [58]. During intramembranous and endochondral bone formation mechanisms, mesenchymal stem cells differentiate into osteoblasts and chondrocytes, respectively, the processes that are controlled by complex transcriptional network that involves HIFs (Fig. 4).
Fig. 4.
Transcriptional network regulating lineage commitment and differentiation of mesenchymal stem cells into various terminal differentiation stages. The fine balance of adipogenic, myogenic, chondrogenic and osteogenic differentiation of MSCs is activated by the action of key transcription factors. The signaling network maintains a delicate differentiation balance through regulating key transcription factors such as MyoD (myoblast), PPARγ (adipocyte), Sox9/5/6 trio (chondrocyte) and Runx2/osterix (osteoblast). In osteoblast differentiation, high levels of Runx2 and β-catenin are necessary to suppress the chondrogenic potential of uncommitted progenitors, such as the proposed osteochondroprogenitor and mesenchymal stem cell progenitors. Osterix is required for the final commitment of progenitors to preosteoblasts. While Sox9 along with Sox5 and Sox6 are essential for early chondrocyte differentiation, HIF1α and Dlx are known to play key role in the development of hypertrophic chondrocytes. Besides, HIF1α has also been predicted to be involved in the transdifferentiation of chondrocytes towards osteoblast lineage.
Chondrocytes are the only resident cell types of cartilage, which are responsible for the synthesis and maintenance of the extracellular matrix. The formation and maintenance of a proper cartilage matrix is dependent on the phenotypic stability and survival of the chondrocytes [59]. Cartilage development occurs in a hypoxic environment and its proximity to vasculature appears to be a key determining factor in the formation of bone over cartilage [[60], [61], [62]]. Articular cartilage does not have blood vessels and is developed and maintained in a hypoxic microenvironment both during normal development as well as regeneration [63]. Chondrocytes residing in articular cartilage are, therefore, exposed to an environment with reduced oxygen tension, ranging from 7 to 10% in the superficial zone and as low as 1% in the deep zones [64,65]. Anaerobic glycolysis contributes up to 75% of their ATP requirement in the articular cartilage chondrocytes, underlining their adaptation to a low O2 environment [66]. Adaptation of chondrocytes to low oxygen tension plays an important role in both the development and maintenance of chondrocyte phenotypes and is known to be primarily mediated through the HIF signaling pathway involving many cellular targets. During degenerative joint diseases like osteoarthritis (OA), the O2 gradient across articular cartilage may be altered due to changes in articular cartilage phenotype such as cartilage thinning and erosion, changes in extracellular matrix composition, and the development of cartilage fissures [67,68]. Indeed, cultures of articular chondrocytes in a low O2 environment have been shown to lose their articular chondrocyte phenotype by their increased propensity to express markers of chondrocyte differentiation [69].
PHD2 is believed to be the crucial oxygen sensor during normoxia and mild hypoxia, which is emphasized by the fact that mice with global knockout of the Phd2 gene results in embryonic lethality by E14.5 [70]. By contrast, mice with global knockout of Phd1 or Phd3 gene develop normally [71]. Because global knockout Phd2 gene results in embryonic lethality, conditional knock out approach has been used to disrupt Phd2 gene specifically in various bone cell types. In our studies, we crossed Phd2 floxed mice with Col1α2-iCre mice in which Cre expression is under the control of the entire regulatory region of Col1α2 gene to disrupt Phd2 gene in osteoblast-lineage cells. We found that mice with conditional deletion of Phd2 gene in Col1α2 expressing osteoblasts developed short stature and died prematurely at 12–14 weeks of age [26]. Analyses of skeletal phenotype by microCT and bone histomorphometry revealed that both bone size as well as trabecular bone mass were reduced in the long bones of Phd2 conditional knockout mice that were caused by reduced osterix expression, bone formation rate and mineral apposition rate, but not due to changes in bone resorption (Table 1). The HIF hydroxylases are dependent on ascorbate, and activation of PHDs in ascorbic acid-treated cells results in ubiquitin-mediated proteosomal degradation of prolyl hydroxylated HIF1α. In our study, we found that treatment of osteoblasts with dimethyloxallyl glycine and ethyl 3, 4-dihydroxybenzoate, known inhibitors of PHD, completely blocked ascorbic acid effect on expression levels of markers of osteoblast differentiation. Furthermore, knockdown of Phd2 expression by lentivirus-mediated shRNA abolished ascorbic acid effects on osteoblast differentiation, thus suggesting that a role for PHD2-mediated mechanism in ascorbic acid regulation of osteoblast functions.
Table 1.
Summary of skeletal phenotypes in mice with conditional disruption of phd1/2/3 in various bone cell types (See the text for details).
To examine the intrinsic effect of PHD2 in osteoblasts, Rauner et al [72] characterized the skeletal phenotype of mice with conditional loss of Phd2 expression in Osx expressing cells and found increased bone density in both lumbar and femur of 12-week-old conditional knockout mice compared to control mice. It is known that Osx is expressed in chondrocytes besides osteoblasts and mice with conditional disruption of Osx expression in Col2α1 expressing chondrocytes died immediately after birth and exhibited severe skeletal growth retardation [73]. Thus, it remains to be determined if the increased bone density in Phd2 conditional knockout mice using Osx-Cre can be explained on the basis of loss of Phd2 expression in chondrocytes since loss of chondrocyte Phd2 expression increases bone density (see below).
