Abstract
Zyxin is a zinc-binding phosphoprotein known to regulate cell migration, adhesion, and cell survival. Zyxin also plays a role in signal transduction between focal adhesions and the nuclear compartment. However, the mechanism of Zyxin shuttling to nucleus is still unclear. Here, we identify that the GlcNAc transferase (O-linked GlcNAc [O-GlcNAc] transferase) can O-GlcNAcylate Zyxin and regulate its nuclear localization. We show that O-GlcNAc transferase O-GlcNAcylates Zyxin at two residues, serine 169 (Ser-169) and Ser-246. In addition, O-GlcNAcylation of Ser-169, but not Ser-246, enhances its interaction with 14-3-3γ, which is a phosphoserine/threonine-binding protein and is reported to bind with phosphorylated Zyxin. Furthermore, we found that 14-3-3γ could promote the nuclear localization of Zyxin after Ser-169 O-GlcNAcylation by affecting the function of the N-terminal nuclear export signal sequence; functionally, UV treatment increases the O-GlcNAcylation of Zyxin, which may enhance the nuclear location of Zyxin. Finally, Zyxin in the nucleus maintains homeodomain-interacting protein kinase 2 stability and promotes UV-induced cell death. In conclusion, we uncover that the nuclear localization of Zyxin can be regulated by its O-GlcNAcylation, and that this protein may regulate UV-induced cell death.
Keywords: OGT, O-GlcNAcylation, Zyxin, 14-3-3γ, nuclear localization signal, UV
Abbreviations: GST, glutathione-S-transferase; HEK293T, human embryonic kidney 293T cell line; HIPK2, homeodomain-interacting protein kinase 2; IF, immunofluorescence; LIM, LIN-11, Isl1, and MEC-3; NES, nuclear export signal; OGA, O-GlcNAcase; O-GlcNAc, O-linked GlcNAc; OGT, O-GlcNAc transferase; qPCR, quantitative PCR; Ser, serine; SFB, S protein-Flag-Streptavidin binding peptide; SIAH1, seven in absentia homolog 1; SIAH2, seven in absentia homolog 2; TG, thiamet-G; VASP, vasodilator-stimulated phosphoprotein; YAP, Yes-associated protein
Zyxin is a member of the LIN-11, Isl1, and MEC-3 (LIM) domain protein family, primarily localizing in focal adhesion plaques (1). LIM domains have been shown to participate in specific protein–protein interactions or protein–DNA interactions. Zyxin plays important roles in the actin cytoskeleton. Zyxin has an α-actinin-binding domain at the N terminus, which specifically interacts with α-actinin (2, 3, 4, 5). In addition, the N terminus of Zyxin can also bind to vasodilator-stimulated phosphoprotein (VASP), and the interaction between Zyxin and VASP mainly functions in the assembly of stress fibers (6, 7, 8). Notably, Zyxin is closely related to the genesis and development of tumors. For instance, recent studies have shown that Zyxin is a positive regulator of Hippo–Yes-associated protein (YAP) signaling pathway. Zyxin acts as a scaffold protein to form ternary complex with seven in absentia homolog 2 (SIAH2) and large tumor suppressor kinase 2 (LATS2) regulating the activity of YAP and the growth of cells (9). It has also been shown that Zyxin is phosphorylated by cyclin-dependent kinase 1 (CDK1) during mitosis, which is necessary for cell growth, suggesting that Zyxin promotes colon cancer tumorigenesis through cyclin-dependent kinase 8 (CDK8)–mediated YAP activation in a mitotic phosphorylation–dependent manner (10).
Zyxin plays important roles in the nuclear–cytoplasmic communication. Zyxin has two nuclear export signal (NES) sequences near the N terminus and C terminus, respectively, which indicates that Zyxin shuttles between the nucleus and the cytoplasm. Addition of nuclear export inhibitor, leptomycin B, leads to nuclear localization of Zyxin, indicating that Zyxin functions both on focal adhesions and nucleus (11, 12). However, as Zyxin lacks canonical nuclear localization signals, the mechanism of Zyxin translocation to nucleus is still unclear (12, 13, 14). Zyxin may regulate gene expression in nucleus through interacting with transcription machinery (15). Interestingly, the nucleoplasm shuttle of Zyxin may also be related to apoptosis. It has been reported that Zyxin serves as an important regulator in homeodomain-interacting protein kinase 2 (HIPK2)–p53 signaling axis in DNA damage response (16, 17). Upon UV irradiation, Zyxin modulates the activity of p53 by regulating the stability of HIPK2, which used to be bound and degraded by the E3 ubiquitin ligase seven in absentia homolog 1 (SIAH1). Zyxin is identified to stabilize HIPK2, especially with nuclear localization. Zyxin in the nucleus interacts with the SIAH1 and regulates HIPK2 stability through interference with the functional active dimerization of SIAH1, which stabilizes HIPK2 and activates p53 to modulate UV-induced cell death (18).
Protein glycosylation with O-linked GlcNAc (O-GlcNAc) is a post-translational modification that is widely present in various intracellular components (19, 20). O-GlcNAcylation is controlled by a pair of enzymes: O-GlcNAc transferase (OGT) and O-GlcNAc glycosidase (O-GlcNAcase [OGA]) (21). OGT transfers UDP-GlcNAc to the serine (Ser) or threonine (Thr) residues of the target protein, whereas OGA catalyzes the removal of UDP-GlcNAc from protein residues. O-GlcNAcylation appears in various organelles, such as the endoplasmic reticulum, Golgi apparatus, nucleus, and mitochondria, and plays a pivotal role in multiple processes, such as mRNA transcription, cell cycle, neurodevelopment, stress response, and metabolic homeostasis (22, 23, 24, 25, 26). Moreover, it has been reported that O-GlcNAcylation is abundant in cytoskeleton proteins or its regulatory proteins and is closely related to breast cancer metastasis, indicating that O-GlcNAcylation may make a difference in coordinating cell migration (22, 27, 28, 29).
In this study, we showed that Zyxin is O-GlcNAcylated by OGT at Ser-169 and Ser-246. O-GlcNAcylation of Zyxin at Ser-169 could promote the nuclear localization of Zyxin in 14-3-3γ-dependent manner. Furthermore, O-GlcNAcylation of Zyxin may play a critical role in UV-induced cell death through regulating the HIPK2–p53 signaling axis. Our findings provide novel insights into the regulatory mechanisms of Zyxin nuclear shuttling and reveal the possible implications of aberrant Zyxin O-GlcNAcylation in cancer malignancies.
Results
Zyxin is O-GlcNAcylated by OGT
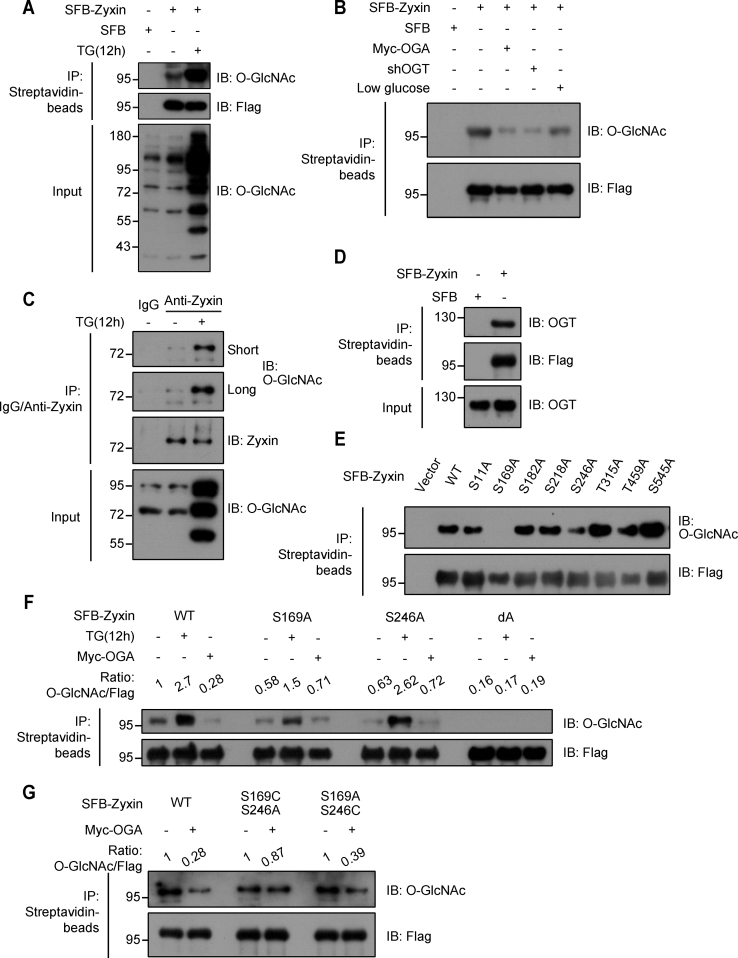
We found that Zyxin could be O-GlcNAcylated by OGT. The S protein-Flag-Streptavidin binding peptide (SFB)-Zyxin was transfected into human embryonic kidney 293T (HEK293T) cells and immunoprecipitated by streptavidin beads in indicated conditions. Then, pan-O-GlcNAcylation antibody CTD110.6 was used to determine whether Zyxin could be O-GlcNAcylated. The results showed that Zyxin could be apparently O-GlcNAcylated, and the level of Zyxin O-GlcNAcylation increased significantly when the cells were treated by thiamet-G (TG, 10 μM), the inhibitor of OGA, or overexpression of Myc-OGT (Figs. 1A and S1A). In contrast, when Myc-OGA was overexpressed to reduce the global O-GlcNAcylation level, the O-GlcNAcylation modification of Zyxin decreased dramatically (Fig. 1B). And we got the same results in cells with endogenous OGT depletion or low glucose treatment (Fig. 1B). To examine the O-GlcNAcylation status of endogenous Zyxin, we immunoprecipitated endogenous Zyxin and found that endogenous Zyxin could also be O-GlcNAcylated (Fig. 1C). Furthermore, Zyxin could interact with OGT (Fig. 1D). These data demonstrated that Zyxin could be O-GlcNAcylated by OGT.
Figure 1.
