Abstract
Expression of the receptor tyrosine kinase ephrin receptor A10 (EphA10), which is undetectable in most normal tissues except for the male testis, has been shown to correlate with tumor progression and poor prognosis in several malignancies, including triple-negative breast cancer (TNBC). Therefore, EphA10 could be a potential therapeutic target, likely with minimal adverse effects. However, no effective clinical drugs against EphA10 are currently available. Here, we report high expression levels of EphA10 in tumor regions of breast, lung, and ovarian cancers as well as in immunosuppressive myeloid cells in the tumor microenvironment. Furthermore, we developed anti-EphA10 monoclonal antibodies (mAbs) that specifically recognize cell surface EphA10, but not other EphA family isoforms, and target tumor regions precisely in vivo with no apparent accumulation in other organs. In syngeneic TNBC mouse models, we found that anti-EphA10 mAb clone #4 enhanced tumor regression, therapeutic response rate, and T cell–mediated antitumor immunity. Notably, the chimeric antigen receptor T cells derived from clone #4 significantly inhibited TNBC cell viability in vitro and tumor growth in vivo. Together, our findings suggest that targeting EphA10 via EphA10 mAbs and EphA10-specific chimeric antigen receptor–T cell therapy may represent a promising strategy for patients with EphA10-positive tumors.
Keywords: ephrin receptor A10, monoclonal antibody, chimeric antigen receptor T cell, triple-negative breast cancer, antitumor immunity, targeted therapy, cancer immunotherapy
Abbreviations: ADCs, antibody–drug conjugates; anti-PD-1/-L1, antiprogrammed cell death 1/ligand 1; CAR, chimeric antigen receptor; CTL, cytotoxic T lymphocyte; ECD, extracellular domain; EphA10, Ephrin receptor A10; ER, estrogen receptor; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; GrB, granzyme; HER2, human epidermal growth factor 2; IF, immunofluorescence; IgG, immunoglobulin G; huEphA10, human EphA10; IHC, immunohistochemistry; mAbs, monoclonal antibodies; MDSCs, myeloid-derived suppressor cells; MOI, multiplicity of infection; PR, progesterone receptor; scFv, single-chain variable fragment; TAMs, tumor-associated macrophages; TME, tumor microenvironment; TNBC, triple-negative breast cancer
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer defined by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor 2 (HER2) expression. Prognosis in patients with advanced TNBC remains poor, in part due to the limited number of effective therapeutic options. In addition, the high risk of recurrence and resistance to chemotherapy in TNBC remains a substantial clinical challenge. Thus, a novel therapeutic strategy for patients with TNBC is considered an unmet medical need (1, 2).
Preclinical and clinical data suggest that antitumor immunity is a critical determinant of outcome in TNBC. Specifically, higher quantities of tumor-infiltrating cytotoxic T lymphocytes (CTLs) have been associated with response to chemotherapy and improved survival in TNBC, suggesting that modulating the tumor microenvironment (TME) is a promising therapeutic strategy in TNBC (3, 4). Indeed, antiprogrammed cell death 1/ligand 1 (anti-PD-1/-L1) immune checkpoint therapy improves survival when used in combination with chemotherapy in patients with PD-L1–positive TNBC (5). However, most patients with advanced TNBC do not benefit from anti-PD-L1 therapy, possibly because of the heterogeneity and heavy N-linked glycosylation of PD-L1 (6, 7, 8). Notably, PD-L1 levels in about 40 to 50% of cancer patients are underestimated as false-negative, as evidenced from immunohistochemistry (IHC) by sample deglycosylation (8, 9). Thus, novel therapeutic strategies aimed at augmenting antitumor immunity are urgently needed.
Ephrin receptors (Ephs), the largest subfamily of receptor tyrosine kinases, are known to modulate cell–cell signaling by interacting with cells’ membrane-bound ephrin ligands on neighboring cells (10, 11), thereby regulating tissue organization, vascular development, and progression of many diseases including cancer (12, 13). The Eph genes were initially identified in several human carcinomas, and many oncogenic processes such as tumor initiation, metastasis, and angiogenesis depend on Eph–ephrin signaling (14, 15). Ephrin receptor A10 (EphA10), a member of the Eph subfamily, is not expressed in normal human adult tissue except in the testis (16, 17). In contrast, EphA10 expression is common in malignant cells across histology of various tumors and has been associated with poor prognosis in patients with breast, prostate, and gallbladder cancers (17, 18, 19, 20, 21). In human breast cancer specimens, expression of EphA10, as assessed by IHC, has been significantly correlated with lymph node metastasis and higher tumor stage (22). The association between EphA10 and poor prognostic indicators as well as its limited expression in normal cells suggests that it is a potentially important driver of the malignant phenotype, making it an attractive target in cancer therapy. Interestingly, higher EphA10 expression has been associated with increased PD-L1 expression and immunosuppression (23). The role of EphA10 in antitumor immunity are limited and worthwhile to pursue.
In this study, we generated monoclonal antibodies (mAbs) with high specificity for EphA10 in vivo. In syngeneic mouse models of TNBC, targeting EphA10 with these therapeutic mAbs markedly inhibited tumor progression and enhanced CTL-mediated antitumor immunity without significant toxicity. In addition, EphA10-specific chimeric antigen receptor (CAR)-T cell-based therapy was also shown to effectively suppress tumor development. Our work provides key data in support of augmenting antitumor immunity in TNBC by targeting EphA10 with newly developed therapeutic mAbs and CAR-T therapy.
Results
EphA10 is highly expressed in cancer cells and immunosuppressive myeloid cells at the tumor region
EphA10 has been suggested to be an ideal target for TNBC since a mAb against EphA10 suppresses tumor growth in a xenograft MDA-MB-435 mouse model (17). Notably, results from a syngeneic 4T1 TNBC mouse model further clarified that genetically blocking EphA10 via a CRISPR–Cas9 knockout system reduces tumor growth and enhances CTL-mediated antitumor immunity (23), strongly suggesting that targeting EphA10 is potentially valuable in cancer therapy. Currently, however, no therapeutic antibodies used in the clinic specifically recognize EphA10, which prompted us to generate anti-EphA10 mAbs that can block the function of EphA10 to promote antitumor immunity in the presence of a functional immune system.
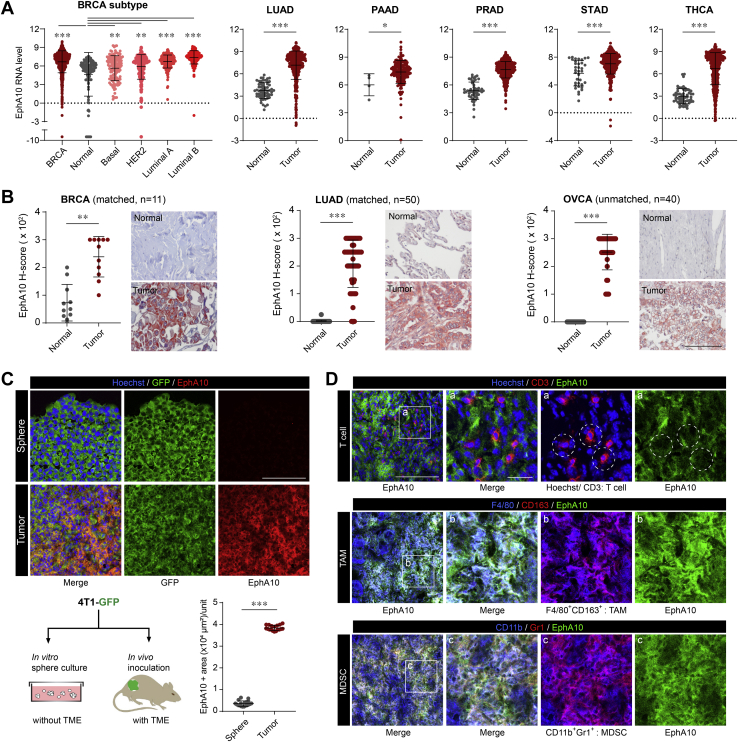
In addition to its role in promoting TNBC, EphA10 upregulation has been shown to promote tumorigenesis in other types of cancer, such as pancreatic (19) and prostate (20) cancers. Indeed, results from The Cancer Genome Atlas RNA-sequencing database showed that the EphA10 mRNA level was significantly higher in tumor tissues than in normal controls in all breast invasive carcinoma subtypes, including basal-like TNBC (Fig. S1), lung adenocarcinoma, pancreatic adenocarcinoma, prostate adenocarcinoma, stomach adenocarcinoma, and thyroid carcinoma (Fig. 1A). We also examined protein levels of EphA10 in breast, lung, and ovarian cancers with use of human tissue microarray by IHC staining and found that levels of EphA10 expression in the tumor regions of all three cancer types were significantly higher than levels in the normal tissues adjacent to the tumors (Fig. 1B). Considering that EphA10 expression is absent in normal tissues except in the male testis (17), researchers theorized that EphA10–based targeted therapy would be more beneficial for female patients with breast cancer and have fewer adverse effects (23). Also of note, IHC results that were obtained in ovarian cancer were similar to those in breast cancer, and EphA10 expression was significantly higher in tumor regions than in normal tissues (Fig. 1B).
Figure 1.
EphA10 is highly expressed in cancer cells and immunosuppressive myeloid cells at the tumor region.A, TCGA database analysis of EphA10 RNA expression in normal and tumor regions of breast invasive carcinoma (BRCA), lung adenocarcinoma (LUAD), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), and thyroid carcinoma (THCA). B, plots of IHC scores (left) and representative images of IHC staining (right) of EphA10 protein expression with anti-EphA10 #8 antibody in adjacent or normal tissues and tumor regions from human patient samples of breast cancer (n = 11, Biomax, #BR251), lung adenocarcinoma (n = 50, Biomax, #LC1504), and ovary cancer (n = 40, Biomax, #OV801a). Original magnification, ×400. C, IF staining of EphA10 expression with anti-EphA10 #4 antibody in sphere-cultured 4T1-GFP cell mass and 4T1-GFP tumor. N= 18 (18 position of sphere sample, 3 position per a tumor from 6 mice). Unit: 214m × 214m = 45,796 μm2. D, IF staining of EphA10 expression with anti-EphA10 #4 antibody in the indicated immune cell populations in the TME, including T cells (CD3+), tumor-associated macrophages (TAMs, F4/80+ and CD163+), and myeloid-derived suppressor cells (MDSCs, CD11b+ and Gr-1+). Circles indicate the location of T cells (red), not merged with EphA10 (green) in tumor regions. Scale bar, 100 μm, magnified: 20 μm. Error bars represent mean ± SD. 0.01<∗p <0.05, 0.001<∗∗p < 0.01, ∗∗∗p < 0.001, unpaired t test. EphA10, Ephrin receptor A10; IHC, immunohistochemistry; IF, immunofluorescence; TME, tumor microenvironment.