In another study, Wu et al [74] have used Osx-Cre mice to disrupt all 3 Phd genes in osteoprogenitors. In this study, the authors found that selective inactivation of Phd1/2/3, Phd1/2 or Phd2/3 resulted in increased trabecular bone mass without altering cortical bone mass. Furthermore, genetic ablation of Phd2 and Phd3 gene together protected mice from ovariectomy-induced trabecular bone loss and that the increased trabecular bone mass in the Phd2/3 conditional knockout mice is predicted to involve osteoblast produced osteoprotegrin to inhibit osteoclastogenesis, and thereby, bone resorption. By contrast to the above findings, Zhu et al [55] reported that the increased trabecular bone mass in mice with deletion of Phd1/2/3 genes in Col1α1 expressing osteoblasts is mainly caused by increased osteoblast function. In this study, bone resorption and osteoclast formation were rather increased in the Phd1/2/3 conditional mice, a finding opposite to that reported by Wu et al [74]. These data suggest that Phds expressed in osteoblast lineage cells regulate osteoblast and osteoclast functions in a complex manner depending on the context of differentiation status of cells as well as the number of Phd genes disrupted.
It is known that the terminally differentiated osteoblasts, osteocytes, reside within a low oxygen microenvironment. To elucidate whether the oxygen sensor PHD2 is critical for osteocyte function, Stegen et al [23] generated osteocyte-specific Phd2 conditional knockout mice by crossing Phd2 floxed mice with Dentin Matrix Protein (Dmp1)-Cre mice and characterized their skeletal phenotype by microCT and bone histomorphometry. Disruption of Phd2 expression in osteocytes increased bone mass in the tibial metaphysis of 8-week-old mice that is caused by increased bone formation and reduced bone resorption. Furthermore, genetic ablation of Phd2 in osteocytes was shown to blunt the bone loss due to estrogen deficiency or mechanical unloading. The anabolic effects of Phd2 disruption in osteocytes is predicted to involve HIF1α-mediated Sitrulin-1-dependent deacetylation of Sost promoter, resulting in decreased screlostin expression and enhanced Wnt signaling. Thus, oxygen sensing by PHD2 in osteocytes appear to negatively regulate bone mass via epigenetic regulation of Sost expression.
It is well established that HIF-1α is a positive regulator of chondrocyte differentiation and ossification as supported by studies showing excessive endochondral bone formation in mice with conditional deletion of the von Hippel-Lindau gene (Vhl), a ubiquitin ligase that promotes proteolysis of HIFs, or overexpression of Vegf, a HIF-1α target [75,76]. To determine the role of Phd2 expressed in chondrocytes, we generated mice with targeted deletion of Phd2 in Col2α1 expressing chondrocytes by crossing Phd2 floxed mice with Col2α1-Cre mice. Our data show that chondrocyte-specific Phd2 conditional knockout mice display a dramatic increase of bone mass in the trabeculae of long bones and spines [11,77]. In addition, loss of Phd2 expression in chondrocytes increased cortical thickness and tissue mineral density at the femoral mid-diaphysis. Disruption of Phd2 expression in chondrocytes caused increased bone formation in the primary spongiosa and reduced resorption in the secondary spongiosa. Based on the findings that expression levels of markers osteoblasts (Osx, Alp, Bsp) were increased in Phd2 knockdown chondrocytes, it is speculated that the increased HIF1α signaling caused by disruption of Phd2 gene in chondrocytes promotes chondrocyte-to-osteoblast transdifferentiation and, thereby, bone formation in the chondrocyte-specific Phd2 conditional knockout mice [26].
Since changes in HIF1α signaling have been implicated in the pathogenesis of osteoarthritis [78], we investigated the consequence of disruption of the Phd2 gene in chondrocytes on articular cartilage phenotype in mice. We found that condition deletion of Phd2 gene in Col2α1 expressing chondrocytes accelerated the progression of progenitors to hypertrophic chondrocytes as revealed by increased chondrocyte hypertrophy and thickness of middle/deep zone and reduced superficial zone thickness of articular cartilage. Immunohistochemistry revealed that while protein levels of markers of articular cartilage progenitors were decreased, but protein levels of hypertrophic markers of chondrocytes were increased. Furthermore, in vitro knockdown of expression of Phd2 using lentiviral shRNA or inhibition of PHD2 activity using chemical inhibitor in articular chondrocytes revealed that PHD2 acts by inhibiting the differentiation of articular cartilage progenitors in part via modulating HIF1α signaling [26].
Loss of Phd2 expression as well as inhibition of PHD2 activity with a chemical inhibitor resulted in dramatic increases in Phd3 expression in chondrocytes. To evaluate if the increased endochondral bone formation in chondrocyte-specific Phd2 conditional knockout mice is in part caused by elevated Phd3 expression, chondrocyte-specific Phd3 conditional knockout mice were generated by crossing Phd3 floxed (Phd3flox/flox) mice with Col2α1-Cre mice. Our data show that neither trabecular bone nor articular cartilage phenotype was affected in the Phd3 conditional knockout mice, thus demonstrating that Phd2 but not Phd3 expressed in chondrocytes regulates endochondral bone formation, and the compensatory increase in Phd3 expression in the chondrocytes of Phd2 conditional knockout mice is not the cause for increased trabecular bone mass in Phd2 conditional knockout mice [79]. Consistent with the lack of skeletal phenotype in mice with disruption of Phd3 gene in chondrocytes, we found that lentiviral shRNA-mediated partial knockdown of Phd3 expression in chondrocytes did not affect expression of markers of chondrocyte differentiation (Col2, Col10, Acan, Sox9) [79]. Based on these data, we conclude that molecular oxygen, through the PHD2/HIF signaling pathway, plays a central role in bone homeostasis by controlling both angiogenesis and osteogenesis. Therefore, therapeutic targeting of PHD2 enzyme can potentially be used for the treatment of bone disorders.
