Abstract
Wild canids serve as reservoir for various vector-borne pathogens of veterinary and medical importance, including the canine heartworm, Dirofilaria immitis. In North and Central America, coyotes (Canis latrans) may be a relevant reservoir host for heartworm transmission. The objective of this study was to determine the occurrence of D. immitis in coyotes across Texas using integrated antigen detection test and molecular assays. Matching whole blood and serum samples were collected from 122 coyotes from different locations across the state of Texas, United States, encompassing nine counties. Collections occurred from February to April 2016, and December 2016. Samples were assessed serologically using a commercial microtiter plate ELISA (DiroCHEK®), and molecularly by conventional PCR targeting the cytochrome oxidase c subunit 1 (cox1) and NADH dehydrogenase subunit 5 (nad5) of the mitochondrial DNA, and via a TaqMan© probe-based real-time PCR protocol, also targeting a fragment of the cox1 gene. Overall, 12 (9.83%) samples tested positive when serological and molecular results were combined. Seven of 122 samples (5.73%) were antigen-positive, 8 (6.55%) were qPCR-positive, and 4 (3.27%) were positive using conventional PCR. Of 12 positive samples, 4 tested antigen-positive by DiroCHEK® but were negative in all molecular tests, another 4 tested positive by at least one of the molecular assays but tested negative by DiroCHEK®, and 3 samples tested positive by both antigen test and at least one of the molecular assays. Two samples (16.67%) tested positive on both the antigen test and both conventional PCR and qPCR. Our study confirmed the presence of D. immitis infection in coyotes from southern and northern Texas. The combination of serologic and molecular diagnostic tests was proven synergistic for the identification of D. immitis infections, including occult dirofilariosis, and revealed a more accurate picture of heartworm occurrence in the sampled coyotes.
Keywords: Heartworm, Immunodiagnostics, Real-time PCR, Vector-borne disease, Wildlife reservoir
Graphical abstract
Highlights
-
•
Coyotes are an important wild reservoir for Dirofilaria immitis in North America.
-
•
We collected 121 matching blood and serum samples of coyotes from Texas, USA.
-
•
Twelve samples (9.92%) tested positive combining serology and molecular tests data.
-
•
Probe-based qPCR was superior than conventional PCR for heartworm diagnosis.
-
•
Combined DiroCHEK® and qPCR data showed a higher prevalence than each test alone.
1. Introduction
Wild canids serve as reservoir for a myriad of vector-borne pathogens of veterinary and medical importance, including the canine heartworm, Dirofilaria immitis (Otranto and Deplazes, 2019). This filarioid nematode is a rather cosmopolitan, and found at high prevalence across tropical, subtropical and temperate areas (McCall et al., 2008). This vector-borne parasite has been reported primarily in canids, including domestic dogs (Canis lupus familiaris), coyotes (Canis latrans), red foxes (Vulpes vulpes), and wolves (Canis lupus), all of which are present in North America. However, heartworm infections have been described in felids, mustelids, ursids, and humans (Crum et al., 1978; Dantas-Torres and Otranto, 2013; Papadopoulos et al., 2017). In the United States (US), D. immitis is distributed in all regions, with highest prevalence in the southeast (Bowman et al., 2009). Numerous factors directly and indirectly related to the parasite, host, and vector have been associated with high heartworm prevalence in the US. One major factor is related to the presence of susceptible host populations in endemic and non-endemic areas, including companion animals, coyotes, and feral dogs (Brown et al., 2012).
The prevalence and potential role of wild carnivores in the transmission and maintenance of D. immitis has been extensively studied (Aher et al., 2016; Magi et al., 2008; Nelson et al., 2003). Coyotes are distributed throughout North and Central America. Due to their opportunistic and adaptable nature, coyotes inhabit a number of diverse environments, and as a response to urbanization and human activities that have altered their wild environment, coyotes have been able to inhabit urban areas (Hody and Kays, 2018; Worsley-Tonks et al., 2021). The vast distribution and variety of environments inhabited by these animals lead to a higher risk of infection due to the close proximities between these reservoirs and pets, especially in the southern region of the US (Gates et al., 2014).
In the US, coyotes are considered the most prominent wild reservoir of D. immitis (Nelson et al., 2003). The expansion of heartworm infections in wild canid populations has highlighted the importance of these animals as sentinels, and the potential risk of D. immitis transmission to domestic animals and humans (Jara et al., 2016; Kotwa et al., 2019; Worsley-Tonks et al., 2021).