Consistent with these histological findings, EphA10 expression appears to be regulated by the TME. Basal levels of EphA10 expression were very low in in vitro culture (Fig. S2_negative control) and sphere culture conditions without TME (Fig. 1C_sphere), but EphA10 levels were highly elevated in tumors with TME (Fig. 1C_tumor). Several studies have suggested that epigenetic crosstalk between TME and cancer cells (24, 25). Interestingly, EphA10 expression was largely increased by treatment with epigenetic modulators such as 5-aza and TSA (Fig. S2). Collectively, EphA10 expression could be epigenetically inhibited in normal tissue except testis and epigenetic suppression of EphA10 may be released by TME. A future study is warranted to investigate the epigenetic regulatory mechanism of EphA10 expression by TME. This characteristic of EphA10 expression provides a great advantage for EphA10 therapy in tumor-specific targeting.
Along with EphA10 being detected on tumor cells, we wondered whether EphA10 is also expressed on immune cells within the TME, which might contribute to its role in cancer immune evasion. To explore this possibility, we performed immunofluorescence (IF) staining with specific antibodies that recognize target antigens for EphA10 (green), tumor-infiltrating T cells (CD3 in red), and immunosuppressive myeloid cells (26) including tumor-associated macrophages (TAMs in magenta; F4/80 in blue and CD163 in red) and myeloid-derived suppressor cells (MDSCs in magenta; CD11b in blue and Gr-1 in red) (Fig. 1D). EphA10 co-localized with TAMs and MDSCs, as indicated by merged signals (lower two panels in white, Fig. 1D), but not with T cells (white circles in the top panel, Fig. 1D). Both TAMs and MDSCs are well known to suppress antitumor immunity (27, 28). Taken together, EphA10 was highly expressed in tumor regions as well as in some immune cell populations such as TAMs and MDSCs and targeting EphA10 by an EphA10 therapeutic antibody in these EphA10-positive malignancies, including breast cancer, may lead to effective cancer treatments through the restoration of antitumor immune surveillance.
Generation and validation of mAbs targeting EphA10
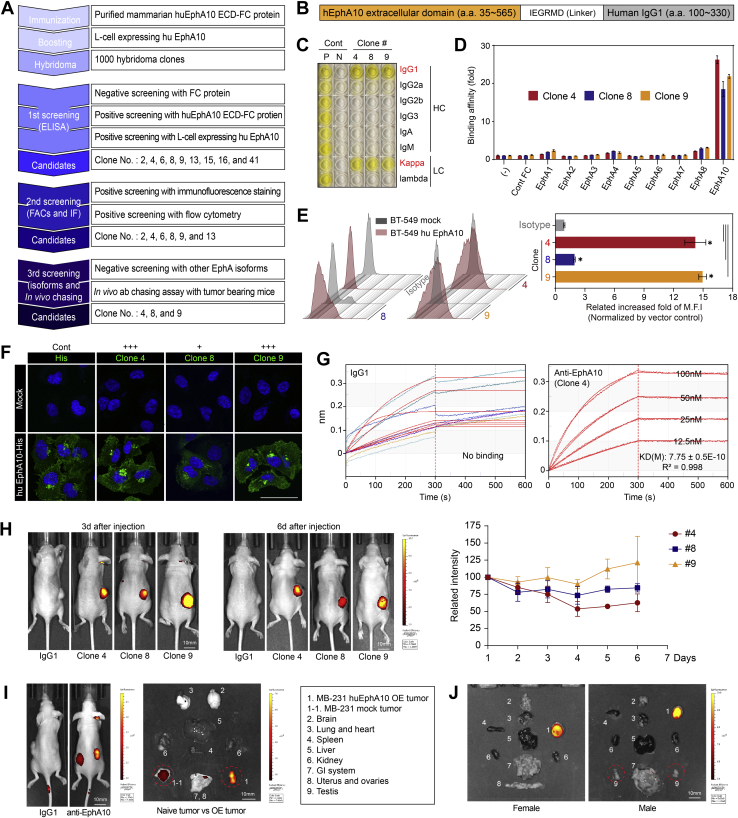
To generate and validate mAbs targeting EphA10, we used hybridoma technology (29, 30) (Fig. 2A). First, BALB/c mice were immunized with the extracellular domain (ECD) of recombinant human EphA10 (huEphA10) Fc chimera protein derived from NS0 cells (Fig. 2B). After the initial immunization with purified huEphA10 ECD-Fc fusion protein to provoke an immune response, mice then received a booster injection with L cells expressing huEphA10 by a whole-cell immunization method, which maintains the intact structure of antigens with native conformation and modification during antibody selections (31). After completing the immunization and boosting steps, we isolated the responding B cells from the spleen and fused them with immortalized myeloma cells, producing large numbers of hybridoma clones that persistently secreted the desired anti-EphA10 mAbs.
Figure 2.
Generation and validation of monoclonal antibodies targeting EphA10.A, workflow for generating and validating EphA10 mAbs. Key activities at each stage are listed in the text boxes. B, antigen source from recombinant human EphA10 Fc chimera protein derived from NS0 mouse myeloma cells. C, determination of the IgG isotypes of anti-EphA10 clones #4, #8, and #9. Cont, control; N, negative; P, positive; HC, heavy chain; LC, light chain. D, ELISA of binding affinity of the indicated antibodies toward a series of EphA family member proteins. Negative control, no protein added (–) or Cont Fc protein. E, representative images (left) and quantification (right) of flow cytometry analysis measuring fluorescence intensity of the indicated antibodies in BT-549 cells expressing huEphA10 or mock control. Isotype serves as a negative control. F, IF staining of EphA10 expression with the indicated antibodies in BT-549 cells expressing huEphA10 or mock control. Anti-His antibody was applied as a positive control. Scale bar, 50 μm. G, binding affinity (KD) analysis of mIgG1 and anti-EphA10 clone #4 by Octet. H, Left, representative images of in vivo antibody chasing assay using anti-EphA10 antibodies and IgG1 as a negative control. Signal intensity and location of the indicated antibodies labeled with Alexa-647 fluorescence dye were examined with use of the IVIS spectrum system at days 3 and 6 after intraperitoneal injection in nude mice bearing huEphA10-expressing MDA-MB-231 cells. Right, quantification of signal intensity of anti-EphA10 antibodies at the indicated time points. I, representative images of tumors and organs by in vivo antibody chasing assay, which were dissected at day 6 after injection of anti-EphA10 antibody into nude mice bearing both naïve MDA-MB-231 cells (left) and huEphA10-expressing MDA-MB-231 cells (right). J, representative images of tumors and organs by in vivo antibody chasing assay, which were dissected at day 6 after injection of anti-EphA10 antibody into female and male nude mice bearing huEphA10-expressing MDA-MB-231 cells. Circles indicate no signal in the testes in male mouse. Error bars represent mean ± SD. ∗p < 0.05, unpaired t test. ECD, extracellular domain; EphA10, Ephrin receptor A10; ELISA, enzyme-linked immunosorbent assay; Flow, flow cytometry; IF, immunofluorescence; IgG, immunoglobulin G; MFI, mean fluorescence intensity.
Among 1000 hybridoma clones, we selected the ones through the first screening that recognized huEphA10 ECD-Fc and L cells expressing huEphA10 proteins but not Fc proteins only. As validated by the sequential second and third screening of biological activity assays, e.g., flow cytometry, IF, enzyme-linked immunosorbent assay (ELISA), and in vivo antibody chasing assay, we selectively obtained three candidates, clones #4, #8, and #9, with high antigen specificity for EphA10 (Fig. 2A). Because EphA10 is highly conserved in species between human and mouse with 91% identity at the amino acid level (16), we also functionally selected the candidate clones from the first screening that recognized not only the intact human but also mouse EphA10 ECD; this would allow the mAbs to be tested in the mouse tumor models.
For choosing an appropriate strategy for antibody purification, we characterized the candidates’ isotypes of immunoglobulin G (IgG) and determined their heavy and light chains to be the IgG1 and Kappa subclasses, respectively (Fig. 2C). Next, we performed a plate–based ELISA to assess the binding affinity of candidate clones to EphA10 as well as to other EphA family members (EphA1−EphA8) known to share a similar architecture with EphA10 (32) and found that they were specifically bound to EphA10, not to other isoforms (Fig. 2D). Furthering the validation of these anti-EphA10 antibodies, we examined their fluorescence intensity by a flow cytometry–based assay to evaluate their recognition abilities bound to cell-surface EphA10 between BT-549 cells expressing huEphA10 and mock control (Fig. 2E, left). Statistical significance was observed in all three candidates compared with the isotype, whereas a higher fluorescence intensity was observed in clones #4 and #9 than in clone #8 (Fig. 2E, right). Similar cell membrane–bound results were obtained by IF since all three candidates had the ability to recognize the intact structure of EphA10 on the cell surface; among them, clones #4 and #9 displayed stronger fluorescence intensity than did clone #8 (Fig. 2F). Further flow cytometry–based analysis using 293T cells expressing mouse EphA10 and mock showed that these anti-EphA10 antibodies were also able to recognize mouse EphA10 (Fig. S3). To determine the kinetics of binding affinity, octet analysis was performed. mIgG1 did not bind EphA10 antigen, whereas clone #4 showed binding stability with specific binding and slow dissociation at serial concentrations of antigen (Fig. 2G).
Finally, to evaluate the half-lives and the targeting regions of these anti-EphA10 candidate clones in vivo, we labeled anti-EphA10 clones with Alexa-647 fluorescent dye and performed an antibody chasing assay in nude mice bearing huEphA10-expressing MDA-MB-231 tumors (Fig. 2, H–J). The fluorescence signals of these clones were maintained in tumors for 6 days without reduced intensity (Fig. 2H), suggesting that these anti-EphA10 antibodies exhibited similar pharmacokinetics within a normal range comparable to that of other IgG1 therapeutics antibodies in a mouse model (33). Of note, the signals of EphA10 mAb accumulated specifically in regions of tumor according to EphA10 levels (Fig. 2I), not other organs isolated from both female and male tumor-bearing mice (Fig. 2J). Collectively, through a series of characterization and validation of the generated hybridoma clones, anti-EphA10 mAbs #4, #8, and #9 exhibited the potential to develop therapeutic antibodies specifically targeting EphA10 in varying degrees.
