Abstract
Background
The colposcopy-conization inconsistency is common in women with cervical intraepithelial neoplasia grade 3 (CIN3). No adequate method has been reported to identify the final pathology of conization. In this study, we explored the ability of PAX1 and ZNF582 methylation to predict the pathological outcome of conization in advance.
Methods
This was a multicenter study and included 277 histologically confirmed CIN3 women who underwent cold knife conization (CKC) from January 2019 to December 2020. The methylation levels of PAX1 (PAX1m) and ZNF582 (ZNF582m) were determined by quantitative methylation-specific polymerase chain reaction (qMSP) and expressed in ΔCp. Receiver operating characteristic curves were used to evaluate predictive accuracy.
Results
The final pathological results in 48 (17.33%) patients were inflammation or low-grade squamous intraepithelial lesion (LSIL), 190 (68.59%) were high-grade squamous intraepithelial lesion (HSIL), and 39 (14.08%) were squamous cervical cancer (SCC). PAX1m and ZNF582m increased as lesions progressed from inflammation/LSIL, HSIL, to SCC. PAX1 and ZNF582 methylation yielded better prediction performance compared with common screening strategies, whether individually or combined. A 4.33-fold increase in the probability of inflammation/LSIL was observed in patients with lower ZNF582 methylation levels (ΔCpZNF582 ≥ 19.18). A 6.53-fold increase in SCC risk was observed in patients with elevated ZNF582 methylation (ΔCpZNF582 < 7.09).
Conclusions
DNA methylation would be an alternative screening method to triage and predict the final outcome of conization in CIN3 cases.
Keywords: cervical intraepithelial neoplasia grade 3, DNA methylation, PAX1, triage, ZNF582
Thirty percent of untreated patients with cervical intraepithelial neoplasia grade 3 (CIN3) progress to cervical cancer in 30 years [1]. Once detected by colposcopically directed biopsy (CDB), the standard treatment for CIN3 is excisional procedures including conization [2]. It has been reported that 4.6%–17.4% of patients with CIN3 on CBD had disease regression on conization [3, 4]. CIN3 is most prevalent among women of reproductive age, and with the changing of China’s family planning policy in recent years, an increasing number of women of childbearing age have fertility requirements. However, conization is associated with complications including fertility loss, premature labor, premature rupture of membranes, and low birth weight [5, 6]. Therefore, the necessity of the procedure should be carefully discussed among this population. Meanwhile, studies have shown that 2.7%–24.2% [3, 4] of patients with CIN3 on CDB are diagnosed with invasive cervical cancer on CKC specimens and that immediate treatment should be performed in these cases. To date, there is still a lack of evidence on how to identify patients with cervical cancer and triage accordingly.
The limited accuracy of cytology and colposcopy calls for an individualized post-CDB triage strategy [7]. Previous studies have proposed human papillomavirus (HPV) viral load as a post-CDB triage tool for CIN2+ lesions [8, 9]. However, few studies have addressed CIN3 in this regard. Abnormal gene methylation is present throughout the entire process of CIN progression [10–12]. A plethora of studies have shown that methylation has a high screening accuracy for lesions of cervical intraepithelial neoplasia grade 2 or worse (CIN2+) and that this screening could be used as a triage method in women with HPV infection [13–18]. In particular, the efficacy of Paired boxed gene 1 (PAX1) and Zinc finger protein 582 (ZNF582) methylation as biomarkers for detection of CIN3 or worse (CIN3+) has been demonstrated in our previous studies [19, 20]. Similar studies have also explored the combination of PAX1/ZNF582 methylation with other triage tools including HPV genotyping, and the results were consistent [21, 22].
To explore the clinical application of PAX1 and ZNF582 methylation in CIN3 triage, we conducted a multicenter study on the prediction of the pathological diagnosis of the specimen of conization and compared their performance with common screening strategies. We further verified the correlation between methylation level and the severity of conization pathological diagnosis.
METHODS
Study Design and Cohort Characteristics
This multicenter study was led by Xiangya Hospital of Central South University and completed in cooperation with 4 other municipal hospitals of 4 cities in Hunan Province, including Yongzhou, Xiangtan, Yiyang, and Chenzhou. From January 2019 to December 2020, 348 women diagnosed with CIN3 who were subsequently advised to undergo cold knife conization (CKC) were included in the study. Exfoliated cervical cell samples were collected on all patients within 7 days before surgery. Exclusion criteria include the following: (1) inability to undergo CKC, (2) inadequate DNA concentration in cell samples or sample confusion, (3) adenocarcinoma in situ (AIS) or adenocarcinoma. For every patient, information was collected on the age at surgery, high-risk HPV (hrHPV), cytology and colposcopy result pre-CKC, and pathological diagnoses of CDB and CKC. We informed the patients of the research programs and obtained written and verbal consent before CKC. The study was approved by the Institutional Review Board of Xiangya Hospital (2018121117).