The American Heartworm Society guidelines recommend annual heartworm testing using both microfilariae and D. immitis antigen test as primary diagnostic screening for dogs over 7 months of age (Nelson et al., 2018). Molecular approaches have been used as alternative or complement tests in different stages of canine dirofilariosis, especially in cases with inconclusive microfilariae and/or antigen test results (Simón et al., 2012). Among wild animals, the detection of heartworm infection is usually based on the identification of D. immitis adults at necropsy (Agostine and Jones, 1982; Aher et al., 2016; Custer and Pence, 1981; Franson, 1976; Gates et al., 2014; Graham, 1975; King and Bohning, 1984; Monson et al., 1973; Nelson et al., 2003; Pappas and Lunzman, 1985; Sacks and Caswell-Chen, 2003; Thornton et al., 1974). Recently, other studies have demonstrated that commercial immunochromatographic tests used in the diagnosis of canine heartworm disease could be an alternative screening test for D. immitis infections in coyote populations (Kotwa et al., 2019; Paras et al., 2012). However, false-positive results can occur, and currently a specific and sensitive diagnostic screening method for wild animals has not been validated. Therefore, the aim of this study was to determine the occurrence of D. immitis infection in coyotes from Texas, United States, integrating results from a commercial microtiter plate ELISA (DiroCHEK®, Zoetis Diagnostics, Parsippany, NJ, USA), conventional PCR targeting two mitochondrial genes, and a probe-based real-time PCR (TaqMan©) assay.
2. Materials and methods
2.1. Sample acquisition
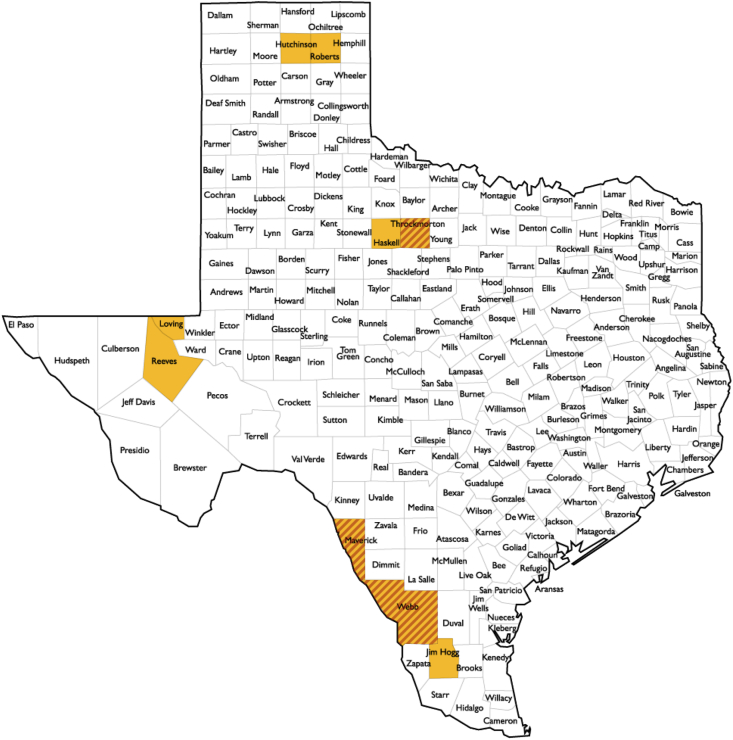
Paired serum samples and DNA extracted from whole blood were prepared from 122 coyotes sampled form February through December of 2016. A total of 71 males and 51 females (119 adults, one juvenile, and two animals that were classified as “Age Not Available”) were sampled. Samples were obtained from coyotes from 9 counties across Texas, including Webb (n = 70), Maverick (n = 20), Jim Hogg (n = 10), Haskell (n = 2), Throckmorton (n = 13), Hutchinson (n = 1), Roberts (n = 2), Reeves (n = 1), and Loving (n = 3) (Fig. 1). Genomic DNA was extracted from of 200 μL of coyote EDTA-whole blood samples using the High Pure PCR Template Preparation Kit (Roche, Indianapolis, IN, USA) with minor modifications. The protocol was modified by inclusion of Proteinase K before the binding buffer reagent as described by Yu et al. (2020). Both DNA and serum samples were stored at −20 °C until use. All samples analyzed were from a previous study designed to evaluate the molecular prevalence of tick-borne pathogens in coyote populations across Texas (Yu et al., 2020).
Fig. 1.