OGT O-GlcNAcylated Zyxin at Ser-169 and Ser-246.A, empty vector or SFB-Zyxin was transfected into HEK293T cells, and cells were treated with DMSO or TG for 12 h. SFB-Zyxin was immunoprecipitated (IP) using the streptavidin beads and then immunoblotted using the indicated antibodies. B, cells overexpressing SFB-Zyxin were transfected with Myc-OGA or cultured with low glucose medium to decrease endogenous O-GlcNAcylation. The O-GlcNAcylation of Zyxin were detected as in (A). C, endogenous Zyxin was immunoprecipitated using anti-Zyxin antibody, and the O-GlcNAcylation of Zyxin was determined. D, Zyxin interacted with OGT. HEK293T cells overexpressing empty vector or SFB-Zyxin were immunoprecipitated using streptavidin beads and immunoblotted using indicated antibodies. E, identification of O-GlcNAcylation modification sites. HEK293T cells were transfected with either wildtype SFB-Zyxin or indicated mutants, and the O-GlcNAcylation of Zyxin was determined. SFB-Zyxin was immunoprecipitated using the streptavidin beads and then immunoblotted using the indicated antibodies. F and G, the O-GlcNAcylation of wildtype Zyxin or mutants was determined after indicated treatments. To quantify the O-GlcNAcylation level of Zyxin, the intensities of anti-O-GlcNAc and anti-FLAG bands were compared and shown on the top and converted into rate measurements. DMSO, dimethyl sulfoxide; HEK293T, human embryonic kidney 293T cell line; OGA, O-GlcNAcase; OGT, O-GlcNAc transferase; Ser, serine; SFB, S protein-Flag-Streptavidin binding peptide; TG, thiamet-G.
To identify the O-GlcNAcylated sites on Zyxin, the protein sequence of Zyxin was analyzed using YinOYang 1.2 Server (http://www.cbs.dtu.dk/services/YinOYang/) to find the possible O-GlcNAcylation sites (Fig. S1, B and C). We then constructed the indicated Zyxin mutations based on the predicted potential of O-GlcNAcylation to determine the real O-GlcNAcylation sites on Zyxin (Figs. 1E and S1D). Zyxin mutations were transfected into HEK293T cells, and the O-GlcNAcylation modification was determined as in Figure 1A. The results show that S169A or S246A mutation could obviously affect the O-GlcNAcylation of Zyxin (Figs. 1E and S1D). To further confirm this finding, we constructed the Zyxin mutants in which both Ser-169 and Ser-246 sites were replaced with alanine (Ala), called Zyxin-dA. Wildtype, S169A, S246A, or dA was expressed in HEK293T cells, and the O-GlcNAcylation of each mutant was determined. These cells were treated with TG to inhibit endogenous OGA to enhance O-GlcNAcylation signals or cotransfected with Myc-OGA to reduce O-GlcNAcylation. The results showed that the O-GlcNAcylation of S169A or S246A mutant decreased significantly in contrast to wildtype Zyxin (Fig. 1F). Intriguingly, the O-GlcNAcylation of dA mutant was completely abolished (Fig. 1F), suggesting that Ser-169 and Ser-246 are the major O-GlcNAcylation sites on Zyxin.
To further confirm the presence of O-GlcNAcylation on Zyxin, we used purified glutathione-S-transferase (GST)-OGT and His-Zyxin to perform in vitro O-GlcNAcylation assay to determine the O-GlcNAcylation on Zyxin. The result showed that Zyxin was directly O-GlcNAcylated by GST-OGT, and the O-GlcNAcylation level of wildtype Zyxin was much higher than that of S169A mutant (Fig. S1E), suggesting that Ser-169 of Zyxin could be directly O-GlcNAcylated by OGT. Furthermore, we examined the O-GlcNAcylation of Zyxin and S169A mutant using another O-GlcNAcylation recognition antibody, RL2, which confirmed that Ser-169 was the real O-GlcNAcylation site on Zyxin (Fig. S1F). Thus, we demonstrated that Ser-169 of Zyxin could be O-GlcNAcylated by OGT.
It has recently been reported that OGT could recognize cysteine (Cys) and generate a thio-linked GlcNAc, which is hydrolytically stable and accurate structural mimic of O-GlcNAc (30, 31, 32). To examine the site-specific O-GlcNAcylation of Zyxin, we constructed SFB-Zyxin mutants in which Ser-169 or Ser-246 sites was replaced by Cys. The results showed that O-GlcNAcylation of wildtype Zyxin was reduced when OGA was overexpressed (Fig. 1G). Intriguingly, the thio-linked GlcNAc modification on the S169C–S246A mutant slightly decreased in contrast to S246C–S169A mutant, which showed significant decrease with OGA overexpression, indicating that Ser-169 is the major O-GlcNAcylation site of Zyxin instead of Ser-246.
OGT promotes migration of breast cancer cell in a Zyxin-dependent manner
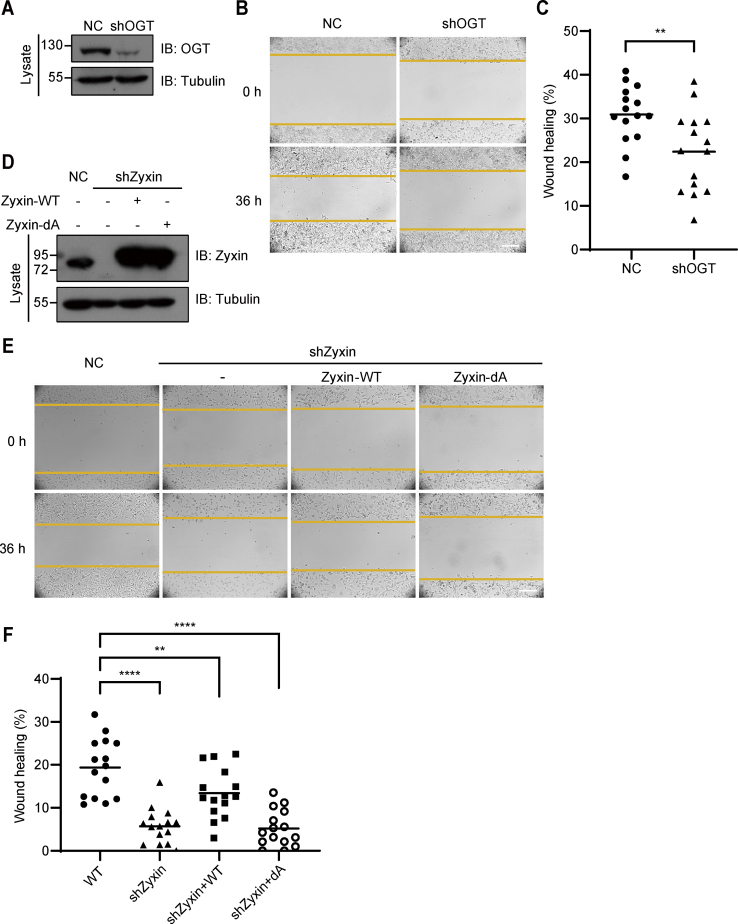
OGT could O-GlcNAcylate actin cytoskeletal regulatory proteins (28), such as cofilin (27), paxillin (33), and others, and regulate cell mobility (27, 28, 29). To confirm this phenomenon, we depleted endogenous OGT using shRNA and assessed the cell mobility by wound healing assay. The cell mobility dramatically declined when OGT was depleted (Fig. 2, A–C). As Zyxin is an actin-binding protein and plays an important role in cell migration, we assumed that the O-GlcNAcylation of Zyxin may also regulate cell migration. To test this hypothesis, we depleted endogenous Zyxin in MDA-MB-231 cells through shRNA and rescued cells with wildtype Zyxin or dA mutant, respectively (Fig. 2D). We found that the mobility of Zyxin-depleted cells was impaired. Interestingly, the cell motility defects in Zyxin-depleted cells could be rescued by wildtype Zyxin but not dA mutant, indicating that the O-GlcNAcylation of Zyxin promotes cell migration (Fig. 2, E and F). These data suggested that O-GlcNAcylation of Zyxin was also involved in the regulation of cell mobility.
Figure 2.
O-GlcNAcylation of Zyxin mediates the migration of breast cancer cells.A, negative control or OGT shRNA was transfected into MDA-MB-231 cells, and OGT protein level was determined. B and C, schematic (B) and quantification (C) of the wound heal assay in measuring the ability of migration in cell lines in (A). D, immunoblot analysis showing the Zyxin protein level in indicated cell lines. E and F, schematic (E) and quantification (F) of the wound heal assay in measuring the ability of migration in cell lines in (D). All error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, NS. Two-tailed unpaired Student’s t test. The scale bars represent 200 μm. NS, not significant; OGT, O-GlcNAc transferase.
The O-GlcNAcylation of Zyxin Ser-169 enhances its interaction with 14-3-3γ
The N terminus of Zyxin contains four proline-rich ActA repeats that mediate the interaction between Zyxin and the actin regulator VASP (6, 7). We wondered whether O-GlcNAcylation of Zyxin, which also locates at the N terminus, would affect the interaction between Zyxin and VASP. We examined the interactions between Zyxin and VASP at different O-GlcNAcylation levels and found that there was no significant change (Fig. S2), indicating that Zyxin O-GlcNAcylation did not affect ActA repeat–mediated protein interactions. Meanwhile, we found that Zyxin O-GlcNAcylation did not affect its interaction with SIAH2 neither (Fig. S2), which mediates the role of Zyxin in regulating Hippo pathway (9).