6. PHD-based therapies for bone repair/regeneration
The process of bone repair and regeneration is unique and is closely associated with that of vessel ingrowth that supply oxygen and nutrients to the regenerating bone. There is now accumulating evidence in the literature that suggest a key role for HIF pathway in angiogenic-osteogenic coupling during bone repair and regeneration [80]. Therefore, upregulation of HIF1α signaling, through PHD inhibition, is considered as a viable strategy to promote vascular in growth and, thereby, fracture repair and regeneration. PHD catalytic activity can be pharmacologically inhibited by PHD-inhibitors (PHIs), such as FG-4592 (Roxadustat), the pan-hydroxylase inhibitor dimethyloxalylglycine (DMOG), or ethyl-3,4-dihydroxybenzoate (EDHB) [81]. Especially, the novel inhibitors of PHD, such as TM6008, TM6089, and FK506-binding protein 38 (FKBP38), have been found and used to activate the HIF pathway and acquire beneficial aspects of the HIF system [82,83]. Majority of the PHIs that are currently under investigation for clinical use are 2-oxoglutarate antagonists.
Based on the established role of angiogenesis in new bone formation, a number of studies have examined if fracture repair can be promoted via manipulation of HIF pathway activity. Rios et al [84] found that using siRNA against PHD2 to activate the HIF-1 pathway supports bone regeneration in an in vivo sheep model. In this study, chambers containing silk fibroin-chitosan scaffolds with siRNA against PHD2 were implanted over the periosteum. After 70 days, it was found that the mean bone volume was significantly increased in animals implanted with silk fibroin-chitosan containing Phd2 siRNA compared to controls, suggesting that PHD2 inhibition supports bone regeneration. In another study, Chen et al. [85] implanted a tissue engineered compound construct consisting of the composite collagen membrane of bone marrow mesenchymal stem cells infected with lentiviral Phd2 shRNA in a rat periodontal fenestration defect model and found that Phd2 gene silencing promoted repair and reconstruction of the periodontal tissue defects. Similarly, inhibition of PHD2 function in human periodontal ligament cells via lentiviral-mediated RNA interference facilitated cell osteogenic differentiation and periodontal repair in Sprague-Dawley rats [86]. Consistent with the findings that knockdown of Phd2 expression promoted bone regeneration in various models, Shen et al [87] found that inhibition of PHD2 action by small molecule PHD inhibitors, diferrioxamine (DFO) and dimethyloxalylglycine (DMOG), at fracture sites in a stabilized murine femur fracture model not only increased vascularity at 14 days but also increased the callus size at 28 days as assessed by microCT, suggesting that HIF activation is a viable approach to increase vascularity and bone formation following skeletal trauma.
Distraction osteogenesis (DO) [88] is a surgical method of endogenous bone tissue engineering used to lengthen limbs. Based on the genetic mouse model lacking pVHL in osteoblasts to activate HIF signaling pathway promoted osteogenesis in DO, Wan et al [89] tested the effects of PHD inhibitors, DFO and l-mimose (L-mim) in DO model and found both agents increased angiogenesis and bone formation. In a similar study, Donneys et al. [88] reported that DFO significantly increased bone structural and mechanical quality parameters compared to controls during mandible DO. While these studies are very encouraging in the use of PHD inhibitors to promote bone regeneration, one caveat is that the PHD inhibitors used are mostly nonspecific. Therefore, further research using specific PHD2 inhibitors are needed to evaluate the safety and efficiency of these inhibitors in promoting bone regeneration and repair.
7. Non-canonical roles of PHD2
The traditional well established canonical role of PHD2 is to regulate HIF protein levels and, thereby, modulate HIF signaling in target tissues. In this regard, hydroxylation of proline residues with in HIFα molecule leads to targeting of HIFα to proteosomes for ubiquitination and subsequent degradation (Fig. 5). In addition to HIF proteins, PHD2 can also regulate hydroxylation of proline residues in other proteins such as and modulate their stability. In this regard, recent studies have revealed some novel PHD2 substrates beyond HIF, thus emphasizing that PHD2 may regulate multiple signaling pathways in normal development and diseases. For example, a recent study by Fan et al [90] revealed that conserved proline residues within the LAP/LAP -like motifs of TET2/3 are hydroxylated by PHD2 resulting in pVHL-mediated proteosomal degradation of TET2/3 and reduced DNA hydroxymethylation. In another study, Erber et al [91] identified PHD2 as a key regulatory enzyme of bromodomain-containing protein-4 (BRD4) prolyl hydroxylation. Furthermore, it has been shown that PHD2-mediated prolyl hydroxylation significantly affects BRD4 interaction with key transcription factors, CDK9 and CCNT1, and thereby, regulate RNA polymerase II-mediated transcriptional activity (Fig. 5).
Fig. 5.