Texas locations where samples were collected, including Webb, Maverick, Jim Hogg, Haskell, Throckmorton, Hutchinson, Roberts, Reeves, and Loving counties.
2.2. DiroCHEK®
The coyote serum samples were tested for the presence of D. immitis antigen utilizing a commercial qualitative enzyme-linked immunosorbent assay (ELISA) test kit, DiroCHEK® (Zoetis, Kalamazoo, Michigan, USA), labeled for use in dogs and cats, and according to the manufacturer's protocol. Results for each sample were assessed by visual color change according to manufacturer recommendations followed by optical density (O.D.) reading. Tested samples were considered positive for the detection of D. immitis antigen if a color change was visible after 5 min of incubation and/or O.D. reading was ≥0.069. Wells with no color change were considered no antigen detected (NAD). The O.D. was performed using a spectrophotometer (Epoch, BioTek Instruments Inc., Winooski, VT, USA) at 590 nm.
2.3. Conventional PCR
Mitochondrial DNA (mtDNA) fragments of the cytochrome oxidase c subunit I (cox1) gene were amplified using the primers COIint forward (5′-TGA TTG GTG GTT TTG GTA A-3′) and COIint reverse (5′-ATA AGT ACG AGT ATC AAT ATC- 3′) (Casiraghi et al., 2001). Cycling conditions included an initial denaturation step at 95 °C for 2 min, followed by 40 cycles at 95 °C for 45 s, 52 °C at 45 s, and 72 °C for 90 s, and a final extension step at 72 °C for 5 min. In addition, we amplified a fragment of the mtDNA NADH dehydrogenase subunit 5 (nad5) gene using the ND5OvA forward (5′- TTG GTT GCC TAA GGC TAT GG -3′) and ND5OvC reverse primers (5′- CCC CTA GTA AAC AAC AAA CCA CA -3′) based on previously-published sequences (Morales-Hojas et al., 2006). The thermal cycler program included the annealing step at 95 °C for 30 s, 50 °C for 45 s, and 72 °C for 45 s for 40 cycles. All reactions were performed in 25 μL volumes using 0.25 μM of each primer, 1x GoTaq® Green Master Mix (Promega Corporation, Madison, Wisconsin, USA) and 1 μM of DNA template. PCR products were visualized under UV light after electrophoresis in a 1% agarose gel stained with SYBR® Safe DNA Gel Stain (Thermo Fisher Scientific, Waltham, MA, USA). Setaria equina DNA was used as positive control and nuclease-free water as negative control.
PCR products were purified using the E.Z.N.A.® Cycle Pure Kit (Omega Bio-Tek Inc., Norcross, GA, USA) as per manufacturer's instructions and sequenced in both directions using the original PCR primers in a 3730xl DNA Analyzer at Eurofins Genomics (Louisville, KY, USA). Generated sequences were aligned and compared to homologous sequences of D. immitis available at GenBank using MEGA X software 10.1 (Kumar et al., 2018).
2.4. Real-time PCR
All samples were molecularly screened for the presence of D. immitis DNA by simplex probe-based TaqMan© real-time PCR (qPCR) protocol as previously described by Laidoudi et al. (2020b) with several modifications. The COI gene was amplified using the forward primer Fil.COI.749-F (5′- CAT CCT GAG GTT TAT GTT ATT ATT TT-3′) and reverse Fil.COI.914-R (5′- CWG TAT ACA TAT GAT GRC CYC A-3′), and a specific TaqMan© probe to D. immitis namely D.imm.COI.777-P (6FAM-CGG TGT TTG GGA TTG TTA GTG-TAMRA). The reaction was performed in 20 μL final volume, containing 5 μL of DNA template, 10 μl (2 × ) of TaqMan© Multiplex Master Mix (Applied Biosystems, Waltham, MA), 50 μM of each primer, and 20 μM of probe. The TaqMan© reaction was performed in a QuantStudio 3 Real-Time PCR System (Thermo Fisher Scientific, USA) using TaqMan© universal-cycling conditions. The cycling conditions included two hold steps at 50 °C for 2 min followed by 15 min at 95 °C, and 40 cycles of two steps at 95 °C for 30 s and 60 °C for 30 s. All qPCR runs included a positive and negative control. The positive control consisted of DNA extracted from an adult D. immitis. The non-template negative control consisted of nuclease-free water.