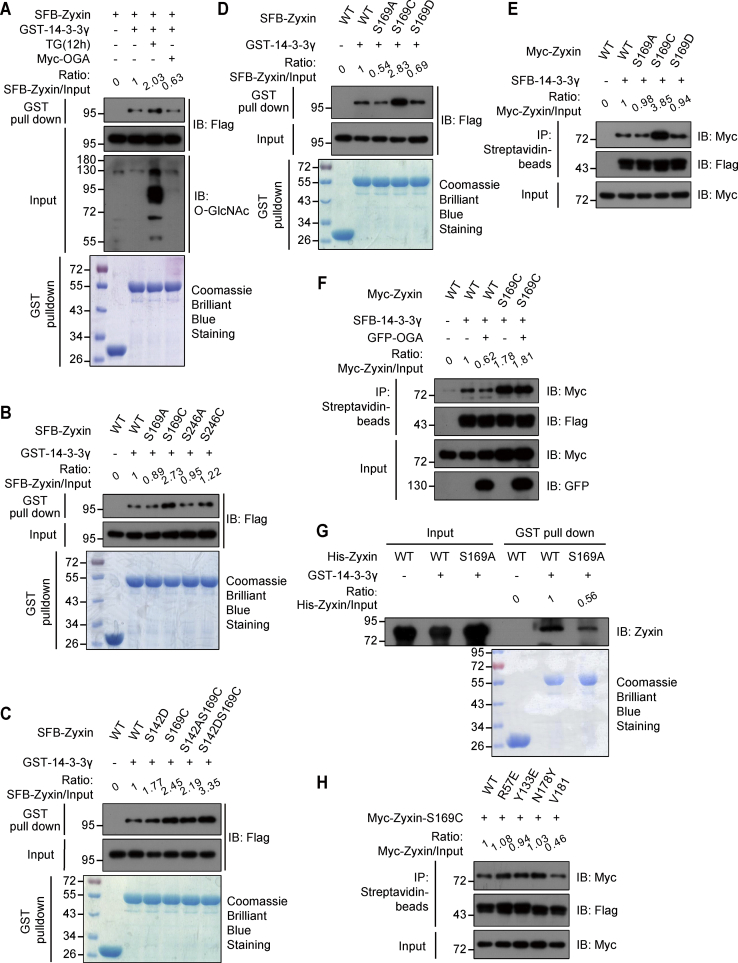
It has been reported that protein O-GlcNAcylation could be recognized by 14-3-3γ (34). As 14-3-3γ is a Zyxin-associated protein, we conjecture whether the O-GlcNAcylation of Zyxin regulates its interaction with 14-3-3γ. To confirm this hypothesis, we purified the GST-tagged 14-3-3γ proteins and found that GST-14-3-3γ could pull down SFB-Zyxin (Fig. 3A). This GST pull-down assay was independently repeated for three times, and the rate measurement of each experiment was shown (Fig. S3A). Interestingly, OGA inhibitor treatment enhanced this interaction, and OGA overexpression decreased this interaction (Figs. 3A and S3A), indicating that the interaction between Zyxin and 14-3-3γ may be regulated by the level of Zyxin O-GlcNAcylation.
Figure 3.
Zyxin interacts with 14-3-3γ through Ser-169 O-GlcNAcylation.A, interaction between Zyxin and 14-3-3γ in different O-GlcNAcylation level. Empty vector or SFB-Zyxin was transfected into HEK293T cells, and cells were treated with TG or Myc-OGA overexpression. Cell lysate was incubated with purified GST-tagged 14-3-3γ proteins. GST pull-down assay was performed using the GST-agarose and immunoblotted using the indicated antibodies. The GST-fused 14-3-3γ proteins were detected by Coomassie blue staining. B–D, interaction between Zyxin and 14-3-3γ was analyzed in different conditions. Indicated SFB-Zyxin mutant was transfected into HEK293T cells. GST pull-down assay was performed using the GST-agarose and immunoblotted using the indicated antibodies. GST-fused 14-3-3γ protein was detected by Coomassie blue staining. E and F, interaction between Zyxin and 14-3-3γ was analyzed in HEK293T cells. SFB-14-3-3γ and Myc-Zyxin mutants were transfected into HEK293T cells. SFB-14-3-3γ was immunoprecipitated using streptavidin beads and analyzed by indicated antibodies. G, OGT-mediated Zyxin O-GlcNAcylation promotes the interaction between His-Zyxin and GST-14-3-3γ. His-OGT was used to O-GlcNAcylate His-Zyxin in vitro. The interaction between the modified His-Zyxin and GST-14-3-3γ was analyzed by GST pull-down assay. H, interaction between Zyxin and 14-3-3γ mutants was analyzed in HEK293T cells. Indicated SFB-14-3-3γ mutants and Myc-Zyxin S169C mutants were transfected into HEK293T cells. SFB-14-3-3γ mutants were immunoprecipitated using streptavidin beads. The results were converted into rate measurements and shown on the top of each figure. GST, glutathione-S-transferase; HEK293T, human embryonic kidney 293T cell line; OGT, O-GlcNAc transferase; Ser, serine; SFB, S protein-Flag-Streptavidin binding peptide; TG, thiamet-G.
To further validate role of Zyxin O-GlcNAcylation on its interaction with 14-3-3γ, we transfected wildtype Zyxin or S169A, S169C, S246A, S246C mutants into HEK293T cells, respectively. GST pull-down assay showed that the interaction between S169C mutant and 14-3-3γ was increased significantly, whereas the interaction between S246C and 14-3-3γ exhibited slight alterations (Figs. 3B and S3B), suggesting that O-GlcNAcylation of Ser-169, but not Ser-246, promoted the association between 14-3-3γ and Zyxin. It has been reported that the phosphorylation of Zyxin Ser-142 was also involved in its interaction with 14-3-3γ (35). We compared the effects of phosphorylation of Ser-142 and O-GlcNAcylation of Ser-169 on the interaction between Zyxin and 14-3-3γ. First, we found that S142D–S169C mutant showed strongest interaction with 14-3-3γ, and S169C mutant also showed stronger interaction with 14-3-3γ than S142D mutant (Figs. 3C and S3C), indicating that the effect of Zyxin O-GlcNAcylation on its interaction with 14-3-3γ may be stronger than Zyxin phosphorylation. Furthermore, we constructed S169C–S142A mutant, which maintains protein O-GlcNAcylation and lacks protein phosphorylation. GST pull-down assay showed that S142A–S169C mutant still have stronger interaction with 14-3-3γ (Figs. 3C and S3C), suggesting that Ser-169 O-GlcNAcylation alone could promote the interaction between Zyxin and 14-3-3γ.
As Ser-169 might also be a phosphorylation site, we examined whether the effect of Ser-169 on the interaction between Zyxin and 14-3-3γ solely depends on Ser-169 O-GlcNAcylation but not Ser-169 phosphorylation. The results showed that S169D mutant, which mimicked the Ser-169 phosphorylation, did not increase the interaction between Zyxin and 14-3-3γ, which was the same as S169A, implying that Ser-169 O-GlcNAcylation could be specifically recognized by 14-3-3γ (Figs. 3D and S3D). We got the same results by coimmunoprecipitation in HEK293T cells overexpressing SFB-14-3-3γ and Zyxin mutants (Figs. 3E and S3E). We also compared the affinity between 14-3-3γ and Zyxin with different Zyxin O-GlcNAcylation level. Decreasing O-GlcNAcylation level by overexpression of OGA reduced the affinity between 14-3-3γ and wildtype Zyxin but not S169C mutant (Figs. 3F and S3F), indicating that Ser-169 O-GlcNAcylation is important for its interaction with 14-3-3γ. To determine whether 14-3-3γ directly bound with Zyxin, we conducted in vitro GST pull-down experiments using GST-14-3-3γ and His-Zyxin protein. We found that GST-14-3-3γ but not GST protein could pull down wildtype His-Zyxin, suggesting that these two proteins did interact directly (Fig. S3H). Furthermore, to explore the role of O-GlcNAcylation on the association between GST-14-3-3γ and Zyxin, we first used His-OGT protein to O-GlcNAcylate wildtype His-Zyxin or His-Zyxin S169A mutant for 4 h. Then, we added GST-14-3-3γ to pull down these His-Zyxin proteins. We found that, in contrast to His-Zyxin S169A, OGT treatment significantly increased the association between GST-14-3-3γ and wildtype His-Zyxin, indicating that the interaction between these two proteins was regulated by O-GlcNAcylation (Fig. 3G). Thus, we demonstrated that O-GlcNAcylation of Zyxin facilitated the direct association between GST-14-3-3γ and His-Zyxin.
Previous study showed that arginine (Arg)-57, tyrosine (Tyr)-133, asparagine (Asn)-178, and valine (Val)-181 are the key binding sites on 14-3-3γ, which is required for phosphorylation (Arg-57 and Tyr-133) or O-GlcNAcylation binding (Asn-178 and Val-181) (34). We constructed these mutants of 14-3-3γ and checked their affinity with Zyxin S169C mutant. The results revealed that V181W, one of O-GlcNAcylated group recognition-defective mutants, obviously decreased its interaction with Zyxin S169C (Figs. 3H and S3G). R57E or Y133E, which affects the recognition of phosphorylation modification by 14-3-3γ, did not affect its interaction with Zyxin. N178Y mutant, which has been shown to be important for its recognition of O-GlcNAcylation modification, did not show obvious change when compared with wildtype 14-3-3γ, suggesting that Val-181 is important for the interaction between 14-3-3γ and Zyxin Ser-169 O-GlcNAcylation. Taken together, these findings suggested Zyxin Ser-169 O-GlcNAcylation was an important regulator of its interaction with 14-3-3γ.