Canonical and non-canonical mechanisms of PHD2 action in target cells. In the presence of O2, Fe2+, vitamin C and α-ketoglutarate, PHD2 hydroxylates key proline residues in the oxygen-dependent degradation domain of HIF1α, and, thereby, induce degradation of HIF1α via ubiquitin proteosomal degradation pathway (canonical signaling). Besides hydroxylating HIF1α, PHD2 can also hydroxylate other targets such as 10–11 translocation dioxygenase (TET)1 and TET2 to induce their degradation (non-canonical). PHD2 hydroxylation of bromodomain-containing protein 4 (BRD4) can modify its interaction with other nuclear proteins to regulate BRD4-mediated transcriptional activity. PHD2 is also known to be localized in the nucleus and induce hydroxylation of methyl cytosine, and, thereby, regulate transcription of target genes epigenetically.
We and others have shown that PHD2 is localized in the nucleus, besides cytoplasm and that PHD2 has significant hydroxylase activity in the nucleus [92]. Ten-eleven translocation hydroxylases (TET), that also belong to the 2OGGD family as PHDs, are known to oxidize 5-methylcytosines and promote locus-specific reversal of DNA methylation and, thereby, regulate gene transcription epigenetically in various cell types. To determine if PHD2 is involved in the regulating oxidation of methyl cytosines, we determined the levels of nuclear 5-hydroxymethylcytosine (5-hmC) in osteoblasts in osteoblast cultures treated with ascorbic acid, an activator of PHD2 activity. We found that while ascorbic acid increased 5-hmC levels in osteoblast-specific genes, Alp, Ihh and Osx. By contrast, inhibition of PHD2 activity with a specific inhibitor, IOX2, decreased 5-hmC levels in the promoters of Hif1α and Vegf in chondrocytes [92]. Furthermore, knockdown of Phd2 expression reduced nuclear 5-hmC levels in chondrocytes. These data are consistent with the possibility that PHD2, like TETs, can regulate transcription of target genes epigenetically by its hydroxylase activity to induce demethylation (Fig. 5).
8. Future research considerations
Despite efforts to understand the role of PHD proteins in the regulation of hypoxia signaling, there are still considerable gaps (see below) in our understanding of the role and molecular mechanism of action of PHD proteins, specifically, PHD2, in regulating various cell types that contribute to development and maintenance of bone.
-
1)
Of the 3 members of PHD family, PHD2 is expressed in more abundance than PHD1 or PHD3 in bone. Accordingly, we found that disruption of Phd2 but not Phd3 expression in chondrocytes exerted significant effect on the skeletal and articular cartilage phenotype in mice [79]. In addition, Wu et al [74] reported that while combined inactivation of Phd1/2/3, Phd2/3 or Phd1/2 in Osx expressing cells caused trabecular bone accumulation, inactivation of Phd1/3 had no effect. However, the issue of whether PHD1 and/or PHD3 expressed in osteoblasts/chondrocytes exerts significant role in bone or cartilage metabolism under conditions where bone metabolism is perturbed (eg, estrogen deficiency, aging, glucocorticoid treatment) remains to be determined.
-
2)
In addition to the autocrine effects of PHD2 expressed in bone cell types, there is evidence that PHD2 expressed in cells of the hematopoietic system exert an effect on the skeletal phenotype via influencing circulating levels of erythropoietin levels. In mice with conditional disruption of Phd2 in erythropoietin producing cells using CD68-Cre, there was a significant reduction in trabecular and cortical bone density due to reduction in osteoblast activity. Based on the findings of elevated serum levels of erythropoietin in the CD68-Cre/Phd2 floxed mice and that erythropoietin inhibits osteoblast functions, it is predicted that loss of PHD2 in erythropoietin producing cells results in reduced osteoblast function and diminished bone formation in part via increasing inhibitory effects of endocrine erythropoietin actions [72]. Future studies should address the issues of whether the effect of hematopoietic cell produced PHD2 on bone formation is primarily through alterations in erythropoietin signaling and if therapeutic approaches to inhibit erythropoietin signaling locally in bone can be used to correct bone formation deficit in conditions where serum levels of erythropoietin are increased.
-
3)
Much of published work is focused on the role of PHD2 in regulating canonical hypoxia signaling in target tissues. While our published data provide experimental evidence a PHD2-mediated mechanism in regulating DNA methylation in the promoter regions of osteoblast-specific genes, further work is needed to establish that PHD2 effect on 5-hmC changes in the promoter regions of target genes are direct and the extent to which this epigenetic mechanism contributes to PHD2 effects on cartilage and bone development [92]. Furthermore, efforts on methylation sequencing and pathway analyses could provide a deeper understanding of which genes are epigenetically regulated by PHD2-mediated mechanism in osteoblasts and chondrocytes.
-
4)
While the consequence of disruption of PHD family members in osteoblasts and chondrocytes has been determined, little is known on the expression patterns of Phd family members in osteoclast-lineage cells and their role in bone modeling and remodeling processes during normal physiological and pathological conditions.
-
5)
Consistent with the findings that genetic ablation of Phd2 in chondrocytes and osteocytes increase bone mass, administration of pan-PHD inhibitor, dimethyloxalylglycine, rescued ovariectomy-induced bone loss [93]. Future studies are warranted to determine the safety and efficacy of specific inhibitors of PHD2 over a long term to treat bone loss.
CRediT author statement
David Wolf: Writing – Original draft. Aruljothi Muralidharan: Writing – Original draft. Subburaman Mohan: Conceptualization, Writing – Review & editing.
Conflicts of interest
The authors declare no competing interests.