Anti-EphA10 antibody therapy inhibits tumor progression in syngeneic TNBC models
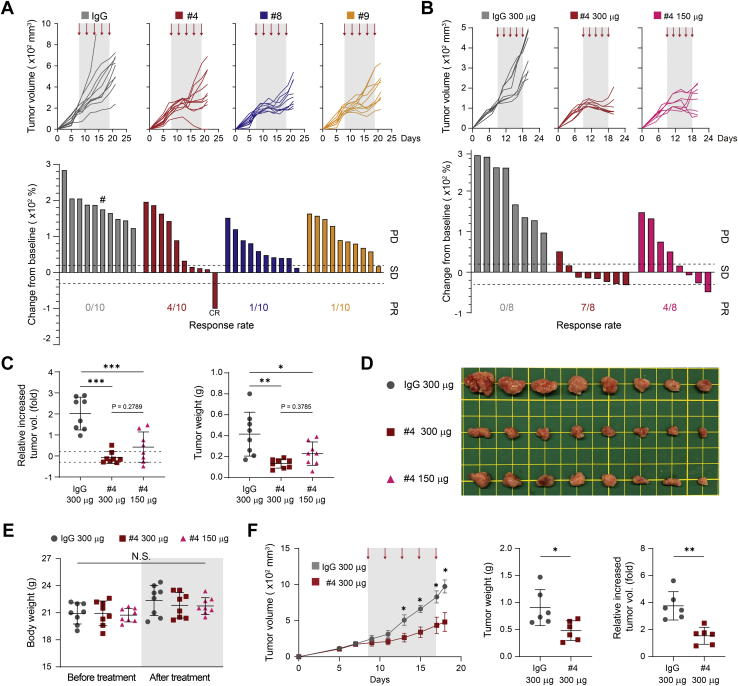
To evaluate the therapeutic efficacy of the above-generated anti-EphA10 antibodies in a syngeneic tumor system, 4T1 TNBC cells were implanted into the mammary fat pad of immunocompetent BALB/c mice, followed by injection of the individual clone #4, #8, #9, or IgG as a negative control (150 μg/mouse); the tumor volume was measured and graphed as a continuous variable in a waterfall plot (Fig. 3A). In 4T1 tumor-bearing mice, treatment with either anti-EphA10 clone impaired tumor growth, compared with the IgG control, with more potent effects from clone #4 than from clones #8 and #9 (Fig. 3A); the overall health of the mice appeared good during the course of treatment since the body weight did not change significantly (Fig. S4). Notably, among the three candidate clones, mice that received clone #4 had the most improved response rate under the therapeutic categories of stable disease and partial response/complete response (34) (Fig. 3A, bottom; 40% for clone #4 versus 10% for clone #8 or #9). Accordingly, we further validated antigen–antibody specificity on clone #4 by comparing its therapeutic efficacy between two doses of treatment (150 and 300 μg/mouse) (Fig. 3,B− E).
Figure 3.
Anti-EphA10 antibody therapy inhibits tumor progression in syngeneic TNBC mouse models.A, tumor growth curves (top) and waterfall plot analysis (bottom) of 4T1 cells in BALB/c mice treated with anti-EphA10 clones #4, #8, #9, or IgG control as indicated (150 μg/mouse). n = 10 mice per group. #: mouse dead at day 15. B, tumor growth curves (top) and waterfall plot analysis (bottom) of 4T1 cells in BALB/c mice treated with various doses of anti-EphA10 #4 antibody (150 and 300 μg/mouse) or IgG control as indicated. n = 8 mice per group. In (A) and (B), tumors were measured at the indicated time points and dissected at the end point. The gray box and arrows in each panel indicate the duration and time of treatment. The waterfall plot generated from the tumor growth curve indicates tumor progression in each mouse under individual antibody treatment. The number of mice that experienced tumor progression in each group is shown in parentheses. C, the increased fold of tumor volume (left) and tumor weight (right) from (B). D, representative images of mice tumors dissected at the end point from. Scale bar, 1 cm. E, measurement of body weight from (B) before and after treatment with the indicated antibodies. Gray box indicates the body weight at the end point at day 21. Error bar, mean ± SD. One-way ANOVA. NS, not significant. F, average tumor growth of EMT6 cells in BALB/c mice treated with anti-EphA10 #4 antibody or IgG control. Tumors were measured at the indicated time points and dissected at the end point. The gray box and arrows indicate the duration and time of treatment. n = 6 mice per group (left). The tumor weight (middle) and the increased fold of tumor volume (right). Error bar, mean ± SD. Mann–Whitney test (C, E, and F). 0.01<∗p <0.05, 0.001<∗∗p < 0.01, ∗∗∗p < 0.001. CR, completed response; EphA10, Ephrin receptor A10; IgG, immunoglobulin G; PD, progressive disease; PR, partial response; SD, stable disease; TNBC, triple-negative breast cancer.
Consistent with previous results (Fig. 3A), lower-dose treatment with clone #4 (150 μg/mouse) in 4T1 tumor-bearing mice led to smaller tumor size and a higher response rate than did the IgG control treatment (Fig. 3B). Strikingly, results from the waterfall plot analysis showed that mice treated with a higher dose of clone #4 had a better response rate than did those that received a lower dose (Fig. 3B, bottom; 87.5% and 50%, 300 μg and 150 μg, respectively). Based on the mouse tumor measurements, including the representative images displayed, higher-dose treatment with clone #4 tended to more inhibit tumor volume and tumor weight, compared with lower-dose treatment (Fig. 3, C and D), suggesting that anti-EphA10 clone #4 dose-dependently may exert its therapeutic efficacy to inhibit tumor progression. In addition, both doses of clone #4 resulted in good overall health in the mice across treatment as their body weight did not significantly differ from that of the IgG control mice (Fig. 3E). By using another syngeneic TNBC model, EMT6, we obtained similar results in response to clone #4 treatment (150 μg/mouse), which showed a significant reduction in tumor volume and tumor weight in EMT6 tumor-bearing mice (Fig. 3F). Together, these results, as evidenced by two syngeneic TNBC models, suggested that our in-house EphA10 antibody clone #4 might be a suitable candidate for further development of therapeutic antibodies against EphA10, which could substantially benefit patients with TNBC (Table S1).
Anti-EphA10 antibody therapy increases the infiltration and activity of CTLs
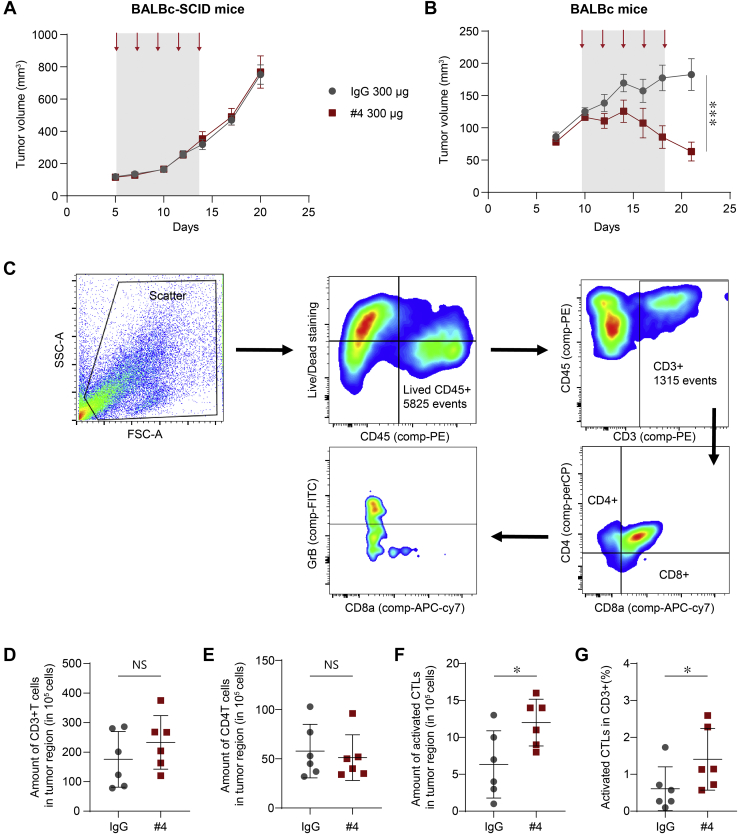
To investigate the role of anti-EphA10 antibody in antitumor immunity, we examined the effects of anti-EphA10 mAb (clone #4) in BALB/c-SCID and BALB/c mice bearing orthotopic injected 4T1 tumor. While anti-EphA10 mAb did not show any antitumor effect in immune deficiency (Fig. 4A_BALB/c-SCID), significant tumor regression was observed without toxicity (Fig. S5) in immunocompetent mice (Fig. 4B_BALB/c), suggesting the antitumor immunity is needed for anti-EphA10 mAb-mediated tumor suppression.
Figure 4.
Anti-EphA10 antibody therapy increases the infiltration and activity of CTLs.A and B, the effect of anti-EphA10 antibody in tumor growth of BALB/c-SCID (A) and BALB/c (B) mice bearing orthotopic injected 4T1 tumor. Mice (n = 6 per group) were treated with anti-EphA10 mAb (clone #4) or IgG control. The gray box and arrows indicate the duration and time of treatment. Error bar represents mean ± SEM. Data were analyzed by two-way ANOVA analysis. ∗∗∗p < 0.001. C, the gating strategy used for flow cytometric analysis of (D–G). Cells were analyzed by forward scatter/side scatter parameters and Zombie Violet− staining to determine total live events, in which various cell populations were identified on the basis of marker expression: CD45+ (immune cell population), CD3+/CD4−/CD8+ (CD8 T cells), and granzyme B+ (GrB) in CD8 T cells. D–G, flow cytometry analysis of the amount of CD45+/CD3+ Pan-T cell (D), the amount of CD3+/CD4+/CD8- T cell (E), the amount of CD8+/GrB+ CTLs (F), and percentage of CD8+/GrB+ CTLs in CD3+ cells (G) in 4T1 tumor region under IgG control or anti-EphA10 #4 antibody treatment (300 μg/mouse). n = 6 mice per group. Data were analyzed by student t test analysis. 0.01 < ∗p < 0.05. CTL, cytotoxic T lymphocyte; EphA10, Ephrin receptor A10; IgG, immunoglobulin G; mAbs, monoclonal antibodies; TNBC, triple-negative breast cancer.
Since numerous studies showed that tumor regression arises from the restoration of antitumor immunity in the host (35), we asked whether treating tumor-bearing mice with anti-EphA10 mAb #4 enhances the activation of CD8+ CTLs, a central effector in antitumor immunity that eliminates malignant cells mainly by releasing granzyme B (GrB) into the target (36), which in turn may lead to its therapeutic effect on tumor shrinkage. To this end, we first digested 4T1 tumors from BALB/c mice treated with anti-EphA10 clone #4 or IgG control into single cells to measure the amount of CD8+/GrB+ cells representing activated CTLs using flow cytometric analysis. The gating strategy started by using forward scatter and side scatter to find viable cell events, and then, cell populations were grouped by different types of markers, including CD45+, CD3+, CD4−, CD8+, and GrB+ (Fig. 4C). Although treatment of anti-EphA10 clone #4 in 4T1 tumor-bearing mice did not increase the total number of pan CD3+ T cells (Fig. 4D) and CD4+ T cells (Fig. 4E) in the tumor, of note, it indeed significantly restored the CTL-mediated adaptive immunity as measured by the total amount of activated CD8+/GrB+ CTLs (Fig. 4F) and percentage of activated CD8+/GrB+ CTLs in CD3+ T cells (Fig. 4G), compared with the control.
Since TAM and MDSC expressed EphA10 (Fig. 1D), we also examined the infiltration and viability of these two immune-suppressive cells in 4T1 syngeneic mouse model treated with anti-EphA10 clone #4 or IgG control. The gating strategy started by using forward scatter and side scatter to find viable cell events, and then cell populations were grouped by different types of markers, including CD45+, CD3-, F4/80+, CD206+, CD11b+, and Gr1+ (Fig. S6A). Although the proportion and viability of intratumoral MDSC tended to decrease in the anti-EphA10 mAb treatment group, no statistically significant results were obtained due to individual variation. Thus, the results showed that treatment of anti-EphA10 clone #4 does not affect the infiltration and viability of TAM and MDSC in tumor region (Fig. S6B).