The O-GlcNAcylation of Zyxin promotes its nuclear localization induced by 14-3-3γ
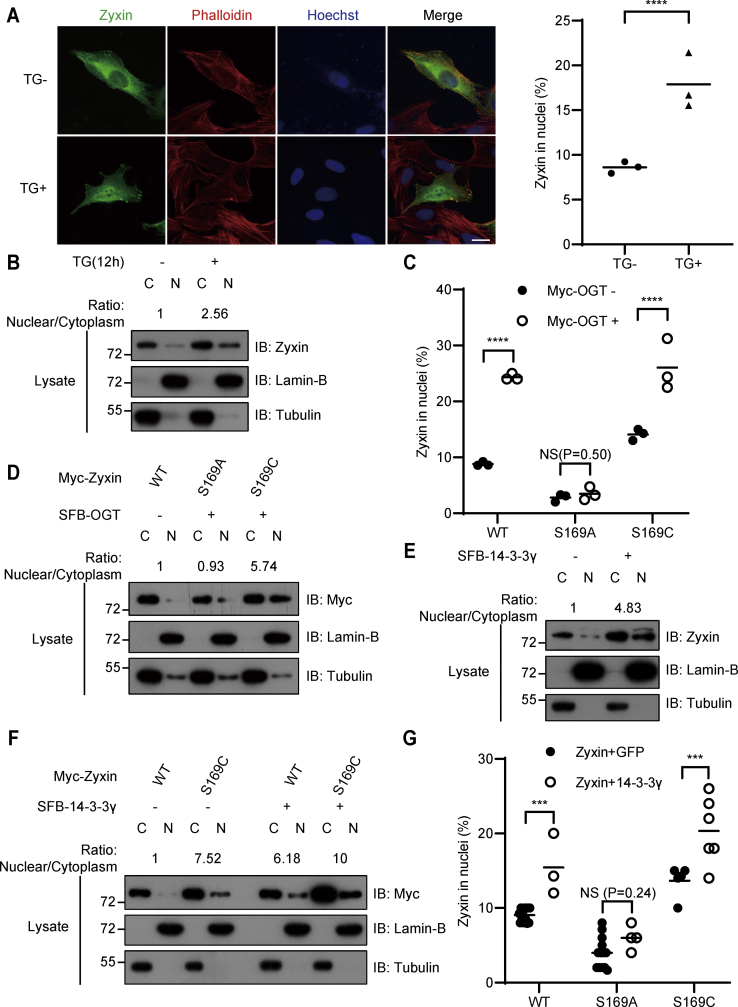
There are two leucine-rich NESs within Zyxin protein, and Zyxin can shuttle between cytoplasm and nucleus. It has been reported that 14-3-3γ triggers Zyxin nuclear localization, which requires Akt-dependent Ser-142 phosphorylation (34). As Zyxin O-GlcNAcylation enhanced the association between 14-3-3γ and Zyxin, we hypothesize that Zyxin O-GlcNAcylation may also promote 14-3-3γ-dependent Zyxin nuclear localization. To verify our speculation, we analyzed the cytoplasmic and nuclear distributions of Zyxin in HCT116 cells treated with TG or dimethyl sulfoxide by immunofluorescence (IF) and Western blot assay. We found that around 18% of cells showed obvious Zyxin nuclear localization with TG treatment in contrast to 9% in dimethyl sulfoxide–treated cells (Fig. 4A). Consistently, nuclear fraction of Zyxin increased remarkably upon TG treatment (Fig. 4B). These results suggested that protein O-GlcNAcylation may promote the nuclear distribution of Zyxin. Next, we compared Zyxin nuclear distribution of wildtype GFP-Zyxin and indicated Zyxin mutants. The nuclear distribution of S169A mutant decreased greatly compared with wildtype Zyxin or S169C mutant (Figs. 4C and S4A). Interestingly, nuclear distribution of Zyxin or S169C increased greatly after OGT overexpression. On the contrary, nuclear distribution of S169A did not change after OGT overexpression (Figs. 4C and S4A), indicating that Zyxin Ser-169 O-GlcNAcylation contributed to the nuclear localization of Zyxin. Consistently, OGT overexpression greatly increased the nuclear localization of S169C but not S169A by analyzing the cytoplasmic and nuclear distribution of Zyxin using Western blot assay (Fig. 4D).
Figure 4.
14-3-3γ promotes Zyxin nuclear localization.A, subcellular localization of Zyxin. GFP-Zyxin was transfected into U2OS cells, the cell nuclei were stained with Hoechst, actin was stained with TRITC–phalloidin, and localization of Zyxin was observed by fluorescence microscopy. The results were converted into rate measurements. B, endogenous Zyxin nuclear localization. HCT116 cells were treated with TG or DMSO for 12 h and separated into cytosolic and nuclear fractions. Immunoblotting was performed using anti-Zyxin antibody. Lamin B together with tubulin acted as a loading control. The results were converted into rate measurements. C, the quantification of Zyxin in nuclei. GFP-Zyxin and Myc-OGT were transfected into U2OS cells. The cell nuclei were stained with Hoechst, and localization of Zyxin was observed by fluorescence microscopy. D–F, Zyxin nuclear localization was analyzed in different conditions. Myc-Zyxin and SFB-14-3-3γ were transfected into HCT116 cells, and cells were separated into cytosolic and nuclear fractions. Immunoblotting was performed using anti-Zyxin and anti-Myc antibody, Lamin B together with tubulin acted as a loading control. The results were converted into rate measurements. G, the quantification of Zyxin in nuclei. mCherry-Zyxin and empty vector or GFP-14-3-3γ were transfected into U2OS cells. The cell nuclei were stained with Hoechst, and localization of Zyxin was observed by fluorescence microscopy. All error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, NS. Two-tailed unpaired Student’s t test. The scale bars represent 20 μm. C, cytoplasm; DMSO, dimethyl sulfoxide; N, nuclear; NS, not significant; OGT, O-GlcNAc transferase; SFB, S protein-Flag-Streptavidin binding peptide; TG, thiamet-G; TRITC, tetramethylrhodamine isothiocyanate.
As Zyxin Ser-169 O-GlcNAcylation promotes its association with 14-3-3γ, we examined whether 14-3-3γ participated in the O-GlcNAcylation-mediated Zyxin nuclear localization. Overexpression of 14-3-3γ increased the nuclear fraction of endogenous Zyxin (Fig. 4E), which is consistent with previous report. Then, we compared the effect of 14-3-3γ overexpression on wildtype Zyxin or S169C and found that the nuclear distribution of S169C was higher than wildtype Zyxin after 14-3-3γ overexpression (Fig. 4F). After determining the cellular distribution of Zyxin by IF, we found that overexpression of 14-3-3γ did enhance the nuclear localization of wildtype Zyxin or S169C but not S169A (Figs. 4G and S4B), indicating that 14-3-3γ was involved in Zyxin Ser-169 O-GlcNAcylation-mediated nuclear localization.
Ser-169 O-GlcNAcylation regulates the Zyxin nuclear localization through affecting the function of N-terminal NES
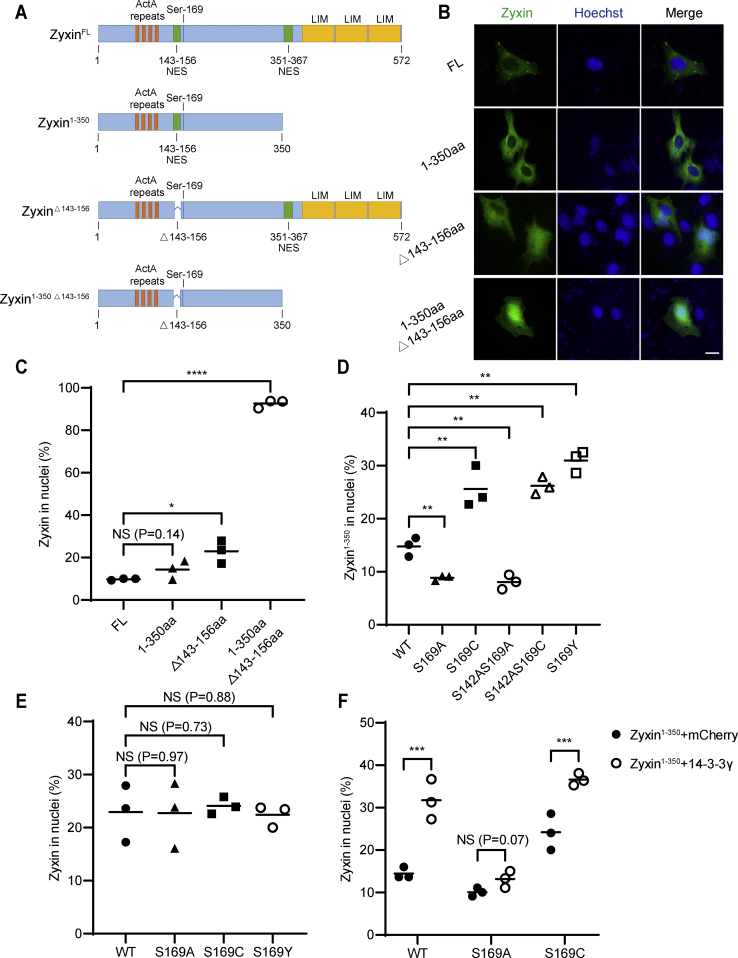
Zyxin has two NES motifs, and the two NES sequences are located at 143 to 156 amino acids (NES1) and 351 to 367 amino acids (NES2), respectively (36, 37). The position of the first NES was predicted by bioinformatics analysis (38), and it is not clear whether NES1 is functional. We constructed deletion variants of Zyxin (Fig. 5A) and compared the subcellular distributions of full-length Zyxin and several deletion variants. The results showed that the nuclear distribution of Zyxin (1–350 amino acids) (15%), which lacks the NES2, and Zyxin (Δ143–156 amino acids) (20%), which lacks NES1, was slightly increased, compared with full-length Zyxin (9%). When two NES sequences were deleted simultaneously, the nuclear localization rate of Zyxin increased significantly (90%) (Fig. 5, B and C), implying that deletion of both NESs totally abolished the nuclear export of Zyxin. These results indicate that both NES sequences of Zyxin are functional.
Figure 5.
Ser-169 O-GlcNAcylation regulates Zyxin nuclear shuttling through affecting the function of NES1.A, scheme of full-length Zyxin and C-terminal deletion variants. Representation of Zyxin domains according to Sabino et al. (36). B and C, GFP-Zyxin was transfected into U2OS cells, the cell nuclei were stained with Hoechst, and localization of Zyxin was observed by fluorescence microscopy (B). Quantification of cells with nuclear Zyxin was shown (C). D, the quantification of Zyxin1–350 in nuclei. GFP-Zyxin1–350 was transfected into U2OS cells. The cell nuclei were stained with Hoechst, and localization of Zyxin was observed by fluorescence microscopy. Quantification of cells with nuclear Zyxin was shown. E, the quantification of ZyxinΔ143–156 in nuclei. GFP-ZyxinΔ143–156 was transfected into U2OS cells. The cell nuclei were stained with Hoechst, and localization of Zyxin was observed by fluorescence microscopy. F, GFP-Zyxin1–350 and empty vector or mCherry-14-3-3γ were transfected into U2OS cells. The cell nuclei were stained with Hoechst, and localization of Zyxin was observed by fluorescence microscopy. Quantification of cells with nuclear Zyxin was shown. All error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, NS. Two-tailed unpaired Student’s t test. The scale bars represent 20 μm. NES1, nuclear export signal 1; NS, not significant; Ser, serine.