Acknowledgments
This study was supported by funding from Veterans Administration BLR&D merit review grant 1-101-BX-005262 to Subburaman Mohan, and the National Institutes of Arthritis and Musculoskeletal Disease RO1 grant AR048139 to Subburaman Mohan. Subburaman Mohan is a recipient of Senior Research Career Scientist Award from the Veterans Administration. ORCID David Wolf: 0000-0001-5934-0737. Aruljothi Muralidharan: 0000-0001-8939-0276. Subburaman Mohan: 0000-0003-0063-986X.
Footnotes
Peer review under responsibility of The Korean Society of Osteoporosis.
References
- 1.Lindsey R.C., Rundle C.H., Mohan S. Role of IGF1 and EFN-EPH signaling in skeletal metabolism. J Mol Endocrinol. 2018;61:T87–T102. doi: 10.1530/JME-17-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black D.M., Rosen C.J. Postmenopausal Osteoporosis. N Engl J Med. 2016;374:2096–2097. doi: 10.1056/NEJMc1602599. [DOI] [PubMed] [Google Scholar]
- 3.Baylink D.J., Strong D.D., Mohan S. The diagnosis and treatment of osteoporosis: future prospects. Mol Med Today. 1999;5:133–140. doi: 10.1016/s1357-4310(98)01426-9. [DOI] [PubMed] [Google Scholar]
- 4.Arnett T.R., Gibbons D.C., Utting J.C., Orriss I.R., Hoebertz A., Rosendaal M., et al. Hypoxia is a major stimulator of osteoclast formation and bone resorption. J Cell Physiol. 2003;196:2–8. doi: 10.1002/jcp.10321. [DOI] [PubMed] [Google Scholar]
- 5.Muzylak M., Price J.S., Horton M.A. Hypoxia induces giant osteoclast formation and extensive bone resorption in the cat. Calcif Tissue Int. 2006;79:301–309. doi: 10.1007/s00223-006-0082-7. [DOI] [PubMed] [Google Scholar]
- 6.Bozec A., Bakiri L., Hoebertz A., Eferl R., Schilling A.F., Komnenovic V., et al. Osteoclast size is controlled by Fra-2 through LIF/LIF-receptor signalling and hypoxia. Nature. 2008;454:221–225. doi: 10.1038/nature07019. [DOI] [PubMed] [Google Scholar]
- 7.Kaelin W.G., Jr., Ratcliffe P.J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Minamishima Y.A., Moslehi J., Bardeesy N., Cullen D., Bronson R.T., Kaelin W.G., Jr. Somatic inactivation of the PHD2 prolyl hydroxylase causes polycythemia and congestive heart failure. Blood. 2008;111:3236–3244. doi: 10.1182/blood-2007-10-117812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke K., Kalucka J., Mamlouk S., Singh R.P., Muschter A., Weidemann A., et al. HIF-1alpha is a protective factor in conditional PHD2-deficient mice suffering from severe HIF-2alpha-induced excessive erythropoiesis. Blood. 2013;121:1436–1445. doi: 10.1182/blood-2012-08-449181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng S., Xing W., Pourteymoor S., Mohan S. Conditional disruption of the prolyl hydroxylase domain-containing protein 2 (Phd2) gene defines its key role in skeletal development. J Bone Miner Res. 2014;29:2276–2286. doi: 10.1002/jbmr.2258. [DOI] [PubMed] [Google Scholar]
- 11.Cheng S., Xing W., Pourteymoor S., Schulte J., Mohan S. Conditional deletion of prolyl hydroxylase domain-containing protein 2 (Phd2) gene reveals its essential role in chondrocyte function and endochondral bone formation. Endocrinology. 2016;157:127–140. doi: 10.1210/en.2015-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holland H.D. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361:903–915. doi: 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasting J.F., Howard M.T. Atmospheric composition and climate on the early Earth. Philos Trans R Soc Lond B Biol Sci. 2006;361:1733–1741. doi: 10.1098/rstb.2006.1902. discussion 41-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raymond J., Segrè D. The effect of oxygen on biochemical networks and the evolution of complex life. Science. 2006;311:1764–1767. doi: 10.1126/science.1118439. [DOI] [PubMed] [Google Scholar]
- 15.Gregg L.S. Life with oxygen. Science. 2007;318:62–64. doi: 10.1126/science.1147949. [DOI] [PubMed] [Google Scholar]
- 16.Och L.M., Shields-Zhou G.A. The Neoproterozoic oxygenation event: environmental perturbations and biogeochemical cycling. Earth Sci Rev. 2012;110:26–57. [Google Scholar]
- 17.Lyons T.W., Reinhard C.T., Planavsky N.J. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 18.Knoll A.H., Nowak M.A. The timetable of evolution. Sci Adv. 2017;3 doi: 10.1126/sciadv.1603076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holdsworth M.J., Gibbs D.J. Comparative biology of oxygen sensing in plants and animals. Curr Biol. 2020;30:R362–R369. doi: 10.1016/j.cub.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 20.Carreau A., El Hafny-Rahbi B., Matejuk A., Grillon C., Kieda C. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med. 2011;15:1239–1253. doi: 10.1111/j.1582-4934.2011.01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKeown S.R. Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. Br J Radiol. 2014;87 doi: 10.1259/bjr.20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markolovic S., Wilkins S.E., Schofield C.J. Protein hydroxylation catalyzed by 2-Oxoglutarate-dependent oxygenases. J Biol Chem. 2015;290:20712–20722. doi: 10.1074/jbc.R115.662627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Islam M.S., Leissing T.M., Chowdhury R., Hopkinson R.J., Schofield C.J. 2-Oxoglutarate-Dependent oxygenases. Annu Rev Biochem. 2018;87:585–620. doi: 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- 24.Walport L.J., Schofield C.J. Adventures in defining roles of oxygenases in the regulation of protein biosynthesis. Chem Rec. 2018;18:1760–1781. doi: 10.1002/tcr.201800056. [DOI] [PubMed] [Google Scholar]