Detection and Diagnosis
Detection of CIN3 was according to the consensus guidelines published by the Chinese Society of Colposcopy and Cervical Pathology (CSCCP) [23]. The Cobas 4800 test (Roche, USA), Hybrid capture 2 system (Qiagen, Germany), and HPV Polymerase Chain Reaction (HPV PCR; Genetel Pharmaceuticals, China) were utilized for HPV testing. Cobas 4800 tested HPV16 and HPV18 separately, as well as a pool of 12 other high-risk types (HPV31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68). The HC2 system detected the viral loads of 13 types of hrHPV, and a value >1.0 RLU/PC was defined as positive. HPV PCR, which was developed and validated in China and has been approved by the State Food and Drug Administration (SFDA), detected 13 hrHPV genotypes containing HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68. The testing method was determined by the primary care hospital and gynecologists.
Cytology testing was performed using the ThinPrep cytologic test (TCT, Hologic, USA), and the result was confirmed by 2 cytopathologists. A diagnosis of atypical squamous cells of undetermined significance (ASCUS) or worse (including low-grade squamous intraepithelial lesions [LSIL]; atypical squamous cells cannot exclude high-grade squamous cells of undetermined signification [ASC-H], high-grade squamous intraepithelial lesions [HSIL], atypical glandular cells [AGC], and squamous cell carcinoma [SCC]) was considered abnormal. Patients with abnormal cytologic or hrHPV results underwent colposcopy, and biopsy was indicated when any suspicion of abnormal epithelium occurred. Endocervical curettage (ECC) was performed when the colposcopy was not satisfactory.
According to the World Health Organization (WHO) classification of tumors of female reproductive organs published in 2014, the pathological results of CKC specimens were determined by the most serious lesions including LSIL, HSIL, and cervical cancer [24]. LSIL indicated CIN1, and HSIL included CIN2 and CIN3. A follow-up diagnostic excisional procedure and limited-time surgery were recommended for LSIL, HSIL, and cervical cancer, respectively. As infiltration of neutrophils and lymphocytes was observed in all conization specimens, we used the term “inflammation” to indicate patients without CIN. All the included patients were confirmed negative for bacteria, mycoplasma, and chlamydia infection before conization by vaginal and cervical discharge examination. The primary end point of the study was divided into 3 groups: inflammation/LSIL, HSIL, and SCC [25]. All pathological diagnoses of tissues were evaluated separately and confirmed by 2 experienced pathologists.
Specimen Collection, DNA Preparation, and Methylation Tests
Patients with CIN3 were suggested to undergo CKC. Exfoliated cervical cells were collected within 7 days before CKC and kept in phosphate-buffered saline (PBS) solution at −20°C until testing. The QIAamp DNA Mini Kit (Qiagen, Germany) and NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA) were used for extracting genomic DNA (gDNA) and assessing the purity and concentration of gDNA, respectively.
All the methylation tests were performed by a certified company (HOOMYA, China). Quantitative methylation-specific PCR (qMSP) was performed to determine the methylation level of ZNF582 (ZNF582m) and PAX1 (PAX1m) using the TaqMan-based technology performed in a Lightcycler LC480 system (Roche Applied Science, Germany) with the VIC gene as an internal reference. The crossing point (Cp) value of ZNF582, PAX1, and VIC could be obtained in each sample. The Cp value for VIC should be <35 and otherwise would be retested. DNA methylation status was calculated based on the differences between the Cp values of the tested and referred genes (delta Cp (ΔCp) = CpPAX1 or CpZNF582 – CpVIC), and a low ΔCp value indicated a high methylation level. Caski and C33A cancer cell lines were used as positive and negative controls [21, 26, 27].