Since Ser-169 is close to NES1, we hypothesized that O-GlcNAcylation of Zyxin might regulate its nuclear shuttling by affecting the function of NES1. As Zyxin (1–350 amino acids) only contains NES1, we analyzed the effect of O-GlcNAcylation on Zyxin (1–350 amino acids) nuclear export in the following experiments. The nuclear distribution of Zyxin (1–350 amino acids) was slightly affected by S169A mutant (Figs. 5D and S5A). On the contrary, S169C mutant obviously increased the nuclear distribution of Zyxin (1–350 amino acids) (Figs. 5D and S5A), which is consistent with the effect of Ser-169 O-GlcNAcylation on full-length Zyxin. As Ser-142 phosphorylation has been reported to regulate nuclear distribution of Zyxin through blocking the function of NES1, we analyzed the effect of Ser-169 O-GlcNAcylation on Zyxin (1–350 amino acids) nuclear distribution when Ser-142 was mutated to Ala and got the same result (Figs. 5D and S5A), implying that Ser-142 has nothing to do with Ser-169-mediated regulation of Zyxin nuclear distribution. Meanwhile, S169A mutant decreased the nuclear localization of Zyxin (1–350 amino acids) (Figs. 5D and S5A). In addition, we used the S169Y mutant to mimic Ser-169 O-GlcNAcylation (39) and obtained the same result as S169C mutant (Figs. 5D and S5A). Intriguingly, when we deleted NES1, the effect of S169C or S169Y on nuclear distribution of Zyxin diminished (Figs. 5E and S5B), suggesting that Ser-169 O-GlcNAcylation may regulate Zyxin nuclear distribution by affecting NES1. Furthermore, we also examined the role of 14-3-3γ on the function of NES1 and found that 14-3-3γ promotes nuclear localization of Zyxin (1–350 amino acids) or S169C Zyxin (1–350 amino acids) instead of S169A Zyxin (1–350 amino acids) (Figs. 5F and S5C), indicating that 14-3-3γ may influence the function of NES1 when Ser-169 is O-GlcNAcylated. These results demonstrated that 14-3-3γ is involved in the regulation of Zyxin nuclear localization through affecting the function of NES1, which may be mediated by O-GlcNAcylation of Ser-169.
The O-GlcNAcylation of Zyxin promotes apoptosis through HIPK2–p53 signaling axis pathway
The tumor suppressor p53 is a central player in cellular DNA damage responses. The p53 is a nuclear protein and activated after DNA damage to determine the cell fate, such as cell cycle arrest or apoptosis (40, 41). It has been reported that HIPK2 can phosphorylate p53 at Ser-46 to activate its transcriptional activity after UV irradiation (16, 42), and Zyxin can regulate UV-induced apoptosis by HIPK2–p53 signaling axis (18). After UV treatment, nuclear Zyxin could inhibit the SIAH1 dimer formation, which weakened the degradation of HIPK2 and enhanced the activation of p53 (18). In consistent with these previous reports, we found that UV treatment clearly increased the Ser-46 phosphorylation of p53, and Zyxin depletion significantly reduced this HIPK2-mediated phosphorylation on p53 (Fig. S6A). Meanwhile, we found that there was no obvious interaction between Zyxin and p53 (Fig. S6B).
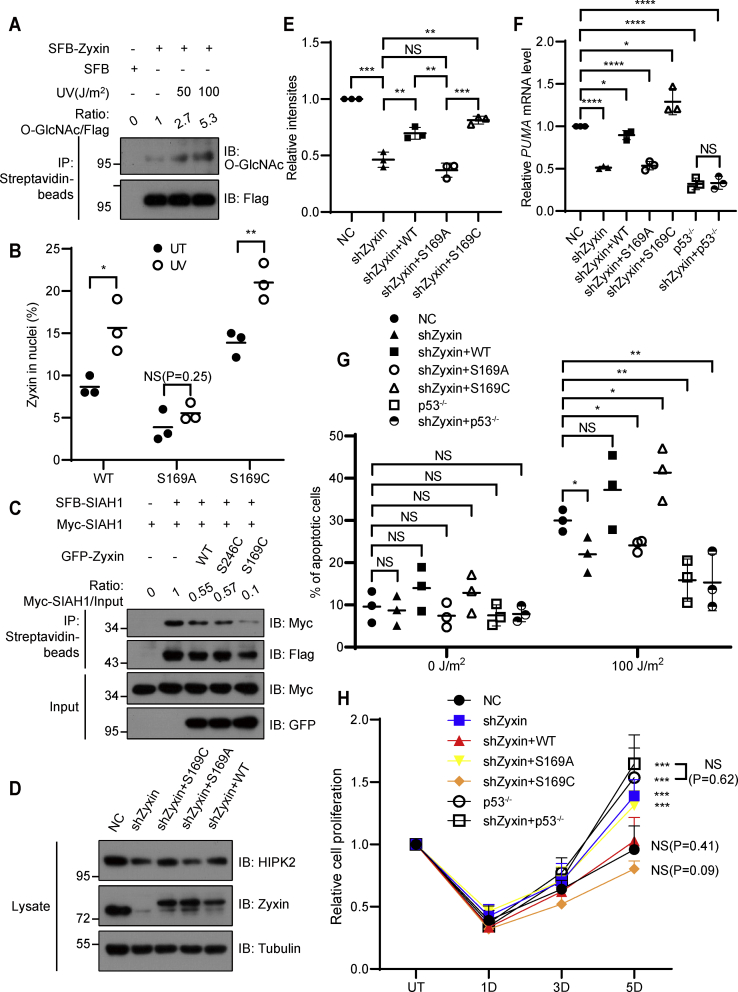
To investigate whether O-GlcNAcylation-mediated Zyxin nuclear localization is involved in the regulation of HIPK2–p53 signaling axis, we first examined Zyxin O-GlcNAcylation in HEK293T cells after UV treatment and found that the O-GlcNAcylation level of Zyxin increased obviously after UV treatment (Fig. 6A). Next, we analyzed the nuclear distribution of Zyxin after UV treatment. Wildtype Zyxin or S169C mutant showed significant nuclear localization of Zyxin in contrast to S169A mutant after UV treatment, indicating that Zyxin O-GlcNAcylation may participate in UV-induced Zyxin nuclear localization (Figs. 6B and S6C). We then examined the dimer formation of SIAH1 under different conditions. Zyxin overexpression decreased the dimer formation after UV treatment, which is consistent with previous report (18). Interestingly, S169C, but not S246C, greatly reduced the dimer formation of SIAH1, demonstrating that Ser-169 O-GlcNAcylation negatively regulated SIAH1 dimer formation (Fig. 6C). As SIAH1 dimer destabilizes HIPK2 after DNA damage, we checked the HIPK2 level after Zyxin depletion. Consistent with previous report, Zyxin depletion decreased HIPK2 protein level (Fig. 6, D and E). Overexpression of wildtype Zyxin or S169C mutant could rescue HIPK2 level, but not S169A (Fig. 6, D and E), implying that the nuclear localization mediated by Ser-169 O-GlcNAcylation may regulate the stability of HIPK2.
Figure 6.
The O-GlcNAcylation of Zyxin impairs cell survival under UV treatment.A, empty vector or SFB-Zyxin was transfected into HEK293T cells, and cells were treated with UV irradiation of corresponding energy, followed by recovering for 2 h. SFB-Zyxin was immunoprecipitated using streptavidin beads and then immunoblotted using the indicated antibodies. B, quantification of Zyxin in the nuclei. U2OS cells transfected with GFP-Zyxin were treated with UV irradiation of corresponding energy and recovered for 2 h. The cell nuclei were stained with Hoechst, and localization of Zyxin was visualized by fluorescence microscopy. Quantification of cells with nuclear Zyxin was shown. C, SFB-SIAH1 and Myc-SIAH1 as well as indicated mutants of Zyxin were transfected into HEK293T cells. SFB-SIAH1 was immunoprecipitated using streptavidin beads and immunoblotted using the indicated antibodies. The results were converted into rate measurements. D and E, immunoblot analysis (D) and quantification (E) of the level of HIPK2 expression in indicated conditions, which were treated with 100 J/m2 UV irradiation and then recovered for 24 h. F, relative mRNA level of PUMA in HCT116 cell lines was determined by RT–qPCR. G, quantification of apoptotic cells in HCT116 cell lines, which were pretreated with 100 J/m2 UV irradiation, then incubated with normal medium for 24 h. Apoptotic cells were analyzed through flow cytometry with propidium iodide and annexin V–APC staining. H, quantification of surviving cells in HCT116 cell lines, which were pretreated with 50 J/m2 UV irradiation and then incubated with normal medium for indicated time. All error bars indicate SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, and NS. Two-tailed unpaired Student’s t test. APC, allophycocyanin; HEK293T, human embryonic kidney 293T cell line; HIPK2, homeodomain-interacting protein kinase 2; NS, not significant; qPCR, quantitative PCR; SFB, S protein-Flag-Streptavidin binding peptide; SIAH1, seven in absentia homolog 1.
To determine whether O-GlcNAcylation-mediated Zyxin nuclear localization influenced the p53 activation after DNA damage, we examined the expression of PUMA, a p53 downstream gene by quantitative PCR (qPCR). Zyxin depletion reduced the expression of PUMA after UV irradiation in HCT116 cells. Wildtype Zyxin and S169C mutant could rescue the expression of PUMA but not S169A mutant in HCT116 cells (Fig. 6F), which was consistent with the role of each construct on HIPK2 stability. To determine these effects was due to p53, we constructed HCT116 p53−/− cell line using CRISPR–Cas9 (Fig. S6, D and E). Zyxin depletion in HCT116 p53−/− cell line did not affect the expression of PUMA (Fig. 6F). We then assayed HCT116 cell lines expressing different Zyxin constructs for the induction of apoptosis after UV treatment using propidium iodide and annexin V–allophycocyanin staining and subsequently flow cytometry. Cells were exposed to 100 J/m2 UV and analyzed 24 h after UV treatment. UV treatment resulted in significant increase of cells undergoing apoptosis, and depletion of Zyxin led to reduced cell apoptosis in HCT116 cells. Intriguingly, overexpression of wildtype or S169C Zyxin in Zyxin-depleted cells could rescue UV-induced apoptosis but not S169A. Meanwhile, Zyxin depletion in p53−/− cell line did not affect UV-induced apoptosis, indicating that the role of Zyxin on UV-induced apoptosis also depended on p53 (Figs. 6G and S6F). In addition, we found that depletion of Zyxin led to enhanced cell viability by analyzing the viability of cells after UV treatment, which could be rescued by wildtype or S169C Zyxin overexpression but not S169A overexpression in HCT116 cells (Fig. 6H). These findings collectively suggested that Zyxin promotes cell apoptosis induced by UV treatment through OGT-dependent Ser-169 O-GlcNAcylation.
