This genomic substudy of a randomized clinical trial assesses whether patients with clonal hematopoiesis of intermediate potential have greater cardiovascular event reduction in response to canakinumab.
Key Points
Question
What is the effect of clonal hematopoiesis of indeterminate potential (CHIP) on the rate of major adverse cardiovascular events in a secondary prevention trial of patients with coronary artery disease and residual inflammatory risk?
Findings
In this exploratory genomic substudy of a randomized clinical trial, compared with published cardiovascular disease cohorts, CHIP was most frequently associated with TET2 variants and patients harboring TET2 variants had numerically fewer major adverse cardiovascular events while taking canakinumab compared with patients without CHIP.
Meaning
In patients with a history of myocardial infarction and elevated high-sensitivity C-reactive protein, the presence of CHIP variant TET2 clones may predispose patients to improved outcomes with targeted anti-inflammatory therapy.
Abstract
Importance
Clonal hematopoiesis of indeterminate potential (CHIP) is associated with increased risk of atherosclerotic cardiovascular disease, and mouse experiments suggest that CHIP related to Tet2 loss of function in myeloid cells accelerates atherosclerosis via augmented interleukin (IL) 1β signaling.
Objective
To assess whether individuals with CHIP have greater cardiovascular event reduction in response to IL-1β neutralization in the Canankinumab Anti-inflammatory Thrombosis Outcomes Trial (CANTOS).
Design, Setting, and Participants
This randomized clinical trial took place from April 2011 to June 2017 at more than 1000 clinical sites in 39 countries. Targeted deep sequencing of genes previously associated with CHIP in a subset of trial participants using genomic DNA prepared from baseline peripheral blood samples were analyzed. All participants had prior myocardial infarction and elevated high-sensitivity C-reactive protein level above 0.20 mg/dL. Analysis took place between June 2017 and December 2021.
Interventions
Canakinumab, an anti–IL-1β antibody, given at doses of 50, 150, and 300 mg once every 3 months.
Main Outcomes and Measures
Major adverse cardiovascular events (MACE).
Results
A total of 338 patients (8.6%) were identified in this subset with evidence for clonal hematopoiesis. As expected, the incidence of CHIP increased with age; the mean (SD) age of patients with CHIP was 66.3 (9.2) years and 61.5 (9.6) years in patients without CHIP. Unlike other populations that were not preselected for elevated C-reactive protein, in the CANTOS population variants in TET2 were more common than DNMT3A (119 variants in 103 patients vs 86 variants in 85 patients). Placebo-treated patients with CHIP showed a nonsignificant increase in the rate of MACE compared with patients without CHIP using a Cox proportional hazard model (hazard ratio, 1.32 [95% CI, 0.86-2.04]; P = .21). Exploratory analyses of placebo-treated patients with a somatic variant in either TET2 or DNMT3A (n = 58) showed an equivocal risk for MACE (hazard ratio, 1.65 [95% CI, 0.97-2.80]; P = .06). Patients with CHIP due to somatic variants in TET2 also had reduced risk for MACE while taking canakinumab (hazard ratio, 0.38 [95% CI, 0.15-0.96]) with equivocal difference compared with others (P for interaction = .14).
Conclusions and Relevance
These results are consistent with observations of increased risk for cardiovascular events in patients with CHIP and raise the possibility that those with TET2 variants may respond better to canakinumab than those without CHIP. Future studies are required to further substantiate this hypothesis.
Trial Registration
ClinicalTrials.gov Identifier: NCT01327846
Introduction
Clonal hematopoiesis of indeterminate potential (CHIP) results from somatic DNA variants occurring in hematopoietic stem cells that promote their selective expansion.1 This process yields clones of variant peripheral leukocytes that can be detected by deep DNA sequencing. The prevalence of CHIP in the general population rises with age: CHIP is uncommon in individuals younger than 50 years but affects up to 50% in those older than 85 years.2 CHIP increases risk for developing hematologic malignant neoplasms, especially myeloid neoplasms, as well as having and being associated with higher overall mortality out of proportion to the development of leukemia.3 Several recent studies have implicated CHIP as a causative risk factor for cardiovascular disease, providing a potential partial explanation for the increased noncancer mortality seen in individuals with CHIP.4,5 The mechanism behind the increased cardiovascular risk likely depends on the variant gene(s) present in leukocytes of patients with CHIP. In the general population, the most common variant gene causing CHIP is DNMT3A, followed by TET2 and then ASXL1.3,4,5,6,7,8 These 3 genes participate in epigenetic regulation of chromatin structure, with DNMT3A and TET2 modulating DNA methylation. Preclinical data show that variants in Dnmt3a and Tet2 lead to an elevated inflammatory response in myeloid cells and both circulating and tissue-infiltrating monocytes with these variants.4,9,10
An extensive literature supports the importance of inflammation as a mediator of cardiovascular disease.11 Epidemiologic studies have correlated the risk of cardiovascular events with elevation in high-sensitivity C-reactive protein (hs-CRP), a general marker of systemic inflammation, and mendelian randomization studies have pointed to the interleukin (IL) 6 pathway as potentially causative for cardiovascular disease.12,13 Further, studies of experimental atherosclerosis in mice have suggested the importance of the IL-1β pathway for the progression of disease.14 The Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) tested the hypothesis that blockade of IL-1β using a specific monoclonal antibody reduces the rate of major adverse cardiovascular events (MACE) in patients with a previous myocardial infarction and evidenced for elevated systemic inflammation as indicated by a hs-CRP level above 0.20 mg/dL (to convert to milligrams per liter, multiply by 10).15 CANTOS randomized patients to placebo, 50 mg of canakinumab, 150 mg of canakinumab, or 300 mg of canakinumab given quarterly. Compared with placebo, patients treated with either 150 or 300 mg of canakinumab showed a 15% relative risk reduction in the combined rate of myocardial infarction, cardiovascular-related death, and stroke. Within CANTOS, IL-1β inhibition was also associated with reduced rates of hospitalization for heart failure.16
Thus, CANTOS provided the opportunity to further characterize the cardiovascular risk of CHIP in a high-risk cardiovascular population with biochemical evidence of inflammation and to explore the response of patients with CHIP to the anti-inflammatory agent canakinumab.