Statistical Analysis
Multivariable logistic regression was used on cohort characteristics to identify potential predictive factors of the final outcome. After adjusting for age, hormonal status, HPV16/18 genotype, cytology, and involvement of glands, odds ratios (ORs) were estimated for each variable. Initial comparisons of PAX1m or ZNF582m among 3 different outcomes were performed using the Student t test. Then we explored whether PAX1m or ZNF582m could be used for prediction of the end point by using specific thresholds confirmed by predictive accuracy and clinical requirements. Subsequently, ΔCp of PAX1 and ZNF582 were considered for inclusion into a multivariable logistic regression model to predict the primary outcome. The model was developed by assessing the area under the receiver operating characteristic curve (AUC) in the training data and 10-fold cross-validation, validated, and adjusted by bootstrap resampling [28, 29]. The final formula was as follows: ΔCpModel = 0.1*ΔCpPAX1 + 0.9*ΔCpZNF582 + 2.032. Receiver operating characteristic (ROC) curves were used to measure the discrimination efficacy of common screening methods including HPV16/18, cytology, colposcopic impression, single PAX1m or ZNF582m, and the model. All data analyses above were performed using Statistical Product and Service Solutions (SPSS), version 23.0, and Stata, version 12.2. A P value of <.05 was considered statistically significant.
RESULTS
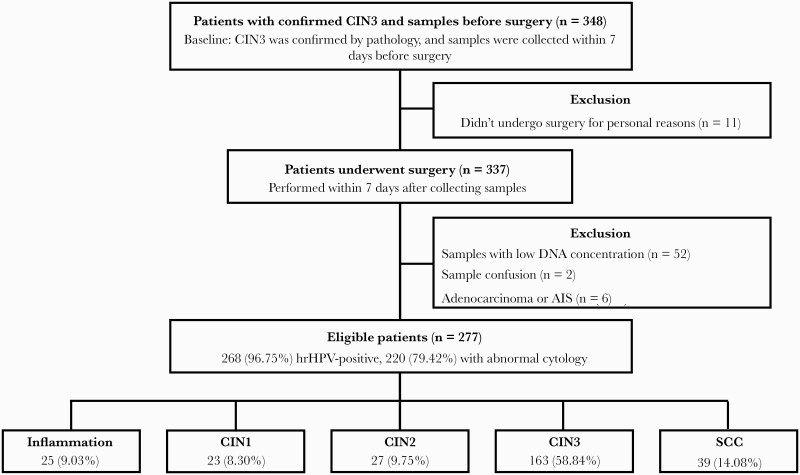
Of the 348 potentially eligible women, exclusions were as follows: 11 for not undergoing surgery for personal reasons, 52 for samples with low concentrations for further methylation testing, 2 for sample confusion, and 6 for adenocarcinoma or adenocarcinoma in situ (AIS). Two hundred seventy-seven women remained in the final study cohort, and the diagnosis of cone samples included 25 (9.03%) women with inflammation, 23 (8.30%) with CIN1, 27 (9.75%) with CIN2, 163 (58.84%) with CIN3, and 39 (14.08%) with SCC. All the included patients had no history of immunodeficiency or cigarette smoking. The flowchart and cohort characteristics are presented in Figure 1 and Supplementary Table 1, respectively. Patients with abnormal cytology results presented the lowest possibility of inflammation/LSIL, with an OR of 0.35 (95% CI, 0.14–0.87), and there was no correlation found in other factors. Gland involvement was associated with higher risk of SCC (OR, 2.77; 95% CI, 1.11–6.89). There was no relationship between HPV16/18 genotype or hormonal status and the final pathological outcome, and the detecting performance is summarized in Supplementary Table 2.
Figure 1.
The flowchart of patients. Abbreviations: AIS, adenocarcinoma in situ; CIN, cervical intraepithelial neoplasia; SCC, squamous cervical cancer.