Discussion
Zyxin, as a member of the LIM protein family, is involved in a range of biological processes, including cytoskeleton function and gene transcription (14, 43, 44, 45). Proteomic studies have shown that LIM protein families (Zyxin, Paxillin, and four and a half LIM domains protein) are sensitive to mechanical stress in the actin cytoskeleton (46, 47, 48, 49, 50). Previous studies have shown that O-GlcNAcylation modification is widely present in a variety of skeleton proteins and plays an important role in cell migration (22, 23, 28, 51). For example, O-GlcNAcylation of cofilin directly affects the cell migration process (27). In this study, we identified that Zyxin can be O-GlcNAcylated by OGT, which also plays an important role in cell migration. This finding suggests that OGT participates in cell migration through O-GlcNAcylation of several different substrates.
Different from other cytoskeleton proteins, Zyxin can shuttle between nucleus and cytoplasm, and the underlying mechanism is still illusive (12, 13, 14). Akt-mediated Zyxin Ser-142 phosphorylation has been reported to promote its nuclear localization through 14-3-3γ binding (35). 14-3-3γ proteins typically bind phosphorylated Ser and Thr consensus-binding motifs. Recently, it has been reported that protein O-GlcNAcylation could also be specifically recognized by 14-3-3γ (34). In this study, we demonstrate that Ser-169 O-GlcNAcylation enhances the association between Zyxin and 14-3-3γ, which promotes Zyxin nuclear localization. In addition, Ser-169 O-GlcNAcylation rather than Ser-142 phosphorylation modification gave rise to much higher affinity of interaction between Zyxin and 14-3-3γ (Fig. 3C), suggesting that O-GlcNAcylation plays an important role in the regulation of Zyxin nuclear localization. Interestingly, Zyxin Ser-246 O-GlcNAcylation is not involved in its association with 14-3-3γ and nuclear localization, suggesting that specific O-GlcNAcylation motifs mediate 14-3-3γ binding, which is the same as phosphorylation-mediated 14-3-3γ binding. The role of O-GlcNAcylation modification at Ser-246 is still unknown, which might participate in the interaction of Zyxin with other proteins.
As Zyxin lacks canonical nuclear localization signal, we speculate that NESs of Zyxin may be affected by O-GlcNAcylation and 14-3-3γ. Zyxin contains two NES motifs. However, whether both NES motifs function independently is not clear. We found that NES1 is functional. One to 350 amino acids of Zyxin, which lacks NES2, localizes in cytoplasm, and depletion of NES1 in 1 to 350 amino acids accumulates in nucleus. Moreover, Ser-169 O-GlcNAcylation affected wildtype Zyxin and 1 to 350 amino acids nuclear localization but not NES1-deleted Zyxin mutant. As the O-GlcNAcylation of Ser-169 is adjacent to NES1, we speculate that O-GlcNAcylation-mediated 14-3-3γ binding can regulate Zyxin nuclear localization through affecting the function of NES1.
Besides its role in cytoskeleton, several studies showed that Zyxin is a positive regulator of Hippo–YAP signaling activity, which plays an important role in tumorigenesis (9, 10). O-GlcNAcylation of Zyxin does not affect its interaction with VASP or SIAH2, suggesting that Zyxin O-GlcNAcylation may not regulate its function in YAP pathway. It has also been reported that nuclear localization of Zyxin regulates apoptosis through the HIPK2–p53 signaling axis by stabilizing HIPK2 (17, 18). We found that Zyxin Ser-169 O-GlcNAcylation promotes its nuclear localization after UV treatment, which stabilized HIPK2 and further affected p53 transcription activity. However, the UV-induced apoptosis in HCT116 p53−/− cell line was not affected by Zyxin depletion, indicating that the role of Zyxin on UV-induced apoptosis depends on p53. Thus, Zyxin O-GlcNAcylation may play an important role in the UV-induced apoptosis through regulating p53 activity. Recently, Wong et al. (52) reported that OGT directly O-GlcNAcylated HIPK2 in Drosophila and human cells, which regulated HIPK2 stability. Thus, there are two pathways through which OGT may regulate HIPK2 stability. It is interesting to elucidate the crosstalk between these two pathways in different physiological conditions. As nuclear Zyxin could regulate gene expression (15, 53, 54, 55, 56), it is also interesting to study whether O-GlcNAcylation-mediated Zyxin nuclear localization affects gene expression in the future.
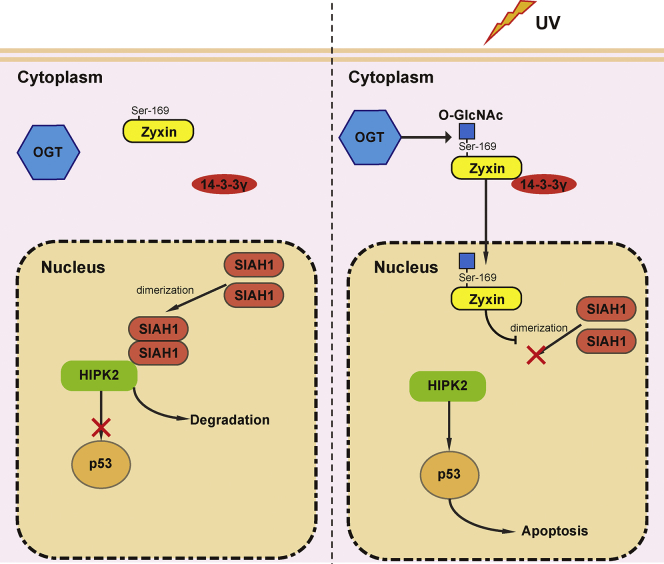
In summary, we propose a new model of the regulation of UV-induced cell death, which is mediated by Ser-169 O-GlcNAcylation modification of Zyxin (Fig. 7). UV irradiation enhances the O-GlcNAcylation of Zyxin, which promotes the interaction between Zyxin and 14-3-3γ. Enhanced interactions lead to nuclear localization of Zyxin and inhibit the dimerization and activation of SIAH1. Inactive SIAH1 fails to mediate the degradation of HIPK2, which promotes apoptosis through the HIPK2–p53 signaling axis.
Figure 7.
Zyxin regulated apoptosis under UV treatment. UV irradiation enhances the O-GlcNAcylation of Zyxin. And O-GlcNAcylation of Zyxin promotes the interaction between Zyxin and 14-3-3γ. The interaction between 14-3-3γ and Zyxin leads to nuclear accumulation of Zyxin, which inhibits the dimerization and activation of SIAH1. Functionally, O-GlcNAcylation-mediated nuclear accumulation of Zyxin enhances the stability of HIPK2 and promotes apoptosis through the HIPK2–p53 signaling axis. HIPK2, homeodomain-interacting protein kinase 2; SIAH1, seven in absentia homolog 1.
Experimental procedures
Cell culture and antibodies
U2OS, HCT116, MDA-MB-231, and HEK293T cells were purchased from the American Type Culture Collection, U2OS cells (HTB-96), HCT116 cells (CCL-247), MDA-MB-231 cells (CRM-HTB-26), and HEK293T cells (CRL-11268). All the cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C and 5% CO2. The following antibodies were used: antibodies against FLAG (Sigma; catalog no.: F3165), Myc (Dia-an; catalog no.: 2097), GFP (Dia-an; catalog no.: 2057), Zyxin (Proteintech; catalog no.: 10330-1-AP), OGT (Proteintech; catalog no.: 11576-2-AP), CTD110.6 (Cell Signaling; catalog no.: 9875S), RL2 (abcam; catalog no.: ab2739), β-tubulin (Proteintech; catalog no.: 11094-1-AP), Phospho-p53 (Ser46) (Cell Signaling; catalog no.: 2521T), and HIPK2 (ABclonal; catalog no.: A9552).
RNA interference and RT–qPCR
For RNA interference, lentiviral packaging plasmids psPAX2 and pMD2.G were cotransfected with the PLKO.1 backbone plasmid into HEK293T cells to produced virus. The following targeting sequences were inserted into the pLKO.1 vector for indicated gene: ZYX, 5’-AGAAGGTGAGCAGTA TTGATT-3’; OGT, 5’-TGCACAATCCTGATAAATTTGA-3’. We used nontarget sequence as negative shRNA control. The sequence is 5’-CAACAAGATGAAGAGCACCAA-3’.
For RT–qPCR, total RNA was extracted using the Trizol RNA extraction reagent (TaKaRa; catalog no.: 9109). Complementary DNA was reverse transcripted using the HIScriptII One Step RT–qPCR Kit (Vazyme; catalog no.: R223-01). Gene expression was analyzed by real-time qPCR using the SYBR Green quantitative PCR Mix (Vazyme; catalog no.: Q311-00). The relative mRNA abundance was calculated by normalization to ACTB mRNA. The following primers were used for RT–qPCR: PUMA (5’-GACCTCAACGCACAGTACGAG-3′ and 5’-AGGAGTCCCATGATGAGATTGT-3′); ACTB (5’-CTCCTTAATGTCACGCACGAT-3′ and 5’-CATGTACGTT GCTATCCAGGC-3′).
Genomic knockout of p53 using CRISPR–Cas9
For p53 knockout, lentiviral packaging plasmids psPAX2 and pMD2.G were cotransfected with the CRISPRv2 backbone plasmid into HEK293T cells to produce virus. The following targeting sequence was inserted into the CRISPRv2 vector: TP53, 5’-CAGAATGCAAGAAGCCCAGA-3’.