Methods
Study Samples
The CANTOS trial was conducted from April 2011 to June 2017 at more than 1000 clinical sites in 39 countries around the globe.15 The trial enrolled 10 061 patients who were followed up for a median of 3.7 years. Patients were randomized into 1 of 4 arms: placebo, 50 mg of canakinumab, 150 mg of canakinumab, or 300 mg of canakinumab delivered subcutaneously every 3 months. Patients in those countries participating in the genomic substudy provided a 10-mL sample of whole blood at baseline visit. Patient demographics, including self-reported race and ethnicity, were collected for all patients at baseline. Local institutional review board approval was required and obtained. Written informed consent was provided by all patients. The trial protocol is available in Supplement 1. This study followed Novartis’s internal reporting guidelines for clinical trials.
Targeted Genomic Sequencing
A total of 3946 DNA samples isolated from whole blood taken from patients were sequenced using SureSelect custom gene sequencing panel (Agilent) on a HiSeq 2500 sequencer (Illumina), which targeted 74 genes screened by Jaiswal et al.4 Of the 3946 samples sequenced, 3923 passed quality control analysis.
CHIP Variant Ascertainment
To identify patients with CHIP, we followed the general framework proposed by Jaiswal et al4 with some modifications. Specifically, we focused on a prespecified list of variants in 74 genes in which variants have recurrently been described in hematological malignant neoplasms. To determine somatic variants, we used a minimum variant read count of 4 for MuTect17 and 6 for Pindel.18 Frameshift variants were excluded if they occurred in the first or last 10% of the gene open reading frame, except for DNMT3A, TET2, ASXL1, and PPM1D, where variants in those regions had been previously reported. We also excluded variants that appeared in more than 20 patients as they are unlikely to arise somatically.2 For reference, the most frequent somatic variant in JAK2, V617F,19 appeared 13 times, much below our threshold of 20. The variant allele frequency was calculated as the number of sequencing reads of the variant allele over the total number of sequencing reads at the variant allele position.
For TET2 and CBL, all missense variants in particular regions20,21 were considered somatic if the variant allele frequency significantly deviated from the expected distribution for a germline allele (defined as a P value from a binomial test of less than .001 assuming a probability of success in a single Bernoulli experiment of .50 and using the alternate allele read count as the number of successes and the alternate allele read count plus the reference allele read count as the number of trials). Additional suspect germline variants observed above variant allele frequency of 0.4 or more were excluded if reported in public germline databases and never observed below variant allele frequency of 0.3 in prior studies of CHIP.
Statistical Analysis
Hazard ratios (HRs) for association between MACE or heart failure and CHIP were determined using a Cox proportional hazards regression adjusted for prespecified confounders from the study reporting and analysis plan: age, time from last myocardial infarction, baseline log2(hs-CRP), sex, type 2 diabetes status, total cholesterol, high-density lipoprotein cholesterol, smoking status, and hypertension. In stratified and interaction analyses of canakinumab response among patients with CHIP, or subsets thereof, a limited set of covariates (age, time from last myocardial infarction, and baseline log2[hs-CRP]) was required to avoid overfitting the small number of patients and events in each strata. Statistical significance of baseline characteristics of patients with and without CHIP were calculated using Fisher exact test for categorical variables and Mann-Whitney test for continuous variables. All analyses were conducted in SAS statistical software version 9.4 TS1M6 (SAS Institute) and R version 4.00 (R Foundation). Analysis took place between June 2017 and December 2021.