Taken together, we successfully developed an EphA10 mAb (clone #4) that could suppress tumor growth in syngeneic TNBC models through a boost of CTL-mediated antitumor immunity, which might provide a therapeutic option, particularly for TNBC patients whose EphA10 is positively expressed.
EphA10-specific CAR-T cells inhibit TNBC growth in vitro and in vivo
Recently, CAR T cell-based therapy with specificity derived from a mAb single-chain variable fragment (scFv) is considered as an emerging and effective cancer immunotherapy (37). To maximize the benefits of CAR-T therapy with less adverse effects, cancer-specific expression of a target protein should be a prerequisite along with tight control of its activity (38). Considering that EphA10 had tumor region specificity (Fig. 1, A and B) without detection in most of normal tissues (16, 17), we speculated that anti-EphA10 mAbs could be applied to the development of EphA10-specific CAR-T therapy.
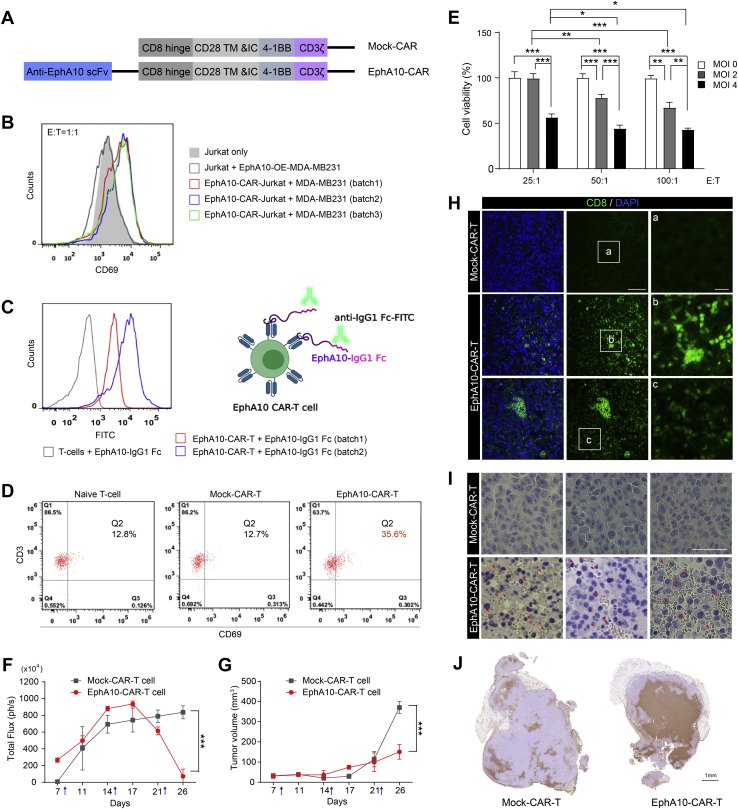
To this end, we cloned the human scFv sequence from anti-EphA10 clone #4 into a third-generation CAR vector (39) (Fig. 5A, EphA10-CAR and Fig. S7A) and transduced EphA10-CAR lentivirus into human T lymphoma Jurkat cells (Fig. 5B, EphA10-CAR-Jurkat) for early assessment of EphA10-CAR. As previous study indicated that Jurkat cells upregulate CD69 upon activation (40), we observed that the expression levels of CD69 were increased after co-culturing CAR-Jurkat cells (but not Jurkat only) with MDA-MB-231 cells (Fig. 5B), suggesting that EphA10-CAR can interact with EphA10 protein on the cell surface of MDA-MB-231 cells. Next, we isolated human CD8+ T cells from healthy volunteers with microbeads and identified the purity of CD8+ T cells near 99% among whole cell population (Fig. S7B, Q2). The CD8+ T cells were transduced with different batches of EphA10-CAR lentivirus (batch 1 and 2) to generate EphA10-CAR-T cells, validated by the interaction with recombinant huEphA10 Fc chimera protein detected by using fluorescein isothiocyanate (FITC)-conjugated anti-human IgG1 Fc antibody (Fig. 5C, right panel). Indeed, the fluorescence signals of recombinant EphA10 proteins were increased in two independent batches of EphA10-CAR-T cells (batch 1: 84.9%; batch 2: 88.8%), compared with the mock-transduced T cells, indicating the successful establishment of EphA10-CAR-T cells in human T cells (Fig. 5C, left panel).
Figure 5.
EphA10-specific CAR-T cells inhibit TNBC growth in vitro and in vivo.A, schematic illustration of the lentiviral mock-CAR (upper panel) and EphA10-CAR (lower panel) derived from clone #4. B, flow cytometry assessment of the sensitivity and specificity of EphA10-CAR construct by CAR-Jurkat cells. Red, blue, and green colors represent different batches of CAR-Jurkat cells as indicated. C, Left panel: Flow cytometry analysis of fluorescein isothiocyanate (FITC) signals of the indicated batches of EphA10-CAR-T cells or mock-CAR-T cells incubated with 0.5 μg of recombinant human EphA10 Fc chimera protein. Right panel: Representative flow cytometry histogram of binding of EphA10-CAR-T cell populations to recombinant human EphA10 Fc chimera protein. D, Flow cytometry assessment of activation of naïve T-cells, mock-CAR-T-cells, and EphA10-CAR-T-cells by mixed culture with MDA-MB-231 cells. E, the MTT assay of EphA10-CAR-T cells against EphA10-positive MDA-MB-231 cells. Data from at least three independent experiments. Error bars present mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way analysis of variance with Newman–Keuls Multiple Comparison Test. All statistical analyses were conducted using Prism 5.01 software (GraphPad Software www.graphpad.com). F, Quantification of signal intensity detected by the IVIS spectrum system of mice bearing MDA-MB-231/luc orthotopic xenograft treated with mock-CAR-T cells (n = 4) or EphA10-CAR-T cells (n = 5) at various time points as indicated. The arrows indicate the time of treatment (day 8, 15, and 22). G, the tumor volume of mice bearing MDA-MB-231/luc orthotopic xenograft treated with mock-CAR-T cells (n = 4) or EphA10-CAR-T cells (n = 5) at various time points as indicated. The arrows indicate the time of treatment (day 8, 15, and 22). Tumor volume (V) was measured two times per week by using the formula V = (L × W2)/2, where L and W represent tumor length and tumor width, respectively. F and G, error bar, mean ± SD. Two-way ANOVA ∗∗∗p < 0.001. H, representative images of IF staining of xenograft sections from the mouse model (G). The xenograft sections were stained with anti-human CD8α-FITC antibody (red arrows indicated the infiltration of EphA10-CAR-T cells). Scale bar: left: 50 μm, right: 10 μm. I, representative images of IHC staining of xenograft sections from the mouse model (G). The xenograft sections were stained with anti-human CD8α antibody and Pierce Peroxidase Detection Kit (red arrows indicated the infiltration of EphA10-CAR-T cells). Scale bar, 50 μm. J, TUNEL assay. The xenograft tumors were resected and preserved in 4% paraformaldehyde at 4 °C, and then paraffin embedded sections of tissue were stained with In Situ Cell Death Detection kit according to the manufacturer’s protocol (Roche, Cat. No. 11684817910). CAR, chimeric antigen receptor; E, effector; EphA10, Ephrin receptor A10; MOI, multiplicity of infection; scFv, single-chain variable fragment; T, target; TNBC, triple-negative breast cancer.
EphA10-CAR-T cells were cocultured with MDA-MB-231 cells for 24 h to explore their activation (Fig. 5D). Our data showed high numbers of CD3+ surface markers (99.3%, 98.9%, and 99.3%) in normal CD8+ T cells, mock-CAR-T cells, and EphA10-CAR-T cells, respectively. The early cell surface activation marker (CD69) increased significantly in EphA10-CAR-T cells (35.6%) compared to mock-CAR-T cells (12.7%) and normal CD8+ T cells (12.8%). These results clearly show that EphA10-CAR-T cells can be specifically activated when recognizing cancer cells expressing EphA10.
To determine the in vitro cytotoxicity of EphA10-CAR-T cells on EphA10-positive human breast cancer cells, we conducted the MTT assay by co-culturing EphA10-CAR-T cells with MDA-MB-231 cells (Fig. 5E). We transduced CD8+ T cells with EphA10-CAR lentivirus, and the multiplicity of infection (MOI) was established according to a previous study (41). We found that the increasing number of EphA10-CAR lentiviral particles per cell during infection significantly decreased cell viabilities of MDA-MB-231 cells (Fig. 5E; MOI 0 versus MOI 2 or MOI 4; MOI 2 versus MOI 4). Consistently, the increasing ratio of effector (E for EphA10-CAR-T cells) to target (T for MDA-MB-231 cells) also correlated to the enhanced cytotoxicity against MDA-MB-231 target cells (Fig. 5E; E:T = 25:1 versus 50:1 or 100:1), further supporting cell growth inhibition by EphA10-specific CAR-T cells.
Finally, we explored the antitumor effect of EphA10-CAR-T cells in vivo by using a mouse model of female CAnN.Cg-Foxn1nu/CrlNarl nude mice bearing orthotopic MDA-MB-231/luc cells xenograft (42, 43). CAR-T cells were then given at day 8, 15, and 22 (1 × 107 cells/injection) through femoral vein. We observed that the bioluminescent signal in the EphA10-CAR-T group started to decrease after the second CAR-T cell injection, demonstrating that the EphA10-CAR-T had a greater tendency to decrease tumor size than the mock-CAR-T (Fig. 5F). Indeed, tumor growth was significantly reduced in the EphA10-CAR-T-treated group compared to the mock-CAR-T-treated group, strongly suggesting the antitumor effect of EphA10-CAR-T upon the TNBC mouse model (Fig. 5G). Consistently, a lot of EphA10-CAR-T cell infiltration was observed in the xenografts of mice treated with EphA10-CAR-T cells, whereas there are no signals of mock-CAR-T cells, demonstrating the specificity of EphA10-CAR-T cell in vivo (Fig. 5,H and I). In addition, we found EphA10-CAR-T cells induced extensive apoptotic cell death in xenograft tumor tissue (Fig. 5J). Although the average body weight of the EphA10-CAR-T group was statistically less than the mock-CAR-T group (Fig. S7C), there were no obvious pathological changes in major organs between the two groups (Fig. S7D), suggesting the negligible on-target off-tumor toxicities. Taken together, our findings indicated that EphA10-CAR-T cells derived from the scFv sequence of anti-EphA10 clone #4 exert a potent antitumor ability in this model.