DNA damage induction
For UV irradiation, cell culture medium was removed, and cells were washed in PBS, cells were exposed to the indicated UV-C dosages using a HL-2000 HybriLinker (UVP). After addition of fresh medium, cells were allowed to recover for the indicated periods.
IF microscopy
Cells were grown to 40 to 60% confluence on coverslips. The cells were washed with PBS and fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. The cells were permeabilized with Hybridlike 0.5% Triton X-100 in PBS for 5 min at 4 °C. The coverslips were incubated with tetramethylrhodamine isothiocyanate–phalloidin (YEASEN; catalog no.: 40734ES75) in PBS. Digital images were captured on an Olympus IX83 microscopy with 60× oil objective lens (numerical aperture, 1.35) and Andor’s Zyla 5.5 sCMOS camera and cellSens Dimension software in each experiment.
Immunoprecipitation and Western blot
For immunoprecipitation, cells were lysed in NETN420 (420 mM NaCl, 20 mM Tris–HCl [pH 8.0], 1 mM EDTA, and 0.5% NP-40) plus protease inhibitor. Cell lysates were incubated with streptavidin beads for 3 h at 4 °C. The beads were washed three times with washing buffer (100 mM NaCl, 20 mM Tris–HCl, pH 8.0, 1 mM EDTA, and 0.5% NP-40). For coimmunoprecipitation experiments, cells were lysed in NETN420 buffer. Cell lysates were incubated with 1 μg anti-Zyxin antibody and Protein-A/G Magnetic Beads (MedChemExpress; catalog no.: HY-K0202). After incubation at 4 °C for 3 h, the beads were washed three times with washing buffer.
For the immunoblotting assays, indicated samples were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% milk in Tris-buffered saline with Tween-20 and probed with indicated primary and then secondary antibodies. Signals were then visualized using ECL (Bio-Rad; catalog no.: 1705061; US EVERBRIGHT; catalog no.: S6008). The following antibodies were used: the dilution of primary antibodies FLAG, Myc, GFP, β-tubulin, Zyxin, OGT, and HIPK2 was 1:3000, and the dilution of primary antibody CTD110.6, RL2, and Phospho-p53 (Ser46) was 1:1500. The secondary antibody horseradish peroxidase–Rb/M was purchased from Jackson ImmunoResearch, and the dilution was 1:20,000. The ImageJ software (National Institutes of Health) was used for image analyses, and the quantification results were normalized to the loading control.
GST pull-down assay
Recombinant GST proteins were purified from Escherichia coli BL21 (DE3) cell by standard protocols. GST (2 μg) or GST fusion proteins (2 μg) were incubated with Glutathione Sepharose 4B (ABclonal; catalog no.: AS044) for 2 h at 4 °C, followed by incubation with cell lysates at 4 °C for 3 h. After 1000 rpm centrifugation, supernatants were collected as input, and the beads were washed three times in washing buffer and resolved by SDS-PAGE. Immunoblotting signals were detected with the indicated antibodies.
Cytoplasmic and nuclear fractionation
HEK293T cells and HCT116 cells were collected and extracted using the nuclear and cytoplasmic protein extraction kit (Beyotime; catalog no.: P0028) according to the manufacturer’s instructions.
Wound healing assay
MDA-MB-231 cell lines were plated in 6-well dishes in triplicates. Cell monolayer was scratched by a P100 pipette tip, then were washed twice with PBS, and incubated with serum-free Dulbecco's modified Eagle's medium for another 36 h. The percentage of wound closure was calculated by using ImageJ software.
Apoptosis assay
An apoptosis assay was performed using annexin V–allophycocyanin/propidium iodide Apoptosis Detection Kit (KeyGEN BioTECH; catalog no.: KGA1030-20) in accordance with the manufacturer’s protocol. The stained apoptotic cells were then examined and quantified by LSRFortessaX20 flow cytometer (BD).
Cell survival assay
HCT116 cell lines (8 × 103) were seeded on 96-well plate in triplicates, further treated with UV irradiation (50 J/m2), and then incubated with normal medium. Cell survival was measured through Cell Counting Kit-8 (Beyotime; catalog no.: C0040) at indicated time points.
In vitro O-GlcNAcylation assay
The following process is quoted from the study of Pei et al. (57), and the details are as follows:
The purified GST-OGT or His-OGT protein (amino acids 323–1041) (3 μg) was incubated with 5 μg of recombinant wildtype His-Zyxin or S169A mutant in 50 μl reactions (50 mM Tris–HCl, 12.5 mM MgCl2, 2 mM UDP-GlcNAc, 1 mM DTT, and pH 7.5) for 4 h at 37 °C, prior to incubation with His-Agarose (ABclonal; catalog no.: AS045) for 3 h at 4 °C. After 1000 rpm centrifugation, supernatants were collected as input, and the beads were washed three times in washing buffer and resolved by SDS-PAGE. Immunoblotting signals were detected with the indicated antibodies.
Statistical analysis
Three independent experiments were performed, and data are presented as means ± SD. Student’s t test was performed, and p values <0.05 were considered statistically significant.
Data availability
All data are available upon request. Sequences of shRNAs, guide RNAs, and primer oligonucleotides are provided in the “Experimental procedures” section.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
Q. C. and Y. Z. conceptualization; Q. C. and Y. Z. methodology; S. Y., X. Z., and S. M. validation; Y. Z. investigation; S. Y., X. Z., and S. M. resources; Y. Z. data curation; Q. C., Y. Z., and J. G. writing–review & editing.
Funding and additional information
Q. C. is supported by grants from the National Key Research and Development Program of China (grant no.: 2018YFC1003400), the National Natural Science Foundation of China (grant nos.: 32170698 and 31770868), Wuhan University (grant no.: 2042018kf0215), and Medical Science Advancement Program (Basic Medical Sciences) of Wuhan University (grant no.: TFJC2018005).
Edited by Phyllis Hanson
Supporting information
References
- 1.Smith M.A., Hoffman L.M., Beckerle M.C. LIM proteins in actin cytoskeleton mechanoresponse. Trends Cell Biol. 2014;24:575–583. doi: 10.1016/j.tcb.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford A.W., Michelsen J.W., Beckerle M.C. An interaction between zyxin and alpha-actinin. J. Cell Biol. 1992;116:1381–1393. doi: 10.1083/jcb.116.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drees B.E., Andrews K.M., Beckerle M.C. Molecular dissection of zyxin function reveals its involvement in cell motility. J. Cell Biol. 1999;147:1549–1560. doi: 10.1083/jcb.147.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reinhard M., Zumbrunn J., Jaquemar D., Kuhn M., Walter U., Trueb B. An alpha-actinin binding site of zyxin is essential for subcellular zyxin localization and alpha-actinin recruitment. J. Biol. Chem. 1999;274:13410–13418. doi: 10.1074/jbc.274.19.13410. [DOI] [PubMed] [Google Scholar]
- 5.Li B., Trueb B. Analysis of the alpha-actinin/zyxin interaction. J. Biol. Chem. 2001;276:33328–33335. doi: 10.1074/jbc.M100789200. [DOI] [PubMed] [Google Scholar]
- 6.Golsteyn R.M., Beckerle M.C., Koay T., Friederich E. Structural and functional similarities between the human cytoskeletal protein zyxin and the ActA protein of Listeria monocytogenes. J. Cell Sci. 1997;110:1893–1906. doi: 10.1242/jcs.110.16.1893. [DOI] [PubMed] [Google Scholar]
- 7.Drees B., Friederich E., Fradelizi J., Louvard D., Beckerle M.C., Golsteyn R.M. Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 2000;275:22503–22511. doi: 10.1074/jbc.M001698200. [DOI] [PubMed] [Google Scholar]
- 8.Niebuhr K., Ebel F., Frank R., Reinhard M., Domann E., Carl U.D., Walter U., Gertler F.B., Wehland J., Chakraborty T. A novel proline-rich motif present in ActA of Listeria monocytogenes and cytoskeletal proteins is the ligand for the EVH1 domain, a protein module present in the Ena/VASP family. EMBO J. 1997;16:5433–5444. doi: 10.1093/emboj/16.17.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma B., Cheng H., Gao R., Mu C., Chen L., Wu S., Chen Q., Zhu Y. Zyxin-Siah2-Lats2 axis mediates cooperation between Hippo and TGF-beta signalling pathways. Nat. Commun. 2016;7:11123. doi: 10.1038/ncomms11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., Zeng Y., Cui L., Chen X., Stauffer S., Wang Z., Yu F., Lele S.M., Talmon G.A., Black A.R., Chen Y., Dong J. Zyxin promotes colon cancer tumorigenesis in a mitotic phosphorylation-dependent manner and through CDK8-mediated YAP activation. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E6760–E6769. doi: 10.1073/pnas.1800621115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nix D.A., Fradelizi J., Bockholt S., Menichi B., Louvard D., Friederich E., Beckerle M.C. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- 12.Hervy M., Hoffman L., Beckerle M.C. From the membrane to the nucleus and back again: Bifunctional focal adhesion proteins. Curr. Opin. Cell Biol. 2006;18:524–532. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Nix D.A., Beckerle M.C. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: A potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., Gilmore T.D. Zyxin and paxillin proteins: Focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 15.Sabino F., Madzharova E., Auf dem Keller U. Cell density-dependent proteolysis by HtrA1 induces translocation of zyxin to the nucleus and increased cell survival. Cell Death Dis. 2020;11:674. doi: 10.1038/s41419-020-02883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann T.G., Moller A., Sirma H., Zentgraf H., Taya Y., Droge W., Will H., Schmitz M.L. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- 17.Winter M., Sombroek D., Dauth I., Moehlenbrink J., Scheuermann K., Crone J., Hofmann T.G. Control of HIPK2 stability by ubiquitin ligase Siah-1 and checkpoint kinases ATM and ATR. Nat. Cell Biol. 2008;10:812–824. doi: 10.1038/ncb1743. [DOI] [PubMed] [Google Scholar]