PAX1 and ZNF582 methylation levels were significantly different among their groups (Figure 2). An ROC curve indicated a high diagnostic value of methylated PAX1 and ZNF582 in the inflammation/LSIL group compared with the HSIL + SCC group, with AUCs of 0.741 and 0.807, respectively (Figure 3A). When we compared the inflammation/LSIL + HSIL group with the SCC group using PAX1m and ZNF582m, the AUCs were 0.741 and 0.773 for SCC detection (Figure 3B). In addition, the AUCs of PAX1m and ZNF582m could reach 0.884 and 0.915, respectively, in distinguishing inflammation/LSIL from SCC (Figure 3C). Relatively speaking, HPV16/18 genotype, cytology, and colposcopic impression provide lower predictive potential (Figure 3A, B, and C). In order to facilitate clinical application, we confirmed the cutoff value of ΔCp, which was determined by controlling the misdiagnosis rate of inflammation/LSIL or by keeping the missed diagnosis rate of SCC <20%. Thus, we established 2 cutoff values, shown with 2 lines on Figure 2A and B, for each gene. For PAX1m, the sensitivity and specificity were 31.25% (95% CI, 17.65%–44.85%) and 81.66% (95% CI, 76.61%–86.71%) in predicting inflammation/LSIL with ΔCpPAX1 ≥20.00, and the sensitivity and specificity were 84.62% (95% CI, 72.77%–96.46%) and 46.64% (95% CI, 40.43%–53.23%) in predicting SCC with ΔCpPAX1 <8.55. For ZNF582m, the sensitivity (50.00%; 95% CI, 35.33%–64.67%) was better than PAX1m with a capable specificity in predicting inflammation/LSIL with a cutoff of 19.18, and similarly, the specificity (58.82%; 95% CI, 52.53%–65.12%) was higher than PAX1m with a capable sensitivity in predicting SCC with ΔCpZNF582 <7.09 (Table 1). The possibility of inflammation/LSIL would be 4.33-fold (95% CI, 2.25–8.33) when ΔCpZNF582 ≥19.18, and the risk of SCC would increase 6.53 times (95% CI, 2.77–15.40) with a value of ΔCpZNF582 <7.09 (Table 1).
Figure 2.
The methylation value of PAX1 and ZNF582 in patients with different clinical outcomes. A, ΔCpPAX1 according to different grades of lesions. B, ΔCpZNF582 according to different grades of lesions. The dotted line corresponds to the threshold for distinguishing inflammation/LSIL/HSIL from SCC, and the dashed line corresponds to the threshold for distinguishing inflammation/LSIL from HSIL/SCC. Statistically significant (P < .05). Abbreviations: CIN, cervical intraepithelial neoplasia; hrHPV, high-risk HPV; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; SCC, squamous cervical cancer.
Figure 3.
Receiver operating characteristic curves for the performance of PAX1, ZNF582 methylation, and the combination model of the 2 genes. A, inflammation/LSIL vs HSIL/SCC. B, inflammation/LSIL/HSIL vs SCC. C, inflammation/LSIL vs SCC. Abbreviations: AUC, area under the receiver operating characteristic curve; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; SCC, squamous cervical cancer.
Table 1.
The Performance of ZNF582 Methylation and PAX1 Methylation in Detecting Inflammation/LSIL or SCC
| Methylation Marker | Sensitivity, % | Specificity, % | PPV, % | NPV, % | OR | P |
|---|---|---|---|---|---|---|
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI)a | ||
| Inflammation/LSIL | ||||||
| ΔCpPAX1 ≥ 20.00 | 31.25 | 81.66 | 26.32 | 85.00 | 2.02 | .047 |
| (17.65–44.85) | (76.61–86.71) | (14.53–38.10) | (80.24–89.76) | (1.01–4.06)b | ||
| ΔCpZNF582 ≥ 19.18 | 50.00 | 81.22 | 35.82 | 88.57 | 4.33 | .000 |
| (35.33–64.67) | (76.13–86.32) | (24.03–47.60) | (84.23–92.99) | (2.25–8.33)b | ||
| ΔCpModel ≥ 21.15 | 50.00 | 80.35 | 34.78 | 88.46 | 4.09 | .000 |
| (35.73–64.67) | (75.16–85.53) | (23.26–46.31) | (84.08–92.84) | (2.13–7.86)b | ||
| SCC | ||||||
| ΔCpPAX1 < 8.55 | 84.62 | 46.64 | 20.63 | 94.87 | 4.81 | .001 |
| (72.77–96.46) | (40.43–53.23) | (14.29–26.96) | (90.82–98.93) | (1.94–11.90)b | ||
| ΔCpZNF582 < 7.09 | 82.05 | 58.82 | 24.62 | 95.24 | 6.53 | .000 |
| (69.45–94.65) | (52.53–65.12) | (17.11–32.12) | (91.75–98.72) | (2.77–15.40)b | ||
| ΔCpModel < 9.19 | 82.05 | 60.50 | 25.40 | 95.36 | 7.00 | .000 |
| (69.45–94.65) | (54.25–66.76) | (17.69–33.10) | (91.97–98.76) | (2.97–16.52)b | ||
Statistically significant (P < .05).
Abbreviations: LSIL, low-grade squamous intraepithelial lesion; NPV, negative predictive value; PPV, positive predictive value; OR, odds ratio; SCC, squamous cervical cancer.
OR was calculated with other ΔCp values by logistic regression analysis.