Anti-EphA10 antibody clone #9 induces EphA10 internalization
As a tumor-associated antigen like EphA10 could be an ideal candidate for an antibody-based approach, antibody–drug conjugates (ADCs) (44, 45), EphA10–ADCs might exert potent antitumor activity, taking advantage of the delivery of cytotoxic drugs with bystander effect in tumor regions for many types of EphA10-positive cancer. To further validate the properties of EphA10 mAbs suitable for ADC application, we examined the ability of EphA10 antibody internalization, which is required for facilitating the ADC–antigen complex to transport into cancer cells via endocytosis, in turn enhancing the antitumor potency of cytotoxic drugs (46). For this purpose, we first labeled anti-EphA10 clones with a pH-sensitive fluorophore pHrodo Red and incubated the labeled antibodies (anti-EphA10-pHrodo) in BT-549 cells expressing huEphA10 or mock control. The fluorescence of the pHrodo Red dye increases with decreasing pH from neutral (e.g., pH 7.2) to acidic (e.g., pH 4.5), making it reporter detectable in the red range in low-pH environments such as lysosomes, which represents the labeled anti-EphA10 antibody is internalized (Fig. S8A). As a result of examining the internalization activity of each clone, anti-EphA10 clone #9 was clearly internalized in the lysosome of BT-549 huEphA10 cells in a dose- and time-dependent manner compared to the mock transfected control. (Fig. S8, B–D). Results from IF analysis further demonstrated the co-localization of clone #9 (red) with LysoTracker (green), indicated by yellow merged signals (Fig. S8E). These results revealed that anti-EphA10 clone #9 possessed internalization properties from the cell surface to be located to the lysosomal compartment. Since cargo proteins can be internalized by diverse mechanisms of endocytosis, such as clathrin-mediated endocytosis (clathrin-ME) or/and caveolae–ME (47), we next asked whether these endocytic pathways are involved in EphA10 antibody internalization. To this end, we assessed the internalization ability of the pHrodo-labeled clone #9 in BT-549 huEphA10 cells pretreated with various inhibitors of clathrin–ME (β-CD and PitStop), caveolae–ME (Genistein and Filipin III), or dual pathways (Dynasore). We found that inhibitors that prevent caveolae–ME, but not clathrin–ME, effectively blocked clone #9 internalization, suggesting that anti-EphA10 clone #9 was internalized into the cells through caveolae-ME (Fig. S8F).
Finally, in order to better understand the antibody–antigen complex of the EphA10 model, we used a molecular docking program (ZODCK) (48) to computationally predict the epitope-located region recognized by the anti-EphA10 mAb #9 candidate (Fig. S8G; highlighted in magenta). The top three ranking outcomes given by the docking program indicated a comparable region, comprising the residues from the second fibronectin (FN)-III domain and the linker between the two FN-III domains (Fig. S8G). A similar result was given by an online epitope prediction software (EpiPred) (49). The EphA10 3D model shown in gray was obtained by SWISS-MODEL (50) by using an EphA4 crystal structure as a template with 47% sequence identity. The model of antibody Fab molecule (Fig. S8G; heavy chain in green; light chain in blue) containing merely the variable region was obtained by AbodyBuilder (51). Together, the prediction results revealed EphA10 antibody #9 in association with the FN-III within the ECD of EphA10 antigen, which further validated the EphA10 antibody–antigen conformation and might be useful in aiding structure-based design of more drug candidates targeting EphA10. These results suggested that anti-EphA10 clone #9 could be an optimal candidate applied to ADC technology, highlighting the potential for clinical implications of EphA10-targeted therapy in the treatment of cancer.
Discussion
Targeting the receptors that are specifically or broadly expressed on cancer cells has been used as a standard therapeutic strategy for breast cancer. A certain population of breast cancer patients benefits from therapies that target hormone receptors (ER and PR) or HER2 (1). However, more than 20% of patients with breast cancer have TNBC, which is negative for ER, PR, and HER2 expression. TNBC patients have limited therapeutic options, and therapeutic failure due to chemotherapy resistance is a significant challenge. A new therapeutic strategy to conquer TNBC is urgently needed (2).
mAbs in cancer therapy are designed to target the tumor-specific antigens for blocking ligand–receptor signaling and inducing antitumor activity in tumor region in cancer patients (52). EphA10 has been revealed to be a tumor-specific surface protein and a promising therapeutic target for cancer therapy (17, 20, 21). However, EphA10 is a pseudo-kinase (32, 53), and thus mAb-based therapy but not kinase inhibitors may be a more practical approach to target EphA10. Previously, Nagano et al (17). demonstrated tumor suppression in a xenograft TNBC mouse model by administration of an anti-EphA10 mAb; the results were interesting yet could not address whether immune response might contribute to the antitumor activity, which is critical for human clinical application. Our current study developed mAbs with high specificity for EphA10 in tumor regions and provided preclinical data to demonstrate the therapeutic efficacy and safety of these anti-EphA10 antibodies in two syngeneic TNBC models. It is worthy of mentioning that EphA10 therapy can apply to all cancer types that overexpressing EphA10. In this study, we used the TNBC model to show therapeutic efficacy; however, based on The Cancer Genome Atlas database as shown in Figures 1A and S1, the newly developed EphA10 targeting therapy may also be applied to other cancer types including hormone-positive and HER2-positive breast cancer (54, 55). Thus, it is especially interesting to test if the newly developed EphA10 target therapy can overcome those breast cancer patients who are resistant to the FDA-approved drugs such as tamoxifen and Herceptin in the future.
Of note, anti-EphA10 antibody treatment not only significantly reduced tumor growth but also increased the infiltration and activity of CTLs in tumor regions. Interestingly, our results also demonstrated that EphA10 was expressed on tumor cells and immunosuppressive cells, including TAMs and MDSCs, in the TME. These findings suggested that EphA10 inhibits immune surveillance and impair the infiltration and activity of CTLs in the TME, which can be restored with anti-EpHA10 antibody treatment. Recent studies have reported that EphA signaling is closely related to T cell immunity. The binding of ephrin-A1 to EphA receptors on CD4+ and CD8+ T cells is known to stimulate T cell migration and mediate T cell chemotaxis (56, 57, 58). Although EphA10 is now considered to be a pseudo-kinase, most ephrin-A ligands have been shown to bind with EphA10 (16), suggesting that this death receptor may act as an inhibitory regulator of T cell immunity through ligand competition with other EphA receptors. In contrast, the bidirectional signaling transduced by Eph–ephrin interaction plays a critical role in the regulation of cancer and the immune system (10). Interestingly, recent studies suggested that reverse signaling from Eph receptors to ephrin ligands is important for regulating the functions of T cells (59). Thus, it will be interesting to elucidate whether EphA10 transduces the reverse signaling to its ephrin ligands expressed on T cells to inactivate cytotoxic immunity. Future studies will be required to validate such a possibility in the functional mechanism of EphA10 in antitumor immunity.
In addition to inducing antitumor immunity, other critical characteristics of these anti-EphA10 antibodies include their high specificity and ability to mediate the internalization of EphA10; thus, these antibodies hold great potential for the development of antibody-derived therapeutic approaches such as CARs and ADCs. Since CAR-T therapy has adverse effects as potent as its therapeutic effects, cancer-specific expression of the target protein should be a prerequisite along with tight control of its activity (38). EphA10 had high tumor-specific expression and was not expressed in normal tissues except testis, and in vivo antibody tracking analysis showed that even testis could be protected from the antibody by the blood–testis barrier. Therefore, considering that there is currently no CAR-T immunotherapy approved to treat solid tumors, our anti-EphA10 antibodies validated in TNBC models may provide an ideal CAR-T to test in TNBC. Indeed, our results indicated the specificity of EphA10-CAR-T cells derived from anti-EphA10 clone #4 showing TNBC tumor shrinkage and T cell infiltration into tumor tissues in vivo, which may provide implications for clinical applications of EphA10-CAR-T cells in TNBC. On the other hand, ADC-based treatment is one of the fastest-growing approaches to cancer therapy, consisting of a mAb conjugated with a cytotoxic payload. This anticancer drug has been used to treat many types of cancer by selectively releasing cytotoxin in the tumor region and reduce systemic toxicity (44). Recently, sacituzumab govitecan, an ADC with a topoisomerase inhibitor linked to an antibody to the trophoblast antigen Trop-2, was approved by the FDA for metastatic TNBC (2). However, one of the challenges for developing ADC therapy is the specificity of the mAbs. Notably, EphA10 fulfills the key requirements of targeted antigen for ADCs; for example, as a membrane protein, it is highly expressed on cell surfaces in tumor regions but not on healthy tissues. Furthermore, the anti-EphA10 antibodies that were generated in this study precisely recognized EphA10 in the tumor region and accumulated there but did not accumulate in other organs in vivo. In addition, the ability of the antibodies to internalize the EphA10 protein will facilitate the transportation of drugs into cells for their tumor-killing effects. Therefore, augmenting antitumor immunity by targeting EphA10 represents a new potent immunotherapy for TNBC, and studies of therapeutic anti-EphA10 antibodies are worth pursuing to expand the development of more antibody-based therapies such as CARs and ADCs.
Experimental procedures
Cell culture
The BT-549, MDA-MB-231, 4T1, and EMT-6 breast cancer cell lines were obtained from ATCC. Human breast cancer cell lines and mouse cancer cell lines were cultured in Dulbecco's modified Eagle's medium/F12 and Dulbecco's modified Eagle's medium, respectively, supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic mixture. All cell lines tested negative for mycoplasma contamination and validated by short-term repeat DNA fingerprinting with use of the AmpF_STR identifier kit according to the manufacturer’s protocol (Applied Biosystems, #4322288) at The University of Texas MD Anderson Cancer Center.
Generation of huEphA10-expressing cells
The huEphA10 expression vector (pCDH-huEphA10) was generated by inserting full-length EphA10 cDNA (NP_001092909; R&D Systems, #RDC1386) into the lentiviral vector pCDH-CMV-MCS-EF1-puro (System Biosciences, #CD510B-1). To establish MDA-MB-231 and BT-549 stable cells with huEphA10 ectopic expression, we conducted lentiviral packaging via transient transfection of 1 μg of pCDH-huEphA10 with 1 μg of pCMV-VSVG and 0.5 μg of pCMV-dR8.91 expression plasmids in 5 × 105 293T cells. After 72 h, 3 ml of conditioned medium from the transfectants was collected, centrifuged at 6000g for 15 min, and filtered through a 0.45-μm filter, followed by incubation with targeted cells at 5 μg/ml polybrene (EMD Millipore, #TR-1003-G) for lentiviral transduction. After transduction for 16 h, cells were replenished with 3 ml of complete medium for 1 day and subjected to puromycin selection (0.5 μg/ml; InvivoGen Corporation, #ant-pr-1) for another 3 days to generate stable cells. All stable transfectants were selected from a pool of clones. The duration of stable cell selection was at least 4 weeks.