Biomarker Analysis
The methods used to measure hs-CRP, IL-6, and IL-18 in patient samples from the CANTOS trial have been previously described.22,23,24
Results
Baseline Characteristics of CANTOS Patients With CHIP
To determine the prevalence of CHIP among the 3946 patients participating in the CANTOS genomic substudy, we performed targeted sequencing of the coding regions of 74 genes previously associated with CHIP or hematologic malignant neoplasms.4 The mean sequencing depth was 191×, with more than 99% of samples achieving a sequencing depth greater than 100×. Sequence variants in target genes fulfilling the criteria for CHIP were identified in 338 patients (8.6%) of the sequenced population (a list of variants identified is available in eTable 1 in Supplement 2).
The baseline characteristics of patients with CHIP compared with the overall CANTOS population and the sequenced CANTOS population are shown in the Table. Randomization in the genomic subset was similar to the overall study population (placebo, n = 1289; 50 mg, n = 831; 150 mg, n = 950; 300 mg, n = 853). As anticipated, patients with CHIP were older than those without CHIP (mean [SD] age, 66.3 [9.2] vs 61.5 [9.6] years; P < .001), while the prevalence of diabetes and current smoking status were balanced between the groups. Of note, in analyses adjusted for age and sex, the diagnosis of heart failure at study entry was more frequent in patients with CHIP compared with those without CHIP (89 [26.3%] vs 672 [18.7%]; P = .02) and patients with CHIP had a lower median (IQR) estimated glomerular filtration rate (72 [56-84] vs 78 [65-93] mL/min/1.73 m2; P = .001). Baseline levels of median (IQR) hs-CRP and IL-18 in patients with CHIP were similar to patients without CHIP (0.44 [0.30-0.71] vs 0.42 [0.28-0.69] mg/dL; P = .47 and 256 [194-337] vs 262 [203-342] ng/L; P = .40, respectively), while the median (IQR) levels of IL-6 at baseline were higher in patients with CHIP (2.97 [1.86-4.70] vs 2.53 [1.77-4.03] ng/L; P = .003).
Table. Baseline Demographics.
| Variable | No. (%) | CHIP vs no CHIP P valuea | |||
|---|---|---|---|---|---|
| All (N = 10 061) | Genomic (n = 3923) | No CHIP (n = 3585) | CHIP (n = 338) | ||
| Age, mean (SD), y | 61.1 (10.0) | 61.9 (9.7) | 61.5 (9.6) | 66.3 (9.2) | <.001 |
| Female | 2587 (25.7) | 994 (25.3) | 898 (25.1) | 96 (28.4) | .19 |
| Male | 7474 (74.3) | 2929 (74.7) | 2687 (75.0) | 242 (71.6) | |
| Race | |||||
| Asian | 1163 (11.6) | 192 (4.9) | 179 (5.0) | 13 (3.9) | .30 |
| Black | 318 (3.2) | 131 (3.3) | 122 (3.4) | 9 (2.7) | |
| White | 8036 (79.9) | 3537 (90.2) | 3223 (89.9) | 314 (92.9) | |
| Otherb | 544 (5.4) | 63 (1.6) | 61 (1.7) | 2 (0.6) | |
| Current smoker | 2366 (23.5) | 1032 (26.3) | 954 (26.6) | 78 (23.1) | .17 |
| Diabetes at baseline | 4057 (40.3) | 1630 (41.6) | 1494 (41.7) | 136 (40.2) | .64 |
| Hypertension at baseline | 8008 (79.6) | 3187 (81.2) | 2896 (80.8) | 291 (86.1) | .02; .09c |
| Time since qualifying MI, mean (SD), y | 4.57 (5.37) | 5.33 (5.75) | 5.26 (5.70) | 6.0 (6.24) | .11 |
| Prior PCI | 6710 (66.7) | 2720 (69.4) | 2487 (69.4) | 233 (68.9) | .85 |
| Prior CABG | 1411 (14.0) | 678 (17.3) | 611 (17.1) | 67 (19.8) | .20 |
| History of heart failure | 2173 (21.6) | 761 (19.4) | 672 (18.7) | 89 (26.3) | .001; .02c |
| Statin use | 8891 (88.4) | 3447 (87.9) | 3155 (88.0) | 292 (86.4) | .38 |
| RAAS inhibitor use at baseline | 7992 (79.7) | 3140 (80.4) | 2873 (80.5) | 267 (79.2) | .57 |
| Prior anti-ischemic agent use | 9191 (91.4) | 3624 (92.4) | 3301 (92.1) | 323 (95.6) | .02 |
| Use of antithrombotic medication | 9747 (97.0) | 3822 (97.6) | 3491 (97.5) | 331 (97.9) | .85 |
| Total cholesterol, median (IQR), mg/dL | 160.0 (136.1-189.0) | 161.0 (138.0-189.5) | 161.3 (138.1-189.9) | 159.9 (135.0-186.4) | .25 |
| LDL cholesterol derived, median (IQR), mg/dL | 82.4 (63.4-107.1) | 83.5 (65.0-107.0) | 83.5 (65.4-107.1) | 83.9 (63.80-104.0) | .43 |
| eGFR, median (IQR), mL/min/1.73/m2 | 79.0 (64.0-93.0) | 78.0 (64.0-92.0) | 78.0 (65.0-93.0) | 72.0 (56.0-84.0) | <.001; .001c |
| hs-CRP, median (IQR), mg/dL | 4.20 (2.80-7.05) | 4.20 (2.80-6.90) | 4.20 (2.80-6.85) | 4.40 (2.95-7.10) | .47 |
| Interleukin 18, median (IQR), ng/Ld | 260 (201-340) | 262 (202-342) | 262 (203-342) | 256 (194-337) | .40 |
| Interleukin 6, median (IQR), ng/Le | 2.58 (1.78-4.10) | 2.55 (1.77-4.09) | 2.53 (1.77-4.03) | 2.97 (1.86-4.70) | .003 |
Abbreviations: CABG, coronary artery bypass surgery; CHIP, clonal hematopoiesis of indeterminate potential; eGFR, estimated glomerular filtration rate; hs-CRP, high-sensitivity C-reactive protein; LDL, low-density lipoprotein; MI, myocardial infarction; PCI, percutaneous coronary intervention; RAAS, renin-angiotensin-aldosterone system.