3. Results
Overall, D. immitis infections were detected in 9.83% (12/122) of the samples when results of the DiroCHEK® and molecular analyses were combined. Among the positive samples, 9 of them were from adult males (9/71; 12.67%) and 3 from adult females (3/51; 5.88%). Regarding geographic location, the occurrence of D. immitis-positive cases was restricted to 3 counties, including Webb (8/70; 11.42%) and Maverick counties (3/20; 15.00%) in southern Texas, and Throckmorton (1/13; 7.69%) in northern Texas (Table 1). None of the samples from the remaining 6 counties tested positive in any of the assays.
Table 1.
Detection of Dirofilaria immitis DNA and/or circulating antigen in coyote samples from Texas, United States.
| County | Total number | Number of Positives (%) |
|---|---|---|
| Webb | 70 | 8 (11.42) |
| Maverick | 20 | 3 (15.00) |
| Throckmorton | 13 | 1 (7.69) |
| Haskell | 2 | – |
| Loving | 3 | – |
| Reeves | 1 | – |
| Hutchinson | 1 | – |
| Roberts | 2 | – |
| Jim Hogg | 10 | – |
| Total | 122 | 12 (9.83) |
The performance of antigen and molecular assays is summarized in Table 2. Overall, 5.73% (7/122) samples tested antigen-positive for D. immitis. Dirofilaria immitis DNA was detected in 8 out of 122 (6.55%) coyote samples when using the probe-based qPCR assay, and 4 (3.27%) samples were positives when using the conventional PCR targeting both cox1 and nd5 genes. Analysis of the partial cox1 sequences (GenBank accession numbers ON062406-ON062409), showed 100% identity with D. immitis sequences available in the GenBank. The generated nd5 sequence was accessioned in GenBank (ON099432-ON099435) and showed 99.1% maximum identity with 3 base pairs differences from previously reported D. immitis sequence. All the cox1 and nd5 fragments amplified from the 4 D. immitis isolates were identical to each other. Of the 12 samples considered positive for heartworm infection, only 3 (25.00%) samples tested positive by both D. immitis antigen test and at least one of the molecular assays, and only 2 samples (16.67%) tested positive on both the antigen test and both conventional PCR and qPCR. Four samples (33.30%) tested antigen-positive by DiroCHEK® but negative results were obtained in all molecular tests. Additionally, 4 samples (33.30%) tested positive by at least one of the molecular assays and had NAD for D. immitis. Finally, one of the serum samples testing NAD on the DiroCHEK®, did not have the corresponding paired DNA sample; consequently, molecular analyses were not performed.
Table 2.
Comparison of real-time PCR, conventional PCR, and DiroCHEK® Heartworm Antigen Test Kit results from Dirofilaria immitis positive coyotes in Texas, USA. Optical density (O.D.) values (DiroCHEK®) are included.
| Sample | Real-time PCRa | Conventional PCRa,b | DiroCHEK® | O.D. readings |
||
|---|---|---|---|---|---|---|
| Sample | - control | + control | ||||
| 10 | + | - | + | 0.205 | 0.046 | 0.454 |
| 15 | + | – | – | 0.052 | 0.046 | 0.410 |
| 24 | + | + | – | 0.056 | 0.069 | 0.753 |
| 26 | + | – | – | 0.048 | 0.046 | 0.454 |
| 27 | + | – | – | 0.049 | 0.069 | 0.753 |
| 28 | - | - | + | 0.066c | 0.069 | 0.753 |
| 39 | - | - | + | 0.373 | 0.046 | 0.410 |
| 49 | + | + | + | 0.700 | 0.047 | 0.625 |
| 73 | + | + | + | 0.199 | 0.047 | 0.625 |
| 84 | + | + | – | 0.057 | 0.045 | 0.529 |
| 85 | - | – | + | 0.281 | 0.045 | 0.529 |
| 101 | - | – | + | 0.420 | 0.048 | 0.493 |
PCR amplification of the cytochrome oxidase c subunit I gene.
PCR amplification of the NADH dehydrogenase subunit 5 gene.
Antigen was detected by visual color change but did not exceed the cutoff for positive on the spectrophotometric assay used.