ELISA assay for EphA isoforms and Ig isotype
96-well plates were coated with 100 μl of the antigen solution at a concentration of 5 μg/ml in coating buffer (Bio-Rad, #BUF030A) overnight at 4 °C. Purified proteins of FC control (#110-HG-100), EphA1 (#7146-A1), EphA2 (#3035-A2), EphA3 (#6444-A3), EphA4 (#6827-A4), EphA5 (#3036-A5), EphA6 (#5606-A6), EphA7 (#6756-A7), EphA8 (#6828-A8), and EphA10 (#5188-EA) were purchased from R&D Systems. Plates were washed three times in PBST (Tween-20; 0.05%) and then incubated with 150 μl of blocking solution (Bio-Rad, #BUF032) for 1 h at 37 °C. Plates were next washed four times in PBST and then incubated with 100 μl of anti-EphA10 antibody solution (0.5 ug/ml in PBST) for 1 h at 37 °C. Plates were then washed three times in PBST, followed by incubation with 100 μl of anti-mouse IgG, HRP-linked antibody solution (0.5 μg/ml in PBST; CST, #7076S). Finally, plates were washed three more times in PBST; 100 μl of substrate solution (Bio-Rad, #BUF062) was added to each well and incubated at room temperature (RT) and in the dark for 30 min. The reaction was then stopped with 0.2 M sulfuric acid, and absorbance was measured at 450 nm (yellow). For determining the Ig isotype, a rapid ELISA mouse mAb isotyping kit (ThermoFisher, #37503) was used according to the manufacturer’s instructions.
Flow cytometric analysis in vitro and CTL analysis in vivo
Single-cell suspensions were prepared and resuspended in a staining buffer (BD Biosciences, #554656). For the in vitro EphA10 antibodies binding assay, 1 × 106 BT-549 EphA10 overexpressing tumor cells were stained with 0.2 μg of anti-EphA10 #4, #8, #9, and IgG1 isotype control (Bio X cells, #BE0083) in 100 μl staining buffer for 30 min at RT. After the cells were washed by staining buffer three times, they were stained with APC anti-mouse second antibody according to a standard protocol. To analyze CTL profiles in mouse tumor samples, a Mouse Tumor Dissociation Kit (Miltenyi Biotec, #130-096-730) and gentle MACS Octo Dissociator (Miltenyl Biotec, #130-096-427) were used to digest mouse tumors into single cells. After red blood cells were removed and hybridized with TruStain FcX CD16/CD32 antibody (1:50; BioLegend, #101319), single cells were stained for flow cytometry according to standard protocols with antibodies against the following: PE-CD45 (1:200; BioLegend, #103105), APC-CD3ε (1:100; BioLegend, #100311), APC/Cy7-CD8a (1:100; BioLegend, #100713), and PreCP-CD4 (1:100; BioLegend, #100537). For further intracellular staining, cells were fixed, permeabilized, and stained with FITC-Granzyme B (1:50; BioLegend, #515403). Stained cells were analyzed with use of a BD FACSCanto II cytometer (BD Biosciences). Data were processed by using the FlowJo software program.
IF for mouse tumor tissue
After perfusion with 0.1 μM PBS (pH 7.4) under anesthesia, tumors extracted from mice were embedded into OCT block and frozen. Cryostat sections (8 μm thick) on slides were fixed with 4% paraformaldehyde for 15 min at RT. After PBS washing, cryostat sections were incubated in blocking solution (PBS containing 3% donkey serum, 1% bovine serum albumin [BSA], 0.3% Triton X-100, pH 7.4) for 30 min at RT. In antibody reaction buffer (PBS plus 1% BSA, 0.3% Triton X-100, pH 7.4), samples were stained with primary antibodies against CD3 (1:100; R&D Systems, #MAB4841-100), F4/80 (1:200; BioLegend, #123121), CD163 (1:300; Abcam, #Ab182422), CD11b (1:300; Novus, #NB110-89474), and Gr1 (1:200; R&D Systems, #MAB1037-SP) overnight at 4 °C, followed by Alexa 350, 488, 546, and 647 (1:3000; Life Technologies) secondary antibodies at RT for 1 h. Hoechst 33342 (Life Technologies, #H3570) was used for nuclear staining. A confocal microscope (Carl Zeiss, #LSM700) was used for image analysis.
In vivo chasing assay of antibody
Alexa Fluor 647-labeled anti-EphA10 mAbs and isotype control were generated according to the manufacturer’s instructions (Life Technologies, #A20173). For the in vivo chasing assay, MDA-MB-231 cells expressing huEphA10 (5 × 106) were suspended in a 1:1 mixture of PBS and Matrigel and injected into the back of 6- to 8-week-old female or male Nu/J mice. Once the tumor volume reached 500 mm3, each mouse was intraperitoneally injected with 200 μg of Alexa Fluor 647-labeled mAbs. The mice were then imaged daily, and associated fluorescent intensity was measured with use of an IVIS Lumina III in vivo imaging system (PerkinElmer). Tumor accumulation was quantified as the ratio of mean fluorescent intensity in tumors. After 6 days of administration, tumor tissues and other organs were isolated from mice, and their mean fluorescent intensity was detected.
Internalization of antibody
IgG and EphA10 (clone #9) antibodies were labeled with pHrodo Red (Thermo Fisher Scientific) according to the manufacturer. To monitor a dynamic internalization of antibodies, the BT549 cells expressing huEphA10 were incubated with pHrodo Red–labeled antibodies and LysoTracker Green (Thermo Fisher Scientific). Time-lapsed images were obtained by an IncuCyte live-cell analysis system (Essen BioScience). A confocal microscope (Carl Zeiss, #LSM700) was used for image analysis.
In vivo efficacy test of anti-EphA10 mAbs
All animal studies were performed according to guidelines approved by the MD Anderson Institutional Animal Care and Use Committee. To study the therapeutic effects of EphA10 antibodies in preclinical tumor models, 4T1-Luc2 (1 × 105) or EMT6 (1 × 105) cells were suspended in a 1:1 mixture of PBS and Matrigel and injected into the mammary fat pad of 6- to 8-week-old female BALB/c mice. Once the tumor volume reached between 100 and 200 mm3, each tumor-bearing mouse was intraperitoneally injected with IgG control (Bio X cells, #BE0083) or anti-EphA10 antibodies every 2 days for a total of five treatments. Tumor volume (V) was measured three times per week by using the formula V = (L × W2)/2, where L and W represent tumor width and tumor length, respectively.
KD determination by octet
For high-throughput KD screening, EphA10 antigen was loaded to AMC sensor with maximum loading 800 nM solution. The baseline was established in assay buffer, and the sensors were exposed to analyte at a single concentration in assay buffer as an association step. Dissociation was performed and monitored in fresh assay buffer. The loaded sensors were exposed to a series of analyte concentrations, and background subtraction was used to correct for sensor drifting. All experiments were performed with sensor shaking at 1000 rpm. ForteBio data analysis software was used to fit the data to a 1:1 binding model to extract an association rate and dissociation rate. The KD was calculated using the ratio kd:ka.
Patient tissue samples and IHC staining
Human tumor tissue specimens were used according to guidelines approved by the Institutional Reviewed Board at MD Anderson Cancer Center. The tissue specimens (#BR251a, #BR251b, #GI701, #OV801a, and #LC1504) were purchased from Biomax.us. For IHC staining, the tissue specimens were incubated with primary antibodies against EphA10 (clone #8, 4.5 mg/ml, 1:200 dilution) followed by detection with biotin-conjugated secondary antibody and avidin-peroxidase and visualization with aminoethyl carbazole chromogen. The staining intensity and the percentage of positive cells were used to determine the H-score of each specimen.
Construction of the EphA10-CAR vector and lentiviral vector production
Fully human anti-EphA10 scFv sequence (clone #4) was cloned into the third generation CAR lentiplasmid (pCDH-EF1α-MCS) that utilize the CD28 and 4-1BB stimulatory domains, and CD3ζ signaling domain with restriction enzymes: BstBI and XbaI. For lentiviral vector production, HEK-293T cells were seeded onto 10-cm dishes in a density of 3 × 106 cells/dish. Next day, these cells were transfected with EphA10 CAT vector and lentiviral packaging plasmids (LentiArt pHelp1, LentiArt pHelp2 and LentiArt pHelp3, Creative Biogene, #CART-027CL). Twenty four-hour and 48-h supernatants were collected and centrifugated at 2000g, 4 °C for 10 min to remove cell debris. The lentivirus were then concentrated by using Lenti-Pac Lentivirus Concentration Solution (GeneCopoeia, #LT007) and centrifugation and stored at −80 °C.
Generation and early assessment of EphA10-CAR-Jurkat cells and EphA10-CAR-T cells
Human T cell lymphoma Jurkat cell line was purchased from Bioresource Collection and Research Center, Taiwan. For Jurkat cell transduction and expansion, 1 × 106 of Jurkat cells were suspended in 90 μl of RPMI-1640 medium with 10% FBS, and 10 μl of 20 × concentrated EphA10-CAR lentivirus were added into wells as a MOI of 1. After transduction EphA10-CAR lentivirus into Jurkat cells, the EphA10-CAR-Jurkat cells generated from three independent experiments were expanded in RPMI-1640 medium with 10% FBS and subsequently co-cultured with MDA-MB-231 cells for 24 h (Effector cells:Target cells; E:T = 1:1). After incubation, these CAR-Jurkat cells were collected and stained with PE-conjugated anti-human CD69 (R&D Systems, #FAB23591p). The signals of CD69 were detected by a flow cytometry (red, blue, and green colors represented different batches of CAR Jurkat cells). MDA-MB-231 cells (5 × 105 cells/well) were seeded onto a 6-well plate 18 h prior to coculture. Mock CAR-T cells and EphA10-CAR-T cells (5 × 105 cells/well) were inoculated into each well with MDA-MB-231 cells (Effector cells:Target cells; E:T = 1:1). Normal CD8+ T-cells (5 × 105 cells/well) cocultured with MDA-MB-231 cells served as the control. After incubation, these T cells were collected and stained with PE-conjugated anti-human CD69 (R&D Systems, #FAB23591p) and PerCP-Cyanine5.5-conjugated antihuman CD3 (Thermo Fisher Scientific Inc # 45-0036-42). The signals were detected by a flow cytometry and processed by FlowJo software program.
Isolation and verification of human CD8+ T cell and T cell transduction
Human peripheral blood mononuclear cells were collected from 20 ml of whole blood from the elbow vein of healthy donors under a China Medical University and Hospital Research Ethics Committee approved human protocol After collection, the whole blood will be mixed with PBS buffer supplied with 2% FBS in the ratio of 1:1 by volume. Overlay the Ficoll-Paque PLUS solution with the blood/PBS mixture using a serological pipette at the lowest speed setting. Then, centrifuge the sample in the desired swing-out rotor at 400g and 20 °C for 30 min, selecting acceleration/deceleration rates of 9/0. After completing the centrifugation, aspirate the entire lymphocyte layer, while keeping the volume to a minimum, and transfer to a fresh tube. Add at least three volumes of PBS with 2% FBS to the lymphocyte layer and carefully mix by pipetting up and down. Centrifuge at 300g and 20 °C for 8 min and discard the supernatant, then resuspend the PBMCs pellet with CD8+ T cell isolation buffer (phosphate-buffered saline, pH7.2, 0.5% BSA, and 2 mM EDTA) in a concentration of 1 × 107 cells/80 μl. Add 20 μl of CD8 MicroBeads (Miltenyi Biotec, #130-045-201) per 107 total cells and mix gently, then incubate for 15 min in the refrigerator (4 °C). Wash cells by adding 2 ml of buffer per 10⁷ cells and centrifuge at 300g for 10 min. Aspirate supernatant completely. Resuspend up to 108 cells in 500 μl of buffer and follow the manufacturers’ protocol to pass through MS Column (Miltenyi Biotec, #130-042-201) to collect the CD8+ T cells. According our experiences, 6 × 106 of CD8+ T cells can be isolated form 20 ml of whole blood. The in vitro expanded human CD8+ T cells were stained with anti-human CD3 and anti-human CD8 antibodies, and the purity of CD8+ T cell population was examined by a flow cytometry. The CD8+ T cells were cultured in KBM 502 medium and activated with ImmunoCult Human CD3/CD28 T Cell Activator (STEMCELL Technologies Inc, # 10971). For virus transduction, CD8+ T cells were activated for 24 h before adding virus particles. Briefly, activated CD8+ T cells were suspended in 1 ml of KBM 502 medium (KOHJIN BIO, #16025020) in a density of 1 × 106 cells/ml. Twenty microliter or 40 μl of 20 × concentrated EphA10-CAR lentivirus were added into wells as a MOI of 2 or 4. The medium were refreshed next day.