SI conversion factors: To convert cholesterol to millimoles per liter, multiply by 0.0259; hs-CRP to milligrams per liter, multiply by 10.
P values were calculated using Fisher exact test for categorical variables and Mann-Whitney test for continuous variables.
Other included Native American, Pacific Islander, other, or unknown.
P values adjusted for age and sex.
Number of patients analyzed for interleukin 18: all, n = 5071; genomic substudy, n = 3801; patients with CHIP, n = 323.
Number of patients analyzed for interleukin 6: all, n = 5058; genomic substudy, n = 3788; patients with CHIP, n = 322.
CHIP prevalence increases with age.25 Figure 1A shows the age distribution of patients with CHIP in CANTOS (all with established atherosclerosis) in comparison with age distributions previously reported among incident coronary disease cohorts by both Jaiswal et al4 and Genovese et al.6 The most common variant gene leading to CHIP in the CANTOS population was TET2, with variants in DMNT3A and ASXL1 occurring at a lower frequency (Figure 1B). This contrasts with all other reported CHIP populations, in which DMNT3A is the most frequent variant gene (Figure 1C).3,4,5,6,7,8 Increased levels of IL-6 among patients with CHIP were not significantly associated with a specific CHIP variant in comparisons with patients without CHIP (eFigure 1 in Supplement 2).
Figure 1. Somatic TET2 Variants Are Enriched in the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) Population.
A, The prevalence of patients with clonal hematopoiesis of indeterminate potential (CHIP) vs age compared with previous reports.3,6,8 B, A histogram of the observed CHIP variant genes. C, The proportion of total variants in target genes with a variant allele frequency of more than 0.02 from the CANTOS population compared with a coronary artery disease population (BioImage/Malmö Diet and Cancer).4
Given the higher prevalence of heart failure in patients with CHIP compared with patients without CHIP (Table), we examined the rate of heart failure hospitalizations in patients with CHIP. Patients with CHIP had a numerically higher rate of heart failure hospitalizations compared with patients without CHIP (eFigure 2 in Supplement 2), but this finding did not reach statistical significance (HR, 1.36 [95% CI, 0.87-2.14]; P = .18). A numerically higher but nonsignificant increase was also observed in patients with a diagnosis of heart failure at CANTOS enrollment (HR, 1.33 [95% CI, 0.74-2.38]; P = .34).
Patients With CHIP and Incident MACE
To assess the effect of CHIP status on the rate of MACE (the primary end point in the CANTOS trial defined as a combination of myocardial infarction, stroke, or cardiovascular death), we performed a Kaplan-Meier analysis using those patients randomized to the placebo arm in the trial (eFigure 3 in Supplement 2). Patients with CHIP showed a numerically increased rate of MACE compared with patients without CHIP that did not meet statistical significance (HR, 1.32 [95% CI, 0.86-2.04]; P = .21). Further, patients with CHIP who had variants in either TET2 or DNMT3A had a 65% increase in the rate of MACE compared with patients without CHIP that also did not meet statistical significance (HR, 1.65 [95% CI, 0.97-2.80]; P = .06).
The variant allele frequency (proportional to CHIP clone size) ranged from 0.03 to 0.77, with the median (IQR) being 0.086 (0.053-0.180) (eFigure 4 in Supplement 2). Previous reports have suggested that those patients with a higher variant allele frequency have a worse burden of disease or cardiovascular outcome.4,5 In patients with CHIP identified in the CANTOS population, those with a variant allele frequency more than 0.1 had a similar rate of MACE compared with those with a variant allele frequency of less than 0.1 (HR, 1.01 [95% CI, 0.49-2.05]) (eFigure 5 in Supplement 2).