- 25.Myllyharju J. Prolyl 4-hydroxylases, key enzymes in the synthesis of collagens and regulation of the response to hypoxia, and their roles as treatment targets. Ann Med. 2008;40:402–417. doi: 10.1080/07853890801986594. [DOI] [PubMed] [Google Scholar]
- 26.Cheng S., Pourteymoor S., Alarcon C., Mohan S. Conditional deletion of the Phd2 gene in articular chondrocytes accelerates differentiation and reduces articular cartilage thickness. Sci Rep. 2017;7:45408. doi: 10.1038/srep45408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nandal A., Ruiz J.C., Subramanian P., Ghimire-Rijal S., Sinnamon R.A., Stemmler T.L., et al. Activation of the HIF prolyl hydroxylase by the iron chaperones PCBP1 and PCBP2. Cell Metabol. 2011;14:647–657. doi: 10.1016/j.cmet.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aghajanian P., Hall S., Wongworawat M.D., Mohan S. The roles and mechanisms of actions of vitamin C in bone: new developments. J Bone Miner Res. 2015;30:1945–1955. doi: 10.1002/jbmr.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza G.L. The genomics and genetics of oxygen homeostasis. Annu Rev Genom Hum Genet. 2020;21:183–204. doi: 10.1146/annurev-genom-111119-073356. [DOI] [PubMed] [Google Scholar]
- 30.Schofield C.J., Ratcliffe P.J. Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun. 2005;338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 31.Brahimi-Horn M.C., Pouyssegur J. HIF at a glance. J Cell Sci. 2009;122:1055–1057. doi: 10.1242/jcs.035022. [DOI] [PubMed] [Google Scholar]
- 32.Semenza G.L. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 33.Huang L.E., Gu J., Schau M., Bunn H.F. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salceda S., Caro J. Hypoxia-inducible factor 1alpha (HIF-1alpha) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions. Its stabilization by hypoxia depends on redox-induced changes. J Biol Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 35.Kaelin W.G. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 36.Semenza G.L. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 37.Epstein A.C., Gleadle J.M., McNeill L.A., Hewitson K.S., O’Rourke J., Mole D.R., et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 38.Frost J., Frost M., Batie M., Jiang H., Rocha S. Roles of HIF and 2-oxoglutarate-dependent dioxygenases in controlling gene expression in hypoxia. Cancers. 2021;13 doi: 10.3390/cancers13020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schofield C.J., Ratcliffe P.J. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 40.Kuiper C., Vissers M.C. Ascorbate as a co-factor for fe- and 2-oxoglutarate dependent dioxygenases: physiological activity in tumor growth and progression. Front Oncol. 2014;4:359. doi: 10.3389/fonc.2014.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bailey P.S.J., Nathan J.A. Metabolic regulation of hypoxia-inducible transcription factors: the role of small molecule metabolites and iron. Biomedicines. 2018;6 doi: 10.3390/biomedicines6020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martínez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11:102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carey B.W., Finley L.W., Cross J.R., Allis C.D., Thompson C.B. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tretter L., Patocs A., Chinopoulos C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim Biophys Acta. 2016;1857:1086–1101. doi: 10.1016/j.bbabio.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Zdzisinska B., Zurek A., Kandefer-Szerszen M. Alpha-ketoglutarate as a molecule with pleiotropic activity: well-known and novel possibilities of therapeutic use. Arch Immunol Ther Exp. 2017;65:21–36. doi: 10.1007/s00005-016-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vatrinet R., Leone G., De Luise M., Girolimetti G., Vidone M., Gasparre G., et al. The alpha-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer Metabol. 2017;5:3. doi: 10.1186/s40170-017-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirsila M., Koivunen P., Gunzler V., Kivirikko K.I., Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 48.Tarhonskaya H., Hardy A.P., Howe E.A., Loik N.D., Kramer H.B., McCullagh J.S., et al. Kinetic investigations of the role of factor inhibiting hypoxia-inducible factor (FIH) as an oxygen sensor. J Biol Chem. 2015;290:19726–19742. doi: 10.1074/jbc.M115.653014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson J.W., Shakir D., Batie M., Frost M., Rocha S. Oxygen-sensing mechanisms in cells. FEBS J. 2020;287:3888–3906. doi: 10.1111/febs.15374. [DOI] [PubMed] [Google Scholar]
- 50.Lieb M.E., Menzies K., Moschella M.C., Ni R., Taubman M.B. Mammalian EGLN genes have distinct patterns of mRNA expression and regulation. Biochem Cell Biol. 2002;80:421–426. doi: 10.1139/o02-115. [DOI] [PubMed] [Google Scholar]