4. Discussion
Overall, D. immitis infections were detected in coyotes sampled across Texas, with a higher occurrence in southern counties. Our findings are consistent with recent prevalence rates of D. immitis in coyotes in North America (Kotwa et al., 2019, 2020; Paras et al., 2012). However, the prevalence of heartworm in wild canids population in the US has varied widely over the past 40 years ranging from 0.6 to 70.8% (Agostine and Jones, 1982; Aher et al., 2016; Custer and Pence, 1981; Franson, 1976; Gates et al., 2014; Graham, 1975; King and Bohning, 1984; Monson et al., 1973; Nelson et al., 2003; Pappas and Lunzman, 1985; Paras et al., 2012; Sacks and Caswell-Chen, 2003; Thornton et al., 1974; Worsley-Tonks et al., 2021). Several factors have been associated with this wide range of D. immitis prevalences in the US, including differences in geographic location, climate, population sampled, and animal age (Brown et al., 2012). Geographic location seems to be a particularly important risk factor. Epidemiological surveys have shown high prevalence rate of heartworm infection among coyote populations in the southern US, including Arkansas (65.8%) (King and Bohning, 1984), Georgia (51.61%) (Gates et al., 2014), Florida (37.26%) (Aher et al., 2016), and Texas (23.08%) (Thornton et al., 1974). In contrast, lower prevalence rates have been reported in more temperate regions of the northeastern and midwestern US regions,a prevalence of 3.4 and 3.9% in New Hampshire and New York respectively (Agostine and Jones, 1982; Monson et al., 1973), and 0.6% in Kansas (Graham, 1975), 3.6% in Iowa (Franson, 1976), 8.9% in Nebraska (Pappas and Lunzman, 1985), and 15.98% in Illinois (Nelson et al., 2003).
The transmission risk of species of Dirofilaria, including D. immitis, has also been associated with climatic conditions and vector distribution (Genchi et al., 2009). A recent study in the Chicago, Illinois metropolitan area, conducted from 2001 through 2016 reported a high prevalence of heartworm in coyote populations (31.1%) and observed a significant temporal increase of infections, competent vector density, together with shifting climate conditions during this time period (Worsley-Tonks et al., 2021). On the other hand, Worsley-Tonks et al. (2021) observed that over the past decade urbanization of wildlife habitats in combination with climate changes factors have contributed to an increased risk of D. immitis infection in adult coyotes. In our study we observed a high concentration of heartworm positive animals in the southern Texas region. Although canine heartworm prevalence data is not available for Throckmorton County from the Companion Animal Parasite Council website, the current data indicates a lower incidence of D. immitis infections in dogs than in coyote population in southern Texas. Moreover, our findings also suggest a high risk of D. immitis exposure and infection in this area and denotes coyotes as indicators of the relative risk of heartworm infection for dogs and cats.
Several limitations should be considered in our study, in special biases inherent to sample collection, which was more concentrated in southern Texas, and the various steps of sample processing. Hence, data should be interpreted carefully. Additionally, we were not able to collect blood samples from dogs from the same region and time period at which coyote samples were collected; hence, it is not possible to provide direct evidence of the relationship between coyote heartworm infection and risk of canine infection.
Post-mortem examination is currently considered the gold standard diagnostic method for heartworm infection in wild animals and has been used in numerous studies of coyotes in North America (Aher et al., 2016; Gates et al., 2014; Kotwa et al., 2019). Necropsy provides an accurate diagnosis but the procedure is time-consuming, requires additional safety precautions, and specific knowledge, and also this method eliminates the possibility of performing longitudinal studies in particular populations of coyotes. Recent studies have evaluated the performance of commercial membrane-bound ELISA tests for the detection of heartworm in wild canids and have been considered a good alternative for the diagnosis and large-scale screening of D. immitis in coyote populations (Kotwa et al., 2020; Worsley-Tonks et al., 2021). In the study conducted by Kotwa et al. (2020) the ELISA assay presented a high specificity rate of 97.9–98.9% when compared with necropsy. However, there are important limitations to consider when interpreting the results from serologic analyses, including the occurrence of false-positive and false-negative results. Molecular approaches have been developed as an alternative and/or complementary diagnostic method for the detection of D. immitis in companion animals (Laidoudi et al., 2020a, 2020b). However, the diagnostic potential of molecular tools for the screening of heartworm disease in wild carnivores has not been fully tapped. To the authors’ knowledge, this is the first study integrating serological antigen detection and molecular approaches for the detection of D. immitis infection in coyotes. Our findings provide evidence that the combination of serological and molecular assays is an efficient method for the diagnosis of D. immitis infections in archival serum and DNA samples and may assist in retrospective studies to assess heartworm prevalence in wild and domestic animals.