Patients With CHIP Who Have Somatic Variants in TET2 and Response to Canakinumab
An initial evaluation of the response of patients with CHIP to canakinumab examined the levels of hs-CRP and IL-6 following 3 months of treatment with either 50 mg, 150 mg, or 300 mg of canakinumab. There was no significant difference in the reduction at 3 months in hs-CRP in patients without CHIP compared with patients with CHIP (0.16 vs 0.15 mg/dL reduction; P = .85). Similarly, there was also no significant difference in the reduction at 3 months of IL-6 in patients without CHIP vs with CHIP (0.51 vs 0.58 ng/L reduction; P = .35). Nor were there significant differences in the reduction at 3 months of hs-CRP (0.16 vs 0.16 mg/dL reduction; P = .64) or IL-6 (0.48 vs 0.45 ng/L reduction; P = .66) at 3 months between patients without CHIP and those with CHIP who have TET2 variants.
In stratified analysis, patients without CHIP treated with either 50 mg, 150 mg, or 300 mg of canakinumab had a nonstatistically significant 7% relative risk reduction in MACE compared with placebo-treated patients (HR, 0.93 [95% CI, 0.78-1.10]; stratified P = .38) (Figure 2). Patients with CHIP showed an 18% (HR, 0.82 [95% CI, 0.50-1.36]) relative risk reduction that also failed to meet statistical significance (stratified P = .45; P for interaction = .64). Patients with CHIP who had variants in TET2 showed a 62% (HR, 0.38 [95% CI, 0.15-0.96]) relative risk reduction in the incidence of MACE that met nominal statistical significance in stratified but not in interaction analysis (stratified P = .04; P for interaction = .14) (Figure 2C). The smaller number of patients with CHIP who have variants in other genes such as DNMT3A did not show statistically significant enhanced benefit from canakinumab treatment compared with patients without CHIP (Figure 2D; eTable 3 in Supplement 2).
Figure 2. Association of Canakinumab and Placebo With Incident Major Adverse Cardiovascular Events (MACE) According to TET2 Clonal Hematopoiesis of Indeterminate Potential (CHIP) Status.
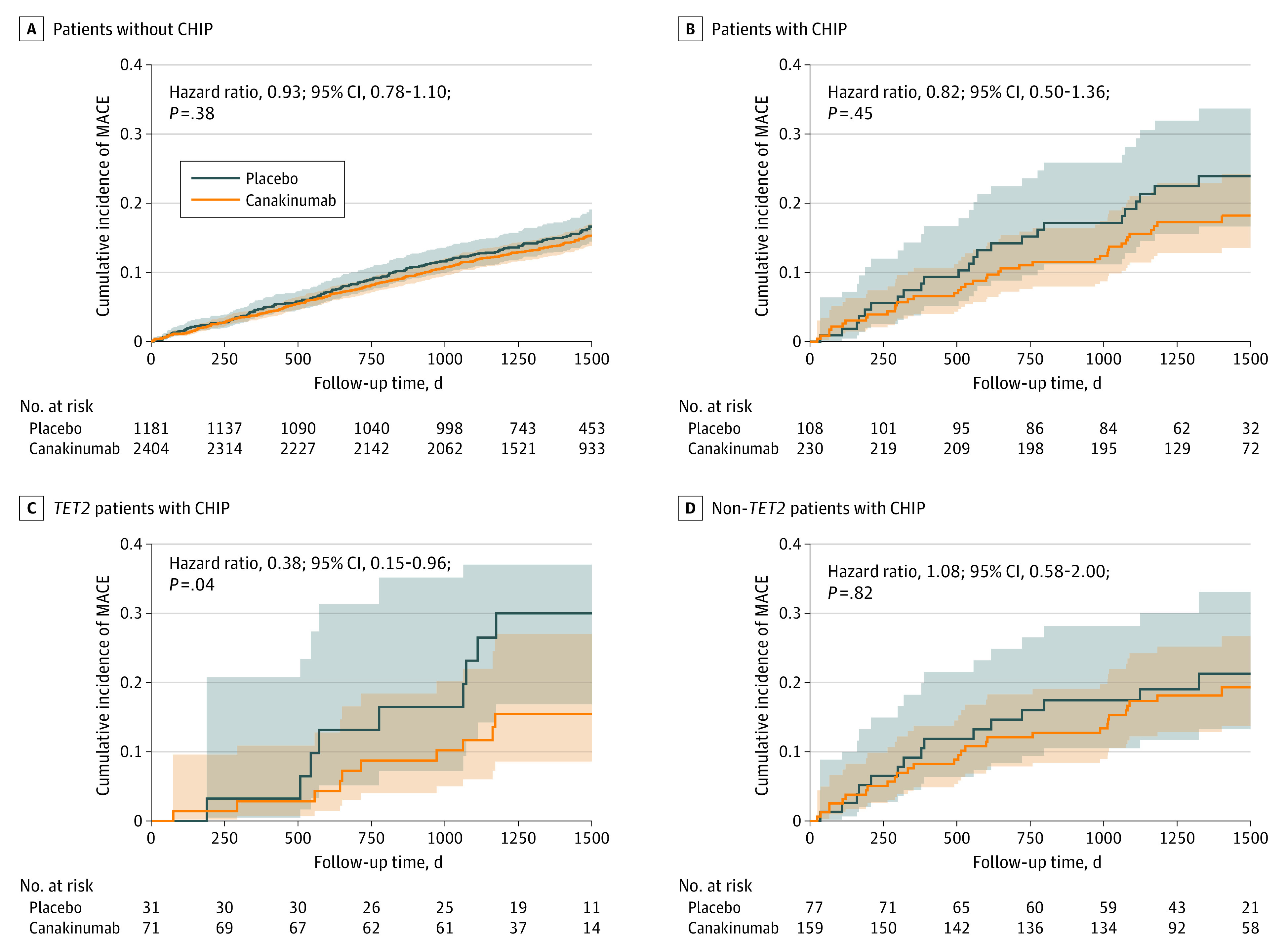
Kaplan-Meier curves are displayed for the rate of MACE in placebo and canakinumab-treated patients (50-, 150-, and 300-mg doses combined) without CHIP (A), patients with CHIP (B), TET2 patients with CHIP (C), or non-TET2 patients with CHIP (D). A Cox proportional hazard model was used to calculate hazard ratios with covariates of age, time to last myocardial infarction, and baseline log2(high-sensitivity C-reactive protein).
Discussion
Individuals with CHIP have an elevated risk for cardiovascular disease.26,27 This report provides further evidence to support this observation and raises the hypothesis that a subset of patients with CHIP may benefit from anti-inflammatory therapy with canakinumab, an anti–IL-1β monoclonal antibody.