- 51.Huang J., Zhao Q., Mooney S.M., Lee F.S. Sequence determinants in hypoxia-inducible factor-1alpha for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J Biol Chem. 2002;277:39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing W., Pourteymoor S., Mohan S. Ascorbic acid regulates osterix expression in osteoblasts by activation of prolyl hydroxylase and ubiquitination-mediated proteosomal degradation pathway. Physiol Genom. 2011;43:749–757. doi: 10.1152/physiolgenomics.00229.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Irwin R., LaPres J.J., Kinser S., McCabe L.R. Prolyl-hydroxylase inhibition and HIF activation in osteoblasts promotes an adipocytic phenotype. J Cell Biochem. 2007;100:762–772. doi: 10.1002/jcb.21083. [DOI] [PubMed] [Google Scholar]
- 54.Marxsen J.H., Stengel P., Doege K., Heikkinen P., Jokilehto T., Wagner T., et al. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381:761–767. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu K., Song P., Lai Y., Liu C., Xiao G. Prolyl hydroxylase domain proteins regulate bone mass through their expression in osteoblasts. Gene. 2016;594:125–130. doi: 10.1016/j.gene.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Zhang G. An evo-devo view on the origin of the backbone: evolutionary development of the vertebrae. Integr Comp Biol. 2009;49:178–186. doi: 10.1093/icb/icp061. [DOI] [PubMed] [Google Scholar]
- 57.Takayama Y., Mizumachi K. Inhibitory effect of lactoferrin on hypertrophic differentiation of ATDC5 mouse chondroprogenitor cells. Biometals. 2010;23:477–484. doi: 10.1007/s10534-010-9291-7. [DOI] [PubMed] [Google Scholar]
- 58.Breeland G., Sinkler M.A., Menezes R.G. StatPearls Publishing Copyright © 2021, StatPearls Publishing LLC.; 2021. Embryology, bone ossification. StatPearls. Treasure island (FL) [PubMed] [Google Scholar]
- 59.Kim H.A., Blanco F.J. Cell death and apoptosis in osteoarthritic cartilage. Curr Drug Targets. 2007;8:333–345. doi: 10.2174/138945007779940025. [DOI] [PubMed] [Google Scholar]
- 60.Schipani E., Ryan H.E., Didrickson S., Kobayashi T., Knight M., Johnson R.S. Hypoxia in cartilage: HIF-1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001;15:2865–2876. doi: 10.1101/gad.934301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goshima J., Goldberg V.M., Caplan A.I. The origin of bone formed in composite grafts of porous calcium phosphate ceramic loaded with marrow cells. Clin Orthop Relat Res. 1991;269:274–283. [PubMed] [Google Scholar]
- 62.Pechak D.G., Kujawa M.J., Caplan A.I. Morphology of bone development and bone remodeling in embryonic chick limbs. Bone. 1986;7:459–472. doi: 10.1016/8756-3282(86)90005-0. [DOI] [PubMed] [Google Scholar]
- 63.Murphy C.L. HIF-2alpha--a mediator of osteoarthritis? Cell Res. 2010;20:977–979. doi: 10.1038/cr.2010.99. [DOI] [PubMed] [Google Scholar]
- 64.Lafont J.E. Lack of oxygen in articular cartilage: consequences for chondrocyte biology. Int J Exp Pathol. 2010;91:99–106. doi: 10.1111/j.1365-2613.2010.00707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou S., Cui Z., Urban J.P. Factors influencing the oxygen concentration gradient from the synovial surface of articular cartilage to the cartilage-bone interface: a modeling study. Arthritis Rheum. 2004;50:3915–3924. doi: 10.1002/art.20675. [DOI] [PubMed] [Google Scholar]
- 66.Lane J.M., Brighton C.T., Menkowitz B.J. Anaerobic and aerobic metabolism in articular cartilage. J Rheumatol. 1977;4:334–342. [PubMed] [Google Scholar]
- 67.Kiaer T., Grønlund J., Sørensen K.H. Subchondral pO2, pCO2, pressure, pH, and lactate in human osteoarthritis of the hip. Clin Orthop Relat Res. 1988;229:149–155. [PubMed] [Google Scholar]
- 68.Grimshaw M.J., Mason R.M. Modulation of bovine articular chondrocyte gene expression in vitro by oxygen tension. Osteoarthritis Cartilage. 2001;9:357–364. doi: 10.1053/joca.2000.0396. [DOI] [PubMed] [Google Scholar]
- 69.Lafont J.E., Talma S., Murphy C.L. Hypoxia-inducible factor 2alpha is essential for hypoxic induction of the human articular chondrocyte phenotype. Arthritis Rheum. 2007;56:3297–3306. doi: 10.1002/art.22878. [DOI] [PubMed] [Google Scholar]
- 70.Berra E., Benizri E., Ginouvès A., Volmat V., Roux D., Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeda K., Ho V.C., Takeda H., Duan L.J., Nagy A., Fong G.H. Placental but not heart defects are associated with elevated hypoxia-inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rauner M., Franke K., Murray M., Singh R.P., Hiram-Bab S., Platzbecker U., et al. Increased EPO levels are associated with bone loss in mice lacking PHD2 in EPO-producing cells. J Bone Miner Res. 2016;31:1877–1887. doi: 10.1002/jbmr.2857. [DOI] [PubMed] [Google Scholar]