In the present study, 12 animals were considered positive for heartworm infection using a combination of the DiroCHEK® and 3 different molecular assays. The discordant results demonstrated between the DiroCHEK® and the different molecular approaches could be explained by the occurrence of false-negative results with both antigen detection and any of the molecular methods. Occult heartworm infections have been reported in dogs, cats, and less frequently in wild canids due to early infection, presence of only adult male D. immitis or immature female worms, or presence of blocked antigen which, usually leading to a NAD result in commercial antigen tests (Kotwa et al., 2020; Little et al., 2014, 2018). Another potential limitation of the ELISA assay is the occurrence of cross-reactivity among D. immitis and other nematodes, especially when immune complex dissociation (ICD) methods are used to prepare the samples prior to running the test (Little et al., 2018; Sobotyk et al., 2021; Venco et al., 2017). Due to the potential risk of cross-reactivity, the American Heartworm Society guidelines do not recommend the use of ICD sample treatment during epidemiological surveys for heartworm infections (Nelson et al., 2018). Consequently, in the present study serum samples were not treated using an ICD method prior to testing. Molecular methods can also result in false-negative results, especially during the prepatent stage associated with amicrofilaremic D. immitis infections. Even though molecular techniques present some limitation, these methods can provide a rapid, sensitive, and accurate diagnoses, and prevent the occurrence of false-positive results due to cross-reactivity. Several studies have reported the use of molecular approaches targeting conserved DNA regions in the diagnosis of numerous filarial nematodes (Laidoudi et al., 2020a, 2020b; Panetta et al., 2021; Rishniw et al., 2006). As expected, with the present study the real-time PCR technique had a higher positive rate than conventional PCR and seems to be an alternative diagnostic method for detecting D. immitis circulating microfilariae in domestic and wild animals.
5. Conclusion
The results of this study demonstrate the detection of D. immitis in coyotes in southern Texas and highlight the need for specific and applicable diagnostic tests for heartworm infection in wild canids to aid in the study of the associated risk of infection in domestic dogs. The diagnostic approach of combining antigen testing and molecular methods allowed for the identification of D. immitis infections in coyotes. This approach is a suitable alternative to necropsy for the diagnosis and epidemiologic surveys of D. immitis infections in wild animals.
Declaration of competing interest
The authors declare no conflict of interest.
References
- Agostine J.C., Jones G.S. Heartworms (Dirofilaria immitis) in coyotes (Canis latrans in New england. J. Wildl. Dis. 1982;18:343–345. doi: 10.7589/0090-3558-18.3.343. [DOI] [PubMed] [Google Scholar]
- Aher A.M., Caudill D., Caudill G., Butryn R.S., Wolf D., Fox M., Blake D.P., Cunningham M.W. Prevalence, genetic analyses, and risk factors associated with heartworm (Dirofilaria immitis) in wild coyotes (Canis latrans) from Florida, USA. J. Wildl. Dis. 2016;52:785–792. doi: 10.7589/2015-09-223. [DOI] [PubMed] [Google Scholar]
- Bowman D., Little S.E., Lorentzen L., Shields J., Sullivan M.P., Carlin E.P. Prevalence and geographic distribution of Dirofilaria immitis, Borrelia burgdorferi, Ehrlichia canis, and Anaplasma phagocytophilum in dogs in the United States: results of a national clinic-based serologic survey. Vet. Parasitol. 2009;160:138–148. doi: 10.1016/j.vetpar.2008.10.093. [DOI] [PubMed] [Google Scholar]
- Brown H.E., Harrington L.C., Kaufman P.E., McKay T., Bowman D.D., Nelson C.T., Wang D., Lund R. Key Factors Influencing Canine Heartworm, Dirofilaria immitis, in the United States. Parasit. Vectors. 2012;5:245. doi: 10.1186/1756-3305-5-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casiraghi M., Anderson T.J.C., Bandi C., Bazzocchi C., Genchi C. A phylogenetic analysis of filarial nematodes: comparison with the phylogeny of Wolbachia endosymbionts. Parasitology. 2001;122:93–103. doi: 10.1017/s0031182000007149. [DOI] [PubMed] [Google Scholar]