Both the prevalence and age dependence of patients with CHIP in CANTOS with established coronary disease resemble that reported for incident coronary disease populations.4,6,8 Furthermore, the observed relationship between patients with CHIP and prevalent heart failure in CANTOS is concordant with recent investigations demonstrating a similar association with incident heart failure.5 In contrast to studies in populations not selected according to the CANTOS inclusion criteria, TET2 is the most common variant gene in patients with CHIP from CANTOS, while in other populations, DNMT3A variants predominate.3,4,5,6,7,8 While this finding has an unclear basis, it may relate to the selection of the CANTOS population or relative differences in sequencing depth (eTable 2 in Supplement 2). In contrast to populations included in previous studies, all CANTOS patients had a previous myocardial infarction, evidence for residual inflammatory risk as indicated by an hs-CRP level more than 0.20 mg/dL at study entry and were receiving guideline-directed secondary prevention therapy.
The results reported here agree with previously published work implicating IL-1β and IL-6 signaling in mediating the effects of Tet2 loss of function in experimental atherosclerosis. Macrophages in atherosclerotic plaques, derived from the myeloid lineage, play an important role in atherosclerotic disease progression and in the development of MACE in part through inflammatory signaling. Indeed, CANTOS demonstrated that inhibiting IL-1β signaling reduces cardiovascular events. Mouse macrophages that lack Tet2 function have enhanced IL-1β and IL-6 expression in response to exposure to low-density lipoprotein, mediators downstream of the NLRP3 inflammasome. IL-1β induces the expression of endothelial-leukocyte adhesion molecules such as vascular cell adhesion molecute-1 that can recruit further monocytes, both clonally derived and wild-type.4,28 Sano and colleagues9 demonstrated that myeloid cells with either Tet2 or Dnmt3a loss of function secrete elevated levels of IL-6 in response to lipopolysaccharide, an inflammatory stimulus. Consistent with this observation, Busque et al7 observed that patients with CHIP and coronary artery disease have elevated levels of the systemic inflammatory marker hs-CRP compared with patients without CHIP. While we did not see differences in hs-CRP in patients with CHIP vs without CHIP in CANTOS, perhaps because participants had elevated hs-CRP (unlike the study by Busque et al7), we did observe increased levels of the downstream inflammatory cytokine IL-6, consistent with the notion that patients with CHIP have a greater systemic inflammatory burden than patients without CHIP. Further, Sano and colleagues9 also demonstrated that only myeloid cells with Tet2 variants, but not Dnmt3a variants, had elevated IL-1β secretion in response to lipopolysaccharide. This finding may explain our observation that patients with CHIP due to variants in TET2 had a greater event rate reduction in response to the anti–IL-1β antibody canakinumab compared with patients with CHIP with variants in other genes, including those in DNMT3A (Figure 2).