Statistically significant OR and CI.
In order to explore whether the combination of PAX1m and ZNF582m could improve accuracy in detecting inflammation/LSIL or SCC, we established a combined model (shown in the “Methods” section) and further confirmed its efficacy. The model had comparable predictive values as single PAX1m or ZNF582m (Figure 2). When the ΔCpModel ≥21.15, the specificity and sensitivity in detecting inflammation/LSIL were 50.00% (95% CI, 35.73%–64.67%) and 80.35% (95% CI, 75.16%–85.53%), and the possibility of inflammation/LSIL increased 4.09-fold (95% CI, 2.13–7.86). When the ΔCpModel <9.19, the sensitivity and specificity were 82.05% (95% CI, 69.45%–94.65%) and 60.50% (95% CI, 54.25%–66.76%) with regard to SCC, and the risk of SCC increased 7.00-fold (Table 1).
Then we further verified the positive correlation between DNA methylation levels and the pathological results of CKC specimens, which repeatedly proved that PAX1m and ZNF582m had potential in CIN3 triage. Exploring whether there were factors related to DNA methylation level, we found that colposcopic impression was correlated with the methylation level of both PAX1m and ZNF582m, as well as the model, showing a positive correlation (Table 2). We further found that the difference in colposcopic impression was limited to the groups of ΔCpPAX1 <8.55 vs ΔCpPAX1 ≥8.55, ΔCpZNF582 <7.09 vs ΔCp ≥7.09, ΔCpModel <9.19 vs ΔCpModel ≥9.19 (P < .05; data not shown).
Table 2.
The Correlation Between DNA Methylation Level and Clinicopathological Features
| Factors | PAX1 | P | ZNF582 | P | Model | P | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔCp ≥ 20.00 | 8.55 ≤ ΔCp < 20.00 | ΔCp < 8.55 | ΔCp ≥ 19.18 | 7.09 ≤ ΔCp < 19.18 | ΔCp < 7.09 | ΔCp ≥ 21.15 | 9.19 ≤ ΔCp < 21.15 | ΔCp < 9.19 | ||||
| Age, y | 44.09 ± 11.19 | 44.75 ± 11.46 | 44.71 ± 9.21 | .916 | 44.52 ± 9.50 | 44.63 ± 9.82 | 44.67 ± 11.68 | .995 | 44.55 ± 9.46 | 43.96 ± 10.28 | 45.41 ± 11.14 | .684 |
| Hormonal status | ||||||||||||
| Premenopause | 41 | 43 | 116 | .991 | 48 | 58 | 94 | .993 | 48 | 61 | 91 | .805 |
| Postmenopause | 16 | 17 | 44 | 19 | 22 | 36 | 21 | 21 | 35 | |||
| HPV16/18 genotype | ||||||||||||
| HPV16(-) and HPV18(-) | 25 | 27 | 57 | .141 | 31 | 29 | 49 | .108 | 33 | 31 | 45 | .052 |
| HPV16(+) or HPV18(+) | 16 | 25 | 73 | 19 | 36 | 59 | 19 | 36 | 59 | |||
| Cytology | ||||||||||||
| Normal | 11 | 11 | 13 | .035a | 13 | 11 | 11 | .069 | 13 | 11 | 11 | .114 |
| ≥ASCUS | 43 | 44 | 133 | 47 | 64 | 109 | 50 | 65 | 105 | |||
| Colposcopic impression | ||||||||||||
| Normal/low grade | 20 | 20 | 30 | .014a | 23 | 28 | 19 | .001a | 24 | 26 | 20 | .006a |
| High grade | 30 | 30 | 104 | 37 | 39 | 88 | 37 | 43 | 84 | |||
| TZ type | ||||||||||||
| Ⅰ–Ⅱ | 9 | 16 | 46 | .195 | 15 | 22 | 34 | .716 | 13 | 24 | 34 | .319 |
| Ⅲ | 41 | 41 | 101 | 46 | 49 | 88 | 49 | 50 | 84 | |||
| Glands | ||||||||||||
| Uninvolved | 21 | 23 | 46 | .275 | 20 | 34 | 36 | .052 | 21 | 33 | 36 | .153 |
| Involved | 34 | 36 | 111 | 46 | 43 | 92 | 47 | 46 | 88 | |||
| Pathological diagnosis of CKC specimen | ||||||||||||
| Inflammation/LSIL | 15 | 22 | 11 | .000a | 24 | 21 | 3 | .000a | 24 | 21 | 3 | .000a |
| HSIL | 39 | 35 | 116 | 40 | 55 | 95 | 41 | 58 | 91 | |||
| SCC | 3 | 3 | 33 | 3 | 4 | 32 | 4 | 3 | 32 | |||
Statistically significant (P < .05).