Interaction between recombinant huEphA10 Fc chimera protein and EphA10-CAR-T cells
Briefly, the EphA10-CAR-T cells were incubated with recombinant huEphA10 Fc chimera protein (R&D SYSTEMS) for 45 min on ice. After incubation, the cells were washed by PBS twice, then FITC-conjugated anti-human IgG Fc antibody (Sigma Aldrich, #F9512) was added into cell suspension for 30 min on ice in the dark. The signals of fluorochrome were detected by a flow cytometry.
In vitro cytotoxicity of EphA10-CAR-T cell against MDA-MB-231 cells
5 × 103 of MDA-MB-231 cells were seeded onto 96-well plate per well. Next day, EphA10-CAR-T cells will be added into wells to coculture with MDA-MB-231 cells for 24 h. After incubation, the CAR-T cells were carefully removed, and MTT reagent was added into each well for 4 h. The violet crystals were dissolved in dimethyl sulfoxide. The absorbance was detected with an ELISA plate reader at the wavelength of 595 nm.
Human MDA-MB-231/luc orthotropic xenograft mouse model
In our study, we use nude mice to establish MDA-MB-231/luc xenografts. Briefly, 2.5 × 106 of MDA-MB-231/luc (stably express luciferase) cells were collected and suspended in 100 μl of PBS/matrigel buffer (PBS:matrigel=1:1). The MDA-MB-231/luc cells were injected into the right fourth fat pad of nude mice, and the bioluminescence was checked with IVIS image system every 3 to 4 days. On day 7, the nude mice were divided into two groups based on the signals of the bioluminescence: mock-transduced mice (n = 4) and EphA10-CAR-T mice (n = 5). The CAR-T cell treatment (1 × 107 of mock-transduced T cells or EphA10-CAR-T cells) were performed every 7 days through left femoral vein on day 8, and total three treatments were conducted (day 8, 15, and 22). In addition, 20,000 IU of human IL-2 was intraperitoneally injected into mice twice a week. Before detecting the bioluminescence of tumor cells, 150 μl of D-luciferin solution (15 mg/ml in PBS) was intraperitoneally injected into mice. The signals of bioluminescence were regularly detected by IVIS spectrum system twice weekly. One week after the last treatment, the animals were sacrificed (day 26). Before sacrifice, the body weights of mice were measured. In the end of experiment, the organs will be resected and analyzed by hematoxylin and eosin stain. The infiltrated T cells were stained with anti-human CD8 antibody and FITC-conjugated secondary antibody while the cell nucleus were stained with DAPI. The signals were detected by a fluorescence microscope. Digital photographs were taken at a magnification of 400×. The animal study protocol was approved by IACUC of China Medical University (CMUIACUC-2021-315).
Statistical analyses
Data are shown as mean ± standard deviation (SD). Statistical analyses were performed with use of the Prism software program (GraphPad Software). Differences between groups were compared by using the unpaired t test, Mann–Whitney test, or one-way ANOVA analysis. The Mann–Whitney test was performed to analyze IHC data. A p value of <0.05 was considered statistically significant for all tests. NS, not significant; ∗0.01 < p < 0.05; ∗∗0.001< p < 0.01; ∗∗∗p < 0.001.
Data availability
All data are contained within the manuscript and supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
J.-H. C., L.-C. C., and M.-C. H. are listed as inventors on a patent application (International Patent Application No. PCT/US2020/070,552 based on U.S. Provisional Patent Application No. 62/903,194 entitled “ANTI-EPHA10 ANTIBODIES AND METHODS OF USE THEREOF”) submitted by The University of Texas MD Anderson Cancer Center. All other authors declare no nonfinancial or financial competing interests.
Acknowledgments
We thank Dr Todd Link at MD Anderson Cancer Center for his scientific input in the Octet mAb binding assay and Monoclonal Antibody Facility at MD Anderson for supporting in generating monoclonal antibodies. We acknowledge the editing service of Tamara K. Locke, Research Medical Library at The University of Texas MD Anderson Cancer Center.
Author contributions
J.-H. C., L.-C. C., Y.-N. W., Y.-Y. C., W.-C. S., S.-P. L., L.-B. J., M.-C. H. conceptualization; J.-H. C., L.-C. C., H.-H. L. methodology; J.-H. C., L.-C. C., Y.-N. W., Y.-Y. C., C.-H. W validation; J.-H. C., L.-C. C., Y.-N. W., Y.-Y. C., C.-H. W., H.-H. L., W. X., J. Y., C.-W. C., F.-R. C., J. L., S.-O. L., W.-H. Y. investigation; J.-H. C., L.-C. C., Y.-N. W., Y.-Y. C., C.-H. W., S.-P. L. writing-original draft; J.-H. C., J. L. H., M.-C. H. writing—review & editing; M.-C. H. project administration; L.-B. J., M.-C. H. funding acquisition; W.-C. S., G. N. H., C.L., L. Y., D. Y., M.-C. H. supervision.
Funding and additional information
This study was supported in part by the following: an MDA Startup Fund; NIH/NCI Cancer Center Support Grant under award number P30CA016672; The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund (to M.-C. H.); Breast Cancer Research Foundation (Grant No. BCRF-21-070 to Y.-N. W., G. N. H., M.-C. H.); Drug Development Center, China Medical University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (to M.-C. H.); Ministry of Science and Technology, Taiwan, R. O. C. (Grant Nos. MOST 110-2639-B-039-001-ASP, MOST 110-2314-B-039-043-MY3, and MOST 109-2314-B-039-025-MY2); China Medical University Hospital (Grant No. C1090615012); The National Research Foundation of Korea grant funded by the Korea government Ministry of Science and ICT (MSIT) (Grant Nos.2020R1C1C1005631 and 2021R1A4A1031856 to J.-H.C.); National Breast Cancer Foundation, Inc; and T32 Training Grant in Cancer Biology (5T32CA186892 to H.-H. L. and L.-C. C.); YingTsai Young Scholar Award (CMU108-YTY-04 to W.-H. Y); Ministry of Science and Technology (MOST 110-2320-B-039-065 to W.-H. Y.); an Innovative Research Grant from the National Health Research Institutes, Taiwan (NHRI-EX110-11010BI to W.-H. Y.).
Edited by Eric Fearon
Contributor Information
Long-Bin Jeng, Email: longbin.cmuh@gmail.com.
Mien-Chie Hung, Email: mhung@cmu.edu.tw, mhung77030@gmail.com.
Supporting information
References
- 1.Harbeck N., Penault-Llorca F., Cortes J., Gnant M., Houssami N., Poortmans P., Ruddy K., Tsang J., Cardoso F. Breast cancer. Nat. Rev. Dis. primers. 2019;5:66. doi: 10.1038/s41572-019-0111-2. [DOI] [PubMed] [Google Scholar]
- 2.Carey L.A. Finding the positive in triple-negative breast cancer. Nat. Cancer. 2021;2:476–478. doi: 10.1038/s43018-021-00211-x. [DOI] [PubMed] [Google Scholar]
- 3.Savas P., Salgado R., Denkert C., Sotiriou C., Darcy P.K., Smyth M.J., Loi S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016;13:228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 4.Gruosso T., Gigoux M., Manem V.S.K., Bertos N., Zuo D., Perlitch I., Saleh S.M.I., Zhao H., Souleimanova M., Johnson R.M., Monette A., Ramos V.M., Hallett M.T., Stagg J., Lapointe R., et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Invest. 2019;129:1785–1800. doi: 10.1172/JCI96313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emens L.A. Immunotherapy in triple-negative breast cancer. Cancer J. 2021;27:59–66. doi: 10.1097/PPO.0000000000000497. [DOI] [PubMed] [Google Scholar]
- 6.Li M., Li A., Zhou S., Xu Y., Xiao Y., Bi R., Yang W. Heterogeneity of PD-L1 expression in primary tumors and paired lymph node metastases of triple negative breast cancer. BMC Cancer. 2018;18:4. doi: 10.1186/s12885-017-3916-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y.N., Lee H.H., Hsu J.L., Yu D., Hung M.C. The impact of PD-L1 N-linked glycosylation on cancer therapy and clinical diagnosis. J. Biomed. Sci. 2020;27:77. doi: 10.1186/s12929-020-00670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H.H., Wang Y.N., Xia W., Chen C.H., Rau K.M., Ye L., Wei Y., Chou C.K., Wang S.C., Yan M., Tu C.Y., Hsia T.C., Chiang S.F., Chao K.S.C., Wistuba I.I., et al. Removal of N-linked glycosylation enhances PD-L1 detection and predicts anti-PD-1/PD-L1 therapeutic efficacy. Cancer Cell. 2019;36:168–178.e164. doi: 10.1016/j.ccell.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mei J., Xu J., Yang X., Gu D., Zhou W., Wang H., Liu C. A comparability study of natural and deglycosylated PD-L1 levels in lung cancer: Evidence from immunohistochemical analysis. Mol. Cancer. 2021;20:11. doi: 10.1186/s12943-020-01304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kullander K., Klein R. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 2002;3:475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 11.Lisabeth E.M., Falivelli G., Pasquale E.B. Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dodelet V.C., Pasquale E.B. Eph receptors and ephrin ligands: Embryogenesis to tumorigenesis. Oncogene. 2000;19:5614–5619. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- 13.Conover J.C., Doetsch F., Garcia-Verdugo J.M., Gale N.W., Yancopoulos G.D., Alvarez-Buylla A. Disruption of Eph/ephrin signaling affects migration and proliferation in the adult subventricular zone. Nat. Neurosci. 2000;3:1091–1097. doi: 10.1038/80606. [DOI] [PubMed] [Google Scholar]
- 14.Pasquale E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandouz M. The Eph/Ephrin family in cancer metastasis: Communication at the service of invasion. Cancer Metastasis Rev. 2012;31:353–373. doi: 10.1007/s10555-012-9352-1. [DOI] [PubMed] [Google Scholar]