Within CANTOS, IL-1β inhibition with canakinumab reduced incident hospitalization for heart failure.16 Preclinical work also indicates that CHIP plays a role in heart failure. In mice with ischemic cardiomyopathy produced by ligation of the left anterior descending coronary artery, bone marrow deficiency in Tet2 yielded a greater reduction in left ventricular ejection fraction and increased cardiomyocyte hypertrophy and fibrosis after 4 weeks compared with wild-type mice.29 Similar detrimental effects in left ventricular remodeling were also seen in mice with Tet2 bone marrow deficiency in both a pressure overload-induced or an angiotensin-induced heart failure in mice.9,29 These results support the previous clinical report that patients with heart failure and CHIP variants in TET2 or DNMT3A have an increased rate of heart failure hospitalizations and an increased mortality compared with patients without CHIP who have heart failure.5 Our results provide further support for this observation, with CANTOS patients with CHIP having a higher prevalence of heart failure at trial entry (Table).
Limitations
The main limitations of this work reside in its exploratory nature and limited number of patients in CANTOS who provided DNA samples and were patients with CHIP, severely reducing the power of this analysis (26% power for incident heart failure hospitalization and 24% power for incident MACE in post hoc analysis). Indeed, CHIP was identified as a cardiovascular risk factor after the design and initiation of CANTOS.30 Furthermore, while we have also observed an enrichment for patients with CHIP across all age groups enrolled in CANTOS, direct comparisons between published CHIP cohorts (Figure 1A and C) remain challenging owing to varied sequencing assays and read depth, which have a large effect on the sensitivity to detect small clones.
Conclusions
This exploratory substudy of the CANTOS trial identified 8.6% of participants with CHIP. The TET2 gene was the most common site for somatic variation leading to CHIP in this population. Patients with somatic variation in TET2 or DNMT3A showed an equivocal increased risk for MACE, and patients with somatic variants in TET2 showed a reduced risk for MACE while taking canakinumab. These results are consistent with observations of increased risk for cardiovascular events in patients with CHIP and raise the possibility that those with TET2 variants may respond better to canakinumab than those without CHIP. Intriguingly, a significant attenuation in cardiovascular events using IL-6 receptor genetic instrument as a proxy for IL-6 inhibition has likewise been reported among individuals with CHIP, further supporting a rationale for targeted anti-inflammatory therapy in high-risk patients.31 Future randomized clinical trials will be necessary to test this hypothesis prospectively.
Trial protocol
eTable 1. List of Gene Variants Identified
eTable 2. Median Sequencing Depth and VAF at Canonical CHIP Loci
eTable 3. Association between CHIP subpopulations, Heart Failure and MACE outcomes
eFigure 1. Baseline IL-6 Concentration by CHIP Status and Gene Variant
eFigure 2. Heart Failure Hospitalizations in Patients with and without CHIP
eFigure 3. Rates of MACE in patients with and without CHIP
eFigure 4. Distribution of Variant Allele Frequency in CHIP(+) Patients
eFigure 5. Rate of MACE in CHIP(+) Patients by Variant Allele Frequency
Data sharing statement
References
- 1.Steensma DP. Clinical consequences of clonal hematopoiesis of indeterminate potential. Blood Adv. 2018;2(22):3404-3410. doi: 10.1182/bloodadvances.2018020222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zink F, Stacey SN, Norddahl GL, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130(6):742-752. doi: 10.1182/blood-2017-02-769869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. doi: 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111-121. doi: 10.1056/NEJMoa1701719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu B, Roberts MB, Raffield LM, et al. ; National Heart, Lung, and Blood Institute TOPMed Consortium . Association of clonal hematopoiesis with incident heart failure. J Am Coll Cardiol. 2021;78(1):42-52. doi: 10.1016/j.jacc.2021.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genovese G, Kähler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. doi: 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busque L, Sun M, Buscarlet M, et al. High-sensitivity C-reactive protein is associated with clonal hematopoiesis of indeterminate potential. Blood Adv. 2020;4(11):2430-2438. doi: 10.1182/bloodadvances.2019000770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie M, Lu C, Wang J, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20(12):1472-1478. doi: 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res. 2018;123(3):335-341. doi: 10.1161/CIRCRESAHA.118.313225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327):842-847. doi: 10.1126/science.aag1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ridker PM. From C-reactive protein to interleukin-6 to interleukin-1: moving upstream to identify novel targets for atheroprotection. Circ Res. 2016;118(1):145-156. doi: 10.1161/CIRCRESAHA.115.306656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarwar N, Butterworth AS, Freitag DF, et al. ; IL6R Genetics Consortium Emerging Risk Factors Collaboration . Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet. 2012;379(9822):1205-1213. doi: 10.1016/S0140-6736(11)61931-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swerdlow DI, Holmes MV, Kuchenbaecker KB, et al. ; Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium . The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379(9822):1214-1224. doi: 10.1016/S0140-6736(12)60110-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J Am Coll Cardiol. 2017;70(18):2278-2289. doi: 10.1016/j.jacc.2017.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridker PM, Everett BM, Thuren T, et al. ; CANTOS Trial Group . Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119-1131. doi: 10.1056/NEJMoa1707914 [DOI] [PubMed] [Google Scholar]
- 16.Everett BM, Cornel JH, Lainscak M, et al. Anti-inflammatory therapy with canakinumab for the prevention of hospitalization for heart failure. Circulation. 2019;139(10):1289-1299. doi: 10.1161/CIRCULATIONAHA.118.038010 [DOI] [PubMed] [Google Scholar]
- 17.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213-219. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25(21):2865-2871. doi: 10.1093/bioinformatics/btp394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McLornan D, Percy M, McMullin MF. JAK2 V617F: a single mutation in the myeloproliferative group of disorders. Ulster Med J. 2006;75(2):112-119. [PMC free article] [PubMed] [Google Scholar]
- 20.Hu L, Li Z, Cheng J, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155(7):1545-1555. doi: 10.1016/j.cell.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 21.Sanada M, Suzuki T, Shih LY, et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460(7257):904-908. doi: 10.1038/nature08240 [DOI] [PubMed] [Google Scholar]
- 22.Ridker PM, Libby P, MacFadyen JG, et al. Modulation of the interleukin-6 signalling pathway and incidence rates of atherosclerotic events and all-cause mortality: analyses from the Canakinumab Anti-Inflammatory Thrombosis Outcomes Study (CANTOS). Eur Heart J. 2018;39(38):3499-3507. doi: 10.1093/eurheartj/ehy310 [DOI] [PubMed] [Google Scholar]
- 23.Ridker PM, MacFadyen JG, Everett BM, Libby P, Thuren T, Glynn RJ; CANTOS Trial Group . Relationship of C-reactive protein reduction to cardiovascular event reduction following treatment with canakinumab: a secondary analysis from the CANTOS randomised controlled trial. Lancet. 2018;391(10118):319-328. doi: 10.1016/S0140-6736(17)32814-3 [DOI] [PubMed] [Google Scholar]
- 24.Ridker PM, MacFadyen JG, Thuren T, Libby P. Residual inflammatory risk associated with interleukin-18 and interleukin-6 after successful interleukin-1β inhibition with canakinumab: further rationale for the development of targeted anti-cytokine therapies for the treatment of atherothrombosis. Eur Heart J. 2020;41(23):2153-2163. doi: 10.1093/eurheartj/ehz542 [DOI] [PubMed] [Google Scholar]
- 25.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366(6465):366. doi: 10.1126/science.aan4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidlow R, Lin AE, Gupta D, et al. The clinical challenge of clonal hematopoiesis, a newly recognized cardiovascular risk factor. JAMA Cardiol. 2020;5(8):958-961. doi: 10.1001/jamacardio.2020.1271 [DOI] [PubMed] [Google Scholar]
- 27.Libby P, Sidlow R, Lin AE, et al. Clonal hematopoiesis: crossroads of aging, cardiovascular disease, and cancer: JACC review topic of the week. J Am Coll Cardiol. 2019;74(4):567-577. doi: 10.1016/j.jacc.2019.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans MA, Sano S, Walsh K. Cardiovascular disease, aging, and clonal hematopoiesis. Annu Rev Pathol. 2020;15:419-438. doi: 10.1146/annurev-pathmechdis-012419-032544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano S, Oshima K, Wang Y, et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1β/NLRP3 inflammasome. J Am Coll Cardiol. 2018;71(8):875-886. doi: 10.1016/j.jacc.2017.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridker PM, Thuren T, Zalewski A, Libby P. Interleukin-1β inhibition and the prevention of recurrent cardiovascular events: rationale and design of the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS). Am Heart J. 2011;162(4):597-605. doi: 10.1016/j.ahj.2011.06.012 [DOI] [PubMed] [Google Scholar]
- 31.Bick AG, Pirruccello JP, Griffin GK, et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141(2):124-131. doi: 10.1161/CIRCULATIONAHA.119.044362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trial protocol
eTable 1. List of Gene Variants Identified
eTable 2. Median Sequencing Depth and VAF at Canonical CHIP Loci
eTable 3. Association between CHIP subpopulations, Heart Failure and MACE outcomes
eFigure 1. Baseline IL-6 Concentration by CHIP Status and Gene Variant
eFigure 2. Heart Failure Hospitalizations in Patients with and without CHIP
eFigure 3. Rates of MACE in patients with and without CHIP
eFigure 4. Distribution of Variant Allele Frequency in CHIP(+) Patients
eFigure 5. Rate of MACE in CHIP(+) Patients by Variant Allele Frequency
Data sharing statement