Abbreviations: ASCUS, atypical squamous cells of undetermined significance; CKC, cold knife conization; HPV, human papillomavirus; LSIL, low-grade squamous intraepithelial lesions; OR, odds ratio; SCC, squamous cervical cancer; TZ, transformation zone.
Statistically significant OR and CI.
DISCUSSION
This is the first study to assess the predictive performance of common screening strategies and DNA methylation in a series of patients with histological CIN3 at baseline. Inflammation/LSIL and SCC on conization were observed in 17% and 12% of our cohort, respectively, which was consistent with previous findings (4.6%–17.4% for inflammation/LSIL and 2.7%–24.2% for SCC) [3, 4]. Most of our patients with CIN3 had fertility requirements. In our cohort, 112/277 (40.43%) patients were age <40 years. Additionally, >1 in 10 patients diagnosed with CIN3 in CBD were found to have SCC in CKC specimens. Therefore, distinguishing lesions helps to reduce overtreatment and misdiagnoses.
Previous studies have reported that the disruption between kinases and phosphatases caused by PAX1 methylation is involved in cervical carcinogenesis [30]. A meta-analysis including 7 studies and 1055 patients found that PAX1 methylation was associated with the transition from normal tissues to CIN and cervical cancer, which could be applied to the identification of lesions that had the potential of becoming SCC [31]. Additionally, PAX1 methylation performed comparably to cytology and had better accuracy than HPV16/18 in the triage of hrHPV infection [32]. ZNF582, previously identified as a tumor suppressor gene in nasopharyngeal carcinoma by regulating the transcription and expression of the adhesion molecules Nectin-3 and NRXN3, has not been thoroughly understood in the carcinogenesis of cervical cancer [33]. In our previous studies, we found that the combination of ZNF582 methylation and HPV16/18 testing was able to eradicate missed diagnoses of cervical cancer and reduce the rate of colposcopy referrals, both in the setting of common screening and the triage of women with abnormal cytological results [21, 22]. A meta-analysis including 7 studies and 1749 patients showed a pooled AUC of ZNF582 methylation in detecting CIN3 or worse (CIN3+) of 0.85, which was higher than a single hrHPV test [20]. Standardized and quantified, PAX1 or ZNF582 methylation could be extremely useful in countries where adequate cytology-based infrastructure is lacking. PAX1 and ZNF582 methylation have yielded excellent results in cervical cancer screening, and their application in the triage of CDB-confirmed CIN3, while promising, needs further evaluation. In our study, the reported accuracy of PAX1 and ZNF582 methylation in predicting inflammation/LSIL on conization was 0.8. In the current study, the accuracy of PAX1 or ZNF582 methylation in predicting the pathological results of CKC ranged from 0.8 to 0.9. However, the clinical performance of combined PAX1 and ZNF582 methylation was controversial [22, 34]. In our study, the combination model had comparable accuracy as a single gene, but further validation is needed.
There were 3 patients with SCC with a ΔCp value >20.00. They were diagnosed with stage IA1 cervical cancer of the endocervical tube on the final pathological report; however, their initial ECC results pre-CKC were CIN3. The discordance can be explained by the presence of type III transformation zone (TZ), which complicates the sampling process and may lead to missed diagnoses. Therefore, for women with unsatisfactory colposcopic examination, adequate sample collection combined with ECC is important. We also observed a positive association between DNA methylation level and the severity of lesions under colposcopy. Vigilance should be raised in patients with both high methylation levels of PAX1 and ZNF582 and high-grade lesions under colposcopy.
The study has several strengths. This is the first multicenter study with a large sample size on the triage strategies of CIN3 patients. We reported excellent performance of PAX1/ZNF582 methylation in predicting the pathological outcome of CDB-confirmed CIN3 and further confirmed the cutoff values of ΔCpPAX1 and ΔCpZNF582 and the relationship between DNA methylation and colposcopic impression. Few studies have addressed the subsequent treatment for CDB-confirmed CIN3, and no studies have indicated that CIN3 could be appropriately delayed or given priority [35]. Our study fulfills these stringent requisites, and the results could thereby potentially stimulate consideration of CIN3 treatment.