- 18.Crone J., Glas C., Schultheiss K., Moehlenbrink J., Krieghoff-Henning E., Hofmann T.G. Zyxin is a critical regulator of the apoptotic HIPK2-p53 signaling axis. Cancer Res. 2011;71:2350–2359. doi: 10.1158/0008-5472.CAN-10-3486. [DOI] [PubMed] [Google Scholar]
- 19.Bond M.R., Hanover J.A. O-GlcNAc cycling: A link between metabolism and chronic disease. Annu. Rev. Nutr. 2013;33:205–229. doi: 10.1146/annurev-nutr-071812-161240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X., Qian K. Protein O-GlcNAcylation: Emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2017;18:452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart G.W., Housley M.P., Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- 22.Zachara N.E., Hart G.W. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Zachara N.E., Hart G.W. Cell signaling, the essential role of O-GlcNAc. Biochim. Biophys. Acta. 2006;1761:599–617. doi: 10.1016/j.bbalip.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Slawson C., Hart G.W. O-GlcNAc signalling: Implications for cancer cell biology. Nat. Rev. Cancer. 2011;11:678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Copeland R.J., Han G., Hart G.W. O-GlcNAcomics--Revealing roles of O-GlcNAcylation in disease mechanisms and development of potential diagnostics. Proteomics Clin. Appl. 2013;7:597–606. doi: 10.1002/prca.201300001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh J.P., Zhang K., Wu J., Yang X. O-GlcNAc signaling in cancer metabolism and epigenetics. Cancer Lett. 2015;356:244–250. doi: 10.1016/j.canlet.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang X., Pan Q., Sun D., Chen W., Shen A., Huang M., Ding J., Geng M. O-GlcNAcylation of cofilin promotes breast cancer cell invasion. J. Biol. Chem. 2013;288:36418–36425. doi: 10.1074/jbc.M113.495713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z., Isaji T., Fukuda T., Wang Y., Gu J. O-GlcNAcylation regulates integrin-mediated cell adhesion and migration via formation of focal adhesion complexes. J. Biol. Chem. 2019;294:3117–3124. doi: 10.1074/jbc.RA118.005923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y., Mi W., Ge Y., Liu H., Fan Q., Han C., Yang J., Han F., Lu X., Yu W. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70:6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- 30.Rao F.V., Dorfmueller H.C., Villa F., Allwood M., Eggleston I.M., van Aalten D.M. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. EMBO J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Leon C.A., Levine P.M., Craven T.W., Pratt M.R. The sulfur-linked analogue of O-GlcNAc (S-GlcNAc) is an enzymatically stable and reasonable structural surrogate for O-GlcNAc at the peptide and protein levels. Biochemistry. 2017;56:3507–3517. doi: 10.1021/acs.biochem.7b00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorelik A., Bartual S.G., Borodkin V.S., Varghese J., Ferenbach A.T., van Aalten D.M.F. Genetic recoding to dissect the roles of site-specific protein O-GlcNAcylation. Nat. Struct. Mol. Biol. 2019;26:1071–1077. doi: 10.1038/s41594-019-0325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwak T.K., Kim H., Jung O., Lee S.A., Kang M., Kim H.J., Park J.M., Kim S.H., Lee J.W. Glucosamine treatment-mediated O-GlcNAc modification of paxillin depends on adhesion state of rat insulinoma INS-1 cells. J. Biol. Chem. 2010;285:36021–36031. doi: 10.1074/jbc.M110.129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toleman C.A., Schumacher M.A., Yu S.H., Zeng W., Cox N.J., Smith T.J., Soderblom E.J., Wands A.M., Kohler J.J., Boyce M. Structural basis of O-GlcNAc recognition by mammalian 14-3-3 proteins. Proc. Natl. Acad. Sci. U. S. A. 2018;115:5956–5961. doi: 10.1073/pnas.1722437115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan C.B., Liu X., Tang X., Fu H., Ye K. Akt phosphorylation of zyxin mediates its interaction with acinus-S and prevents acinus-triggered chromatin condensation. Cell Death Differ. 2007;14:1688–1699. doi: 10.1038/sj.cdd.4402179. [DOI] [PubMed] [Google Scholar]
- 36.Smith M.A., Hoffman L.M., Beckerle M.C. LIM proteins in actin cytoskeleton mechanoresponse. Trends in cell biology. 2014;24:575–583. doi: 10.1016/j.tcb.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabino F., Madzharova E., Auf dem Keller U. Cell density-dependent proteolysis by HtrA1 induces translocation of zyxin to the nucleus and increased cell survival. Cell death & disease. 2020;11:674. doi: 10.1038/s41419-020-02883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siddiqui M.Q., Badmalia M.D., Patel T.R. Bioinformatic Analysis of Structure and Function of LIM Domains of Human Zyxin Family Proteins. International journal of molecular sciences. 2021;22 doi: 10.3390/ijms22052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu J., Wang S., Viollet B., Zou M.H. Regulation of the proteasome by AMPK in endothelial cells: the role of O-GlcNAc transferase (OGT) PloS one. 2012;7:e36717. doi: 10.1371/journal.pone.0036717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Saito S., Yamaguchi H., Higashimoto Y., Chao C., Xu Y., Fornace A.J., Jr., Appella E., Anderson C.W. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 2003;278:37536–37544. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- 41.Liebl M.C., Hofmann T.G. Cell fate regulation upon DNA damage: p53 serine 46 kinases pave the cell death road. Bioessays. 2019;41 doi: 10.1002/bies.201900127. [DOI] [PubMed] [Google Scholar]
- 42.D'Orazi G., Cecchinelli B., Bruno T., Manni I., Higashimoto Y., Saito S., Gostissa M., Coen S., Marchetti A., Del Sal G., Piaggio G., Fanciulli M., Appella E., Soddu S. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 43.Kiss A., Gong X., Kowalewski J.M., Shafqat-Abbasi H., Stromblad S., Lock J.G. Non-monotonic cellular responses to heterogeneity in talin protein expression-level. Integr. Biol. 2015;7:1171–1185. doi: 10.1039/c4ib00291a. [DOI] [PubMed] [Google Scholar]
- 44.Damayanti N.P., Buno K., Narayanan N., Voytik Harbin S.L., Deng M., Irudayaraj J.M.K. Monitoring focal adhesion kinase phosphorylation dynamics in live cells. Analyst. 2017;142:2713–2716. doi: 10.1039/c7an00471k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parshina E.A., Eroshkin F.M., Оrlov E.E., Gyoeva F.K., Shokhina A.G., Staroverov D.B., Belousov V.V., Zhigalova N.A., Prokhortchouk E.B., Zaraisky A.G., Martynova N.Y. Cytoskeletal protein zyxin inhibits the activity of genes responsible for embryonic stem cell status. Cell Rep. 2020;33:108396. doi: 10.1016/j.celrep.2020.108396. [DOI] [PubMed] [Google Scholar]
- 46.Roberts G.C., Critchley D.R. Structural and biophysical properties of the integrin-associated cytoskeletal protein talin. Biophys. Rev. 2009;1:61–69. doi: 10.1007/s12551-009-0009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuo J.C., Han X., Hsiao C.T., Yates J.R., 3rd, Waterman C.M. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mason D.E., Collins J.M., Dawahare J.H., Nguyen T.D., Lin Y., Voytik-Harbin S.L., Zorlutuna P., Yoder M.C., Boerckel J.D. YAP and TAZ limit cytoskeletal and focal adhesion maturation to enable persistent cell motility. J. Cell Biol. 2019;218:1369–1389. doi: 10.1083/jcb.201806065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun X., Phua D.Y.Z., Axiotakis L., Jr., Smith M.A., Blankman E., Gong R., Cail R.C., Espinosa de Los Reyes S., Beckerle M.C., Waterman C.M., Alushin G.M. Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev. Cell. 2020;55:468–482.e7. doi: 10.1016/j.devcel.2020.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belgardt E., Steinberg T., Husari A., Dieterle M.P., Hulter-Hassler D., Jung B., Tomakidi P. Force-responsive Zyxin modulation in periodontal ligament cells is regulated by YAP rather than TAZ. Cell Signal. 2020;72:109662. doi: 10.1016/j.cellsig.2020.109662. [DOI] [PubMed] [Google Scholar]
- 51.Love D.C., Hanover J.A. The hexosamine signaling pathway: Deciphering the "O-GlcNAc code". Sci. STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- 52.Wong K.K.L., Liu T.W., Parker J.M., Sinclair D.A.R., Chen Y.Y., Khoo K.H., Vocadlo D.J., Verheyen E.M. The nutrient sensor OGT regulates Hipk stability and tumorigenic-like activities in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 2020;117:2004–2013. doi: 10.1073/pnas.1912894117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degenhardt Y.Y., Silverstein S. Interaction of zyxin, a focal adhesion protein, with the e6 protein from human papillomavirus type 6 results in its nuclear translocation. J. Virol. 2001;75:11791–11802. doi: 10.1128/JVI.75.23.11791-11802.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suresh Babu S., Wojtowicz A., Freichel M., Birnbaumer L., Hecker M., Cattaruzza M. Mechanism of stretch-induced activation of the mechanotransducer zyxin in vascular cells. Sci. Signal. 2012;5:ra91. doi: 10.1126/scisignal.2003173. [DOI] [PubMed] [Google Scholar]
- 55.Wojtowicz A., Babu S.S., Li L., Gretz N., Hecker M., Cattaruzza M. Zyxin mediation of stretch-induced gene expression in human endothelial cells. Circ. Res. 2010;107:898–902. doi: 10.1161/CIRCRESAHA.110.227850. [DOI] [PubMed] [Google Scholar]
- 56.Youn H., Kim E.J., Um S.J. Zyxin cooperates with PTOV1 to confer retinoic acid resistance by repressing RAR activity. Cancer Lett. 2013;331:192–199. doi: 10.1016/j.canlet.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 57.Peng C., Zhu Y., Zhang W., Liao Q., Chen Y., Zhao X., Guo Q., Shen P., Zhen B., Qian X., Yang D., Zhang J.S., Xiao D., Qin W., Pei H. Regulation of the hippo-YAP pathway by glucose sensor O-GlcNAcylation. Mol. Cell. 2017;68:591–604.e5. doi: 10.1016/j.molcel.2017.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request. Sequences of shRNAs, guide RNAs, and primer oligonucleotides are provided in the “Experimental procedures” section.