- Crum J.M., Nettles V.F., Davidson W.R. Studies on endoparasites of the black bear (Ursus americanus) in the southeastern United States. J. Wildl. Dis. 1978;14:178–186. doi: 10.7589/0090-3558-14.2.178. [DOI] [PubMed] [Google Scholar]
- Custer J.W., Pence D.B. Dirofilariasis in wild canids from the Gulf coastal prairies of Texas and Louisiana, USA. Vet. Parasitol. 1981;8:71–82. doi: 10.1093/jmedent/18.5.409. [DOI] [PubMed] [Google Scholar]
- Dantas-Torres F., Otranto D. Dirofilariosis in the Americas: a more virulent Dirofilaria immitis? Parasit. Vectors. 2013;6:288. doi: 10.1186/1756-3305-6-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franson J.C. Drofilariasis in Iowa coyotes. J. Wildl. Dis. 1976;12:165–166. doi: 10.7589/0090-3558-12.2.165. 162. [DOI] [PubMed] [Google Scholar]
- Gates M., Gerhold R.W., Wilkes R.P., Gulsby W.D., Maestas L., Rosypal A., Miller K.V., Miller D.L. Parasitology, virology, and serology of free-ranging coyotes (Canis latrans) from Central Georgia, USA. J. Wildl. Dis. 2014;50:896–901. doi: 10.7589/2013-10-283. [DOI] [PubMed] [Google Scholar]
- Genchi C., Rinaldi L., Mortarino M., Genchi M., Cringoli G. Climate and Dirofilaria infection in europe. Vet. Parasitol. 2009;163:286–292. doi: 10.1016/j.vetpar.2009.03.026. [DOI] [PubMed] [Google Scholar]
- Graham J.M. Filariasis in coyotes from Kansas and Colorado. J. Parasitol. 1975;61:513–516. [Google Scholar]
- Hody J.W., Kays R. Zookeys 759, 81–97; 2018. Mapping the Expansion of Coyotes (Canis latrans) across North and Central America. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jara R.F., Wydeven A.P., Samuel M.D. Gray wolf exposure to emerging vector-borne diseases in Wisconsin with comparison to domestic dogs and humans. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.W., Bohning A.M. The incidence of heartworm Dirofilaria immitis (Filarioidea), in the wild canids of Northeast Arkansas. SW. Nat. 1984;29:89–92. [Google Scholar]
- Kotwa J.D., Jardine C.M., Berke O., Pearl D.L., Mercer N.J., Peregrine A.S. Prevalence and distribution of Dirofilaria immitis infection in wild canids in southern Ontario. Vet. Parasitol. Reg. Stud. Rep. 2019;18:100349. doi: 10.1016/j.vprsr.2019.100349. [DOI] [PubMed] [Google Scholar]
- Kotwa J.D., Jardine C.M., Pearl D.L., Berke O., Mercer N.J., Peregrine A.S. Evaluation of the SNAP® 4Dx® plus test for the detection of Dirofilaria immitis antigen and characterization of exposure to tick-borne pathogens in wild canids in southern Ontario. Vet. Parasitol. 2020;283:109176. doi: 10.1016/j.vetpar.2020.109176. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidoudi Y., Bedjaoui S., Medkour H., Latrofa M.S., Mekroud A., Bitam I., Davoust B., Otranto D., Mediannikov O. Molecular approach for the diagnosis of blood and skin canine filarioids. Microorganisms. 2020;8 doi: 10.3390/microorganisms8111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidoudi Y., Davoust B., Varloud M., Niang E.H.A., Fenollar F., Mediannikov O. Development of a multiplex qPCR-based approach for the diagnosis of Dirofilaria immitis, D. repens and Acanthocheilonema reconditum. Parasit. Vectors. 2020;13:319. doi: 10.1186/s13071-020-04185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S., Saleh M., Wohltjen M., Nagamori Y. Prime detection of Dirofilaria immitis: understanding the influence of blocked antigen on heartworm test performance. Parasit. Vectors. 2018;11:186. doi: 10.1186/s13071-018-2736-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little S.E., Raymond M.R., Thomas J.E., Gruntmeir J., Hostetler J.A., Meinkoth J.H., Blagburn B.L. Heat treatment prior to testing allows detection of antigen of Dirofilaria immitis in feline serum. Parasit. Vectors. 2014;7:1. doi: 10.1186/1756-3305-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi M., Calderini P., Gabrielli S., Dell'Omodarme M., Macchioni F., Prati M.C., Cancrini G. Vulpes vulpes: a possible wild reservoir for zoonotic filariae. Vector Borne Zoonotic Dis. 2008;8:249–252. doi: 10.1089/vbz.2007.0207. [DOI] [PubMed] [Google Scholar]