One limitation is the study’s self-verification method, as this validated study is still going on. The effect of lesion removal during CBD on the subsequent methylation results was unclear; nevertheless, methylation can be used as an indicator of whether to perform conization surgery. Additionally, ECC was only performed in women with type Ⅲ TZ, and some data related to HPV, cytology, or colposcopy were missing. The accuracy of these common screening methods needs to be reliably reproduced.
CONCLUSIONS
In this exploratory study, methylation of PAX1 and ZNF582 had significant accuracy for CIN3 triage. DNA methylation may be an alternative screening method for triage of controversial cases. Further validation and prospective clinical trials are needed to confirm these findings.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
We appreciate the encouragement in conducting research and assistance in figure preparation of Dong-yang Lou from Sun Yat-sen University. We thank the women who participated in this study and their families. We also thank Juan Yuan, Dong-Zhi Huang, Ju-Ying Xu, and Xiao-Li Hou for sample collection and Fan Zhang and Qiong-Qiong He for assistance with pathological diagnoses. We are grateful to HOOMYA Company for providing us with instruments and equipment.
Financial support. This work was supported by the National Natural Science Foundation of China (grant number U20A20368) and the Natural Science Foundation of Hunan Province (grant number 2020SK2074). Y.Z. was in receipt of the grants.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. Kun Fu: planning, conduct, data analysis, and manuscript writing. Ming Lei: data analysis and manuscript writing. Li-Sha Wu: planning. Jing-Cheng Shi, Si-Yu Yang: statistical analysis. Wen-Qing Yang, Jin-Yun Xu, Ya-Nan Kang, Zhen-Ying Yang, Xuan Zhang, Kang-Ni Huang, Chi Han, Yan Tian: sample collecting. Yu Zhang: design, planning.
Patient consent. We confirm that we obtained the patients’ written consent before proceeding with the study, which was approved by the Institutional Review Board at Xiangya Hospital (2018121117).
References
- 1. Practice Bulletin N. 157: cervical cancer screening and prevention. Obstet Gynecol 2016; 127:e1–20. [DOI] [PubMed] [Google Scholar]
- 2. Perkins RB, Guido RS, Castle PE, et al. 2019 ASCCP risk-based management consensus guidelines for abnormal cervical cancer screening tests and cancer precursors. J Low Genit Tract Dis 2020; 24:102–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giannella L, Mfuta K, Gardini G, et al. High-grade CIN on cervical biopsy and predictors of the subsequent cone histology results in women undergoing immediate conization. Eur J Obstet Gynecol Reprod Biol 2015; 186:68–74. [DOI] [PubMed] [Google Scholar]
- 4. Jung Y, Lee AR, Lee SJ, et al. Clinical factors that affect diagnostic discrepancy between colposcopically directed biopsies and loop electrosurgical excision procedure conization of the uterine cervix. Obstet Gynecol Sci 2018; 61:477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kyrgiou M, Athanasiou A, Kalliala IEJ, et al. Obstetric outcomes after conservative treatment for cervical intraepithelial lesions and early invasive disease. Cochrane Database Syst Rev 2017; 11:CD012847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bevis KS, Biggio JR.. Cervical conization and the risk of preterm delivery. Am J Obstet Gynecol 2011; 205:19–27. [DOI] [PubMed] [Google Scholar]
- 7. Dempster-Rivett K, Innes CR, Simcock BJ, et al. Evaluation of guidelines for observational management of cervical intraepithelial neoplasia 2 in young women. Am J Obstet Gynecol 2020; 223:408.e1–11. [DOI] [PubMed] [Google Scholar]
- 8. Sudenga SL, Shrestha S.. Key considerations and current perspectives of epidemiological studies on human papillomavirus persistence, the intermediate phenotype to cervical cancer. Int J Infect Dis 2013; 17:e216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ostor AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol 1993; 12:186–92. [PubMed] [Google Scholar]
- 10. Szalmas A, Konya J.. Epigenetic alterations in cervical carcinogenesis. Semin Cancer Biol 2009; 19:144–52. [DOI] [PubMed] [Google Scholar]
- 11. Wentzensen N, Sherman ME, Schiffman M, Wang SS.. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol 2009; 112:293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Esteller M. Epigenetics in cancer. N Engl J Med 2008; 358:1148–59. [DOI] [PubMed] [Google Scholar]
- 13. Kong L, Wang L, Wang Z, et al. DNA methylation for cervical cancer screening: a training set in China. Clin Epigenetics 2020; 12:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res (Phila) 2014; 7:1251–7. [DOI] [PubMed] [Google Scholar]