- 73.Cheng S., Xing W., Zhou X., Mohan S. Haploinsufficiency of osterix in chondrocytes impairs skeletal growth in mice. Physiol Genom. 2013;45:917–923. doi: 10.1152/physiolgenomics.00111.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu C., Rankin E.B., Castellini L., Alcudia J.F., LaGory E.L., Andersen R., et al. Oxygen-sensing PHDs regulate bone homeostasis through the modulation of osteoprotegerin. Genes Dev. 2015;29:817–831. doi: 10.1101/gad.255000.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maes C., Goossens S., Bartunkova S., Drogat B., Coenegrachts L., Stockmans I., et al. Increased skeletal VEGF enhances beta-catenin activity and results in excessively ossified bones. EMBO J. 2010;29:424–441. doi: 10.1038/emboj.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weng T., Xie Y., Huang J., Luo F., Yi L., He Q., et al. Inactivation of Vhl in osteochondral progenitor cells causes high bone mass phenotype and protects against age-related bone loss in adult mice. J Bone Miner Res. 2014;29:820–829. doi: 10.1002/jbmr.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheng S., Aghajanian P., Pourteymoor S., Alarcon C., Mohan S. Prolyl hydroxylase domain-containing protein 2 (Phd2) regulates chondrocyte differentiation and secondary ossification in mice. Sci Rep. 2016;6:35748. doi: 10.1038/srep35748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernández-Torres J., Martínez-Nava G.A., Gutiérrez-Ruíz M.C., Gómez-Quiroz L.E., Gutiérrez M. Role of HIF-1α signaling pathway in osteoarthritis: a systematic review. Rev Bras Reumatol Engl Ed. 2017;57:162–173. doi: 10.1016/j.rbre.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 79.Xing W., Pourteymoor S., Gomez G.A., Chen Y., Mohan S. Prolyl hydroxylase domain-containing protein 3 gene expression in chondrocytes is not essential for bone development in mice. Cells. 2021;10 doi: 10.3390/cells10092200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fan L., Li J., Yu Z., Dang X., Wang K. The hypoxia-inducible factor pathway, prolyl hydroxylase domain protein inhibitors, and their roles in bone repair and regeneration. Biomed Res Int. 2014. 2014 doi: 10.1155/2014/239356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fraisl P., Aragones J., Carmeliet P. Inhibition of oxygen sensors as a therapeutic strategy for ischaemic and inflammatory disease. Nat Rev Drug Discov. 2009;8:139–152. doi: 10.1038/nrd2761. [DOI] [PubMed] [Google Scholar]
- 82.Nangaku M., Izuhara Y., Takizawa S., Yamashita T., Fujii-Kuriyama Y., Ohneda O., et al. A novel class of prolyl hydroxylase inhibitors induces angiogenesis and exerts organ protection against ischemia. Arterioscler Thromb Vasc Biol. 2007;27:2548–2554. doi: 10.1161/ATVBAHA.107.148551. [DOI] [PubMed] [Google Scholar]
- 83.Barth S., Nesper J., Hasgall P.A., Wirthner R., Nytko K.J., Edlich F., et al. The peptidyl prolyl cis/trans isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol Cell Biol. 2007;27:3758–3768. doi: 10.1128/MCB.01324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rios C.N., Skoracki R.J., Mathur A.B. GNAS1 and PHD2 short-interfering RNA support bone regeneration in vitro and in an in vivo sheep model. Clin Orthop Relat Res. 2012;470:2541–2553. doi: 10.1007/s11999-012-2475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen C., Li H., Jiang J., Zhang Q., Yan F. Inhibiting PHD2 in bone marrow mesenchymal stem cells via lentiviral vector-mediated RNA interference facilitates the repair of periodontal tissue defects in SD rats. Oncotarget. 2017;8:72676–72699. doi: 10.18632/oncotarget.20243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cui D., Chen C., Luo B., Yan F. Inhibiting PHD2 in human periodontal ligament cells via lentiviral vector-mediated RNA interference facilitates cell osteogenic differentiation and periodontal repair. J Leukoc Biol. 2021;110:449–459. doi: 10.1002/JLB.1MA0321-761R. [DOI] [PubMed] [Google Scholar]
- 87.Shen X., Wan C., Ramaswamy G., Mavalli M., Wang Y., Duvall C.L., et al. Prolyl hydroxylase inhibitors increase neoangiogenesis and callus formation following femur fracture in mice. J Orthop Res. 2009;27:1298–1305. doi: 10.1002/jor.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Donneys A., Deshpande S.S., Tchanque-Fossuo C.N., Johnson K.L., Blough J.T., Perosky J.E., et al. Deferoxamine expedites consolidation during mandibular distraction osteogenesis. Bone. 2013;55:384–390. doi: 10.1016/j.bone.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wan C., Gilbert S.R., Wang Y., Cao X., Shen X., Ramaswamy G., et al. Activation of the hypoxia-inducible factor-1alpha pathway accelerates bone regeneration. Proc Natl Acad Sci U S A. 2008;105:686–691. doi: 10.1073/pnas.0708474105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fan S., Wang J., Yu G., Rong F., Zhang D., Xu C., et al. TET is targeted for proteasomal degradation by the PHD-pVHL pathway to reduce DNA hydroxymethylation. J Biol Chem. 2020;295:16299–16313. doi: 10.1074/jbc.RA120.014538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erber L., Luo A., Chen Y. Targeted and interactome proteomics revealed the role of PHD2 in regulating BRD4 proline hydroxylation. Mol Cell Proteomics. 2019;18:1772–1781. doi: 10.1074/mcp.RA119.001535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindsey R.C., Cheng S., Mohan S. Vitamin C effects on 5-hydroxymethylcytosine and gene expression in osteoblasts and chondrocytes: potential involvement of PHD2. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stegen S., Stockmans I., Moermans K., Thienpont B., Maxwell P.H., Carmeliet P., et al. Osteocytic oxygen sensing controls bone mass through epigenetic regulation of sclerostin. Nat Commun. 2018;9:2557. doi: 10.1038/s41467-018-04679-7. [DOI] [PMC free article] [PubMed] [Google Scholar]