- McCall J.W., Genchi C., Kramer L.H., Guerrero J., Venco L. Advances in Parasitology. Academic Press; 2008. Chapter 4 heartworm disease in animals and humans; pp. 193–285. [DOI] [PubMed] [Google Scholar]
- Monson R., Stone W., Weber B. Heartworms in foxes and wild canids in New York. N. Y. Fish Game J. 1973;20:48–53. [Google Scholar]
- Morales-Hojas R., Checke R.A., Post R.J. Molecular systematics of five Onchocerca species (Nematoda: filarioidea) including the human parasite, O. volvulus, suggest sympatric speciation. J. Helminthol. 2006;80:281–290. [PubMed] [Google Scholar]
- Nelson C.T., McCall J.W., Jones S., Moorhead A. In: Current Canine Guidelines for the Prevention, Diagnosis, and Management of the Heartworm (Dirofilaria immitis) Infection in Dogs. Wilmington, editor. American Heartworm Society; 2018. https://www.heartwormsociety.org/veterinary-resources/american-heartworm-society-guidelines [Google Scholar]
- Nelson T.A., Gregory D.G., Laursen J.R. Canine heartworms in coyotes in Illinois. J. Wildl. Dis. 2003;39:593–599. doi: 10.7589/0090-3558-39.3.593. [DOI] [PubMed] [Google Scholar]
- Otranto D., Deplazes P. Zoonotic nematodes of wild carnivores. Int. J. Parasitol. Parasites Wildl. 2019;9:370–383. doi: 10.1016/j.ijppaw.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panetta J.L., Calvani N.E.D., Orr B., Nicoletti A.G., Ward M.P., Šlapeta J. Multiple diagnostic tests demonstrate an increased risk of canine heartworm disease in northern Queensland, Australia. Parasit. Vectors. 2021;14:393. doi: 10.1186/s13071-021-04896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos E., Komnenou A., Poutachides T., Heikkinen P., Oksanen A., Karamanlidis A.A. Detection of Dirofilaria immitis in a brown bear (Ursus arctos) in Greece. Helminthologia. 2017;54:257–261. [Google Scholar]
- Pappas L.G., Lunzman A.T. Canine heartworm in the domestic and wild canids of southeastern Nebraska. J. Parasitol. 1985;71:828–830. [PubMed] [Google Scholar]
- Paras K.L., Little S.E., Reichard M.V., Reiskind M.H. Detection of Dirofilaria immitis and ehrlichia species in coyotes (Canis latrans), from rural Oklahoma and Texas. Vector Borne Zoonotic Dis. 2012;12:619–621. doi: 10.1089/vbz.2011.0815. [DOI] [PubMed] [Google Scholar]
- Rishniw M., Barr S.C., Simpson K.W., Frongillo M.F., Franz M., Dominguez Alpizar J.L. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet. Parasitol. 2006;135:303–314. doi: 10.1016/j.vetpar.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Sacks B.N., Caswell-Chen E.P. Reconstructing the spread of Dirofilaria immitis in California coyotes. J. Parasitol. 2003;89:319–323. doi: 10.1645/0022-3395(2003)089[0319:RTSODI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Simón F., Siles-Lucas M., Morchón R., González-Miguel J., Mellado I., Carretón E., Montoya-Alonso J.A. Human and animal dirofilariasis: the emergence of a zoonotic mosaic. Clin. Microbiol. Rev. 2012;25:507–544. doi: 10.1128/CMR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobotyk C., Savadelis M.D., Verocai G.G. Detection and cross-reaction of Dirofilaria repens using a commercial heartworm antigen test kit. Vet. Parasitol. 2021;289:109302. doi: 10.1016/j.vetpar.2020.109302. [DOI] [PubMed] [Google Scholar]
- Thornton E., Bell R.R., Reardon M.J. Internal parasites of coyotes in southern Texas. J. Wildl. Dis. 1974;10:232–236. doi: 10.7589/0090-3558-10.3.232. [DOI] [PubMed] [Google Scholar]
- Venco L., Manzocchi S., Genchi M., Kramer L.H. Heat treatment and false-positive heartworm antigen testing in ex vivo parasites and dogs naturally infected by Dirofilaria repens and Angiostrongylus vasorum. Parasit. Vectors. 2017;10:476. doi: 10.1186/s13071-017-2444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley-Tonks K.E.L., Gehrt S.D., Anchor C., Escobar L.E., Craft M.E. Infection risk varies within urbanized landscapes: the case of coyotes and heartworm. Parasit. Vectors. 2021;14:464. doi: 10.1186/s13071-021-04958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S., Modarelli J., Tomeček J.M., French J.T., Hilton C., Esteve-Gasent M.D. Prevalence of common tick-borne pathogens in white-tailed deer and coyotes in south Texas. Int. J. Parasitol. Parasites Wildl. 2020;11:129–135. doi: 10.1016/j.ijppaw.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]