- 16.Aasheim H.C., Patzke S., Hjorthaug H.S., Finne E.F. Characterization of a novel Eph receptor tyrosine kinase, EphA10, expressed in testis. Biochim. Biophys. Acta. 2005;1723:1–7. doi: 10.1016/j.bbagen.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Nagano K., Maeda Y., Kanasaki S., Watanabe T., Yamashita T., Inoue M., Higashisaka K., Yoshioka Y., Abe Y., Mukai Y., Kamada H., Tsutsumi Y., Tsunoda S. Ephrin receptor A10 is a promising drug target potentially useful for breast cancers including triple negative breast cancers. J. Control Release. 2014;189:72–79. doi: 10.1016/j.jconrel.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Li Y., Jin L., Ye F., Ma Q., Yang Z., Liu D., Yang J., Ma D., Gao Q. Isoform expression patterns of EPHA10 protein mediate breast cancer progression by regulating the E-Cadherin and beta-catenin complex. Oncotarget. 2017;8:30344–30356. doi: 10.18632/oncotarget.15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin W.S., Park M.K., Lee Y.H., Kim K.W., Lee H., Lee S.T. The catalytically defective receptor protein tyrosine kinase EphA10 promotes tumorigenesis in pancreatic cancer cells. Cancer Sci. 2020;111:3292–3302. doi: 10.1111/cas.14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagano K., Yamashita T., Inoue M., Higashisaka K., Yoshioka Y., Abe Y., Mukai Y., Kamada H., Tsutsumi Y., Tsunoda S. Eph receptor A10 has a potential as a target for a prostate cancer therapy. Biochem. Biophys. Res. Commun. 2014;450:545–549. doi: 10.1016/j.bbrc.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Yan H.X., Li Q.L., Cai W.W., Tang D., Wang B., Liu X., Liu Z.D., Yang Z.L. EphA10 and EphB3 are prognostic markers in gallbladder squamous cell/adenosquamous carcinomas and adenocarcinomas. Int. J. Clin. Exp. Pathol. 2018;11:512–525. [PMC free article] [PubMed] [Google Scholar]
- 22.Nagano K., Kanasaki S., Yamashita T., Maeda Y., Inoue M., Higashisaka K., Yoshioka Y., Abe Y., Mukai Y., Kamada H., Tsutsumi Y., Tsunoda S. Expression of Eph receptor A10 is correlated with lymph node metastasis and stage progression in breast cancer patients. Cancer Med. 2013;2:972–977. doi: 10.1002/cam4.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang W.H., Cha J.H., Xia W., Lee H.H., Chan L.C., Wang Y.N., Hsu J.L., Ren G., Hung M.C. Juxtacrine signaling inhibits antitumor immunity by upregulating PD-L1 expression. Cancer Res. 2018;78:3761–3768. doi: 10.1158/0008-5472.CAN-18-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klymenko Y., Nephew K.P. Epigenetic crosstalk between the tumor microenvironment and ovarian cancer cells: A therapeutic road less traveled. Cancers (Basel) 2018;10:295. doi: 10.3390/cancers10090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn J., Rao S. Epigenetics and immunotherapy: The current state of play. Mol. Immunol. 2017;87:227–239. doi: 10.1016/j.molimm.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura K., Smyth M.J. Myeloid immunosuppression and immune checkpoints in the tumor microenvironment. Cell Mol. Immunol. 2020;17:1–12. doi: 10.1038/s41423-019-0306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anfray C., Ummarino A., Andon F.T., Allavena P. Current strategies to target tumor-associated-macrophages to improve anti-tumor immune responses. Cells. 2019;9:46. doi: 10.3390/cells9010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostrand-Rosenberg S., Fenselau C. Myeloid-derived suppressor cells: Immune-suppressive cells that impair antitumor immunity and are Sculpted by their environment. J. Immunol. 2018;200:422–431. doi: 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 30.Lu R.M., Hwang Y.C., Liu I.J., Lee C.C., Tsai H.Z., Li H.J., Wu H.C. Development of therapeutic antibodies for the treatment of diseases. J. Biomed. Sci. 2020;27:1. doi: 10.1186/s12929-019-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones M.L., Alfaleh M.A., Kumble S., Zhang S., Osborne G.W., Yeh M., Arora N., Hou J.J., Howard C.B., Chin D.Y., Mahler S.M. Targeting membrane proteins for antibody discovery using phage display. Sci. Rep. 2016;6:26240. doi: 10.1038/srep26240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang L.Y., Patel O., Janes P.W., Murphy J.M., Lucet I.S. Eph receptor signalling: From catalytic to non-catalytic functions. Oncogene. 2019;38:6567–6584. doi: 10.1038/s41388-019-0931-2. [DOI] [PubMed] [Google Scholar]
- 33.Ghetie V., Ward E.S., Vitetta E.S. Handbook of Anticancer Pharmacokinetics and Pharmacodynamics. Springer New York; New York, NY: 2014. Pharmacokinetics of antibodies and immunotoxins in mice and humans; pp. 475–498. [Google Scholar]
- 34.Gillespie T.W. Understanding waterfall plots. J. Adv. Pract. Oncol. 2012;3:106–111. [PMC free article] [PubMed] [Google Scholar]
- 35.Demaria O., Cornen S., Daeron M., Morel Y., Medzhitov R., Vivier E. Harnessing innate immunity in cancer therapy. Nature. 2019;574:45–56. doi: 10.1038/s41586-019-1593-5. [DOI] [PubMed] [Google Scholar]
- 36.Durgeau A., Virk Y., Corgnac S., Mami-Chouaib F. Recent advances in targeting CD8 T-cell immunity for more effective cancer immunotherapy. Front. Immunol. 2018;9:14. doi: 10.3389/fimmu.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S., Yi M., Qin S., Wu K. Next generation chimeric antigen receptor T cells: Safety strategies to overcome toxicity. Mol. Cancer. 2019;18:125. doi: 10.1186/s12943-019-1057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khalil D.N., Smith E.L., Brentjens R.J., Wolchok J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016;13:273–290. doi: 10.1038/nrclinonc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim D.W., Cho J.Y. Recent advances in allogeneic CAR-T cells. Biomolecules. 2020;10:263. doi: 10.3390/biom10020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bloemberg D., Nguyen T., MacLean S., Zafer A., Gadoury C., Gurnani K., Chattopadhyay A., Ash J., Lippens J., Harcus D., Page M., Fortin A., Pon R.A., Gilbert R., Marcil A., et al. A high-throughput method for characterizing novel chimeric antigen receptors in Jurkat cells. Mol. Ther. Methods Clin. Dev. 2020;16:238–254. doi: 10.1016/j.omtm.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tatari N., Maich W.T., Salim S.K., McKenna D., Venugopal C., Singh S. Preclinical testing of CAR T cells in a patient-derived xenograft model of glioblastoma. STAR Protoc. 2020;1:100174. doi: 10.1016/j.xpro.2020.100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Chen M., Wu Z., Tong C., Dai H., Guo Y., Liu Y., Huang J., Lv H., Luo C., Feng K.C., Yang Q.M., Li X.L., Han W. CD133-directed CAR T cells for advanced metastasis malignancies: A phase I trial. Oncoimmunology. 2018;7 doi: 10.1080/2162402X.2018.1440169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xia L., Zheng Z., Liu J.Y., Chen Y.J., Ding J., Hu G.S., Hu Y.H., Liu S., Luo W.X., Xia N.S., Liu W. Targeting triple-negative breast cancer with combination therapy of EGFR CAR T cells and CDK7 inhibition. Cancer Immunol. Res. 2021;9:707–722. doi: 10.1158/2326-6066.CIR-20-0405. [DOI] [PubMed] [Google Scholar]
- 44.Khongorzul P., Ling C.J., Khan F.U., Ihsan A.U., Zhang J. Antibody-drug conjugates: A comprehensive Review. Mol. Cancer Res. 2020;18:3–19. doi: 10.1158/1541-7786.MCR-19-0582. [DOI] [PubMed] [Google Scholar]
- 45.Li C.W., Lim S.O., Chung E.M., Kim Y.S., Park A.H., Yao J., Cha J.H., Xia W., Chan L.C., Kim T., Chang S.S., Lee H.H., Chou C.K., Liu Y.L., Yeh H.C., et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33:187–201.e110. doi: 10.1016/j.ccell.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donaghy H. Effects of antibody, drug and linker on the preclinical and clinical toxicities of antibody-drug conjugates. MAbs. 2016;8:659–671. doi: 10.1080/19420862.2016.1156829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayor S., Parton R.G., Donaldson J.G. Clathrin-independent pathways of endocytosis. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK server: Interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krawczyk K., Liu X., Baker T., Shi J., Deane C.M. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics. 2014;30:2288–2294. doi: 10.1093/bioinformatics/btu190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leem J., Dunbar J., Georges G., Shi J., Deane C.M. ABodyBuilder: Automated antibody structure prediction with data-driven accuracy estimation. MAbs. 2016;8:1259–1268. doi: 10.1080/19420862.2016.1205773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gasser M., Waaga-Gasser A.M. Therapeutic antibodies in cancer therapy. Adv. Exp. Med. Biol. 2016;917:95–120. doi: 10.1007/978-3-319-32805-8_6. [DOI] [PubMed] [Google Scholar]
- 53.Mendrola J.M., Shi F., Park J.H., Lemmon M.A. Receptor tyrosine kinases with intracellular pseudokinase domains. Biochem. Soc. Trans. 2013;41:1029–1036. doi: 10.1042/BST20130104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hsu J.L., Hung M.C. The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev. 2016;35:575–588. doi: 10.1007/s10555-016-9649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortes J., Saura C., Bellet M., Munoz-Couselo E., Ramirez-Merino N., Calvo V., Perez J., Vidal M. HER2 and hormone receptor-positive breast cancer--blocking the right target. Nat. Rev. Clin. Oncol. 2011;8:307–311. doi: 10.1038/nrclinonc.2010.185. [DOI] [PubMed] [Google Scholar]
- 56.Holen H.L., Nustad K., Aasheim H.C. Activation of EphA receptors on CD4+CD45RO+ memory cells stimulates migration. J. Leukoc. Biol. 2010;87:1059–1068. doi: 10.1189/jlb.0709497. [DOI] [PubMed] [Google Scholar]
- 57.Hjorthaug H.S., Aasheim H.C. Ephrin-A1 stimulates migration of CD8+CCR7+ T lymphocytes. Eur. J. Immunol. 2007;37:2326–2336. doi: 10.1002/eji.200737111. [DOI] [PubMed] [Google Scholar]
- 58.Aasheim H.C., Delabie J., Finne E.F. Ephrin-A1 binding to CD4+ T lymphocytes stimulates migration and induces tyrosine phosphorylation of PYK2. Blood. 2005;105:2869–2876. doi: 10.1182/blood-2004-08-2981. [DOI] [PubMed] [Google Scholar]
- 59.Darling T.K., Lamb T.J. Emerging roles for Eph receptors and Ephrin ligands in immunity. Front. Immunol. 2019;10:1473. doi: 10.3389/fimmu.2019.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript and supporting information.