- 15. Rogeri CD, Silveira HCS, Causin RL, et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol Oncol 2018; 150:545–51. [DOI] [PubMed] [Google Scholar]
- 16. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ.. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer 2014; 14:395–405. [DOI] [PubMed] [Google Scholar]
- 17. Bu Q, Wang S, Ma J, et al. The clinical significance of FAM19A4 methylation in high-risk HPV-positive cervical samples for the detection of cervical (pre)cancer in Chinese women. BMC Cancer 2018; 18:1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lendvai A, Johannes F, Grimm C, et al. Genome-wide methylation profiling identifies hypermethylated biomarkers in high-grade cervical intraepithelial neoplasia. Epigenetics 2012; 7:1268–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fang C, Wang SY, Liou YL, et al. The promising role of PAX1 (aliases: HUP48, OFC2) gene methylation in cancer screening. Mol Genet Genomic Med 2019; 7:e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li N, He Y, Mi P, Hu Y.. ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse: a meta-analysis of related studies in Chinese population. Medicine (Baltim) 2019; 98:e14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liou YL, Zhang Y, Liu Y, et al. Comparison of HPV genotyping and methylated ZNF582 as triage for women with equivocal liquid-based cytology results. Clin Epigenetics 2015; 7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liou YL, Zhang TL, Yan T, et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin Epigenetics 2016; 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao Y, Wei LH.. CSCCP guideline on issues related to abnormal cervical cancer screening management in China. J Pract Obstetr Gynecol 2018; 34:101–4. [Google Scholar]
- 24. Lu Z, Chen J.. Introduction of WHO classification of tumours of female reproductive organs, fourth edition [in China]. Zhonghua Bing Li Xue Za Zhi 2014; 43:649–50. [PubMed] [Google Scholar]
- 25. Massad LS, Einstein MH, Huh WK, et al. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstet Gynecol 2013; 121:829–46. [DOI] [PubMed] [Google Scholar]
- 26. Huang J, Liou YL, Kang YN, et al. Real-time colorimetric detection of DNA methylation of the PAX1 gene in cervical scrapings for cervical cancer screening with thiol-labeled PCR primers and gold nanoparticles. Int J Nanomedicine 2016; 11:5335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parashar G, Capalash N.. Promoter methylation-independent reactivation of PAX1 by curcumin and resveratrol is mediated by UHRF1. Clin Exp Med 2016; 16:471–8. [DOI] [PubMed] [Google Scholar]
- 28. Yu X, Gao S, Xue Q, et al. Development of a nomogram for predicting the operative mortality of patients who underwent pneumonectomy for lung cancer: a population-based analysis. Transl Lung Cancer Res 2021; 10:381–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. van Straalen JW, Giancane G, Amazrhar Y, et al. A clinical prediction model for estimating the risk of developing uveitis in patients with juvenile idiopathic arthritis. Rheumatology (Oxford) 2021; 60:2896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Su PH, Lai HC, Huang RL, et al. Paired Box-1 (PAX1) activates multiple phosphatases and inhibits kinase cascades in cervical cancer. Sci Rep 2019; 9:9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luan T, Hua Q, Liu X, et al. PAX1 methylation as a potential biomarker to predict the progression of cervical intraepithelial neoplasia: a meta-analysis of related studies. Int J Gynecol Cancer 2017; 27:1480–8. [DOI] [PubMed] [Google Scholar]
- 32. Chang CL, Ho SC, Su YF, et al. DNA methylation marker for the triage of hrHPV positive women in cervical cancer screening: real-world evidence in Taiwan. Gynecol Oncol 2021; 161:429–35. [DOI] [PubMed] [Google Scholar]
- 33. Zhao Y, Hong XH, Li K, et al. ZNF582 hypermethylation promotes metastasis of nasopharyngeal carcinoma by regulating the transcription of adhesion molecules Nectin-3 and NRXN3. Cancer Commun (Lond) 2020; 40:721–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang H, Li GL, Liu J, et al. The application value of PAX1 and ZNF582 gene methylation in high grade intraepithelial lesion and cervical cancer. Clin Transl Oncol 2021; 23:283–8. [DOI] [PubMed] [Google Scholar]
- 35. Louvanto K, Aro K, Nedjai B, et al. Methylation in predicting progression of untreated high-grade cervical intraepithelial neoplasia. Clin Infect Dis 2020; 70:2